Abstract
Toxoplasma gondii undergoes many phenotypic changes during its life cycle. The recent identification of AP2 transcription factors in T. gondii has provided a platform for studying the mechanisms controlling gene expression. In the present study, we report that a recombinant protein encompassing the TgAP2XI-4 AP2 domain was able to specifically bind to a DNA motif using gel retardation assays. TgAP2XI-4 protein is localised in the parasite nucleus throughout the tachyzoite life-cycle in vitro, with peak expression occurring after cytokinesis. We found that the TgAP2XI-4 transcript level was higher in bradyzoite cysts isolated from brains of chronically infected mice than in the rapidly replicating tachyzoites. A knock-out of the TgAP2XI-4 gene in both T. gondii virulent type I and avirulent type II strains reveals its role in modulating expression and promoter activity of genes involved in stage conversion of the rapidly replicating tachyzoites to the dormant cyst forming bradyzoites. Furthermore, mice infected with the type II KO mutants show a drastically reduced brain cyst burden. Thus, our results validate TgAP2XI-4 as a novel nuclear factor that regulates bradyzoite gene expression during parasite differentiation and cyst formation.
Introduction
Toxoplasma gondii is a unicellular eukaryotic pathogen infecting humans and livestock. It belongs to the apicomplexan phylum of parasites that encompasses many deadly pathogens such as Plasmodium, the cause of malaria, and Cryptosporidium, responsible for cryptosporidiosis. T. gondii is an obligate intracellular parasite that is a leading cause of focal central nervous system infections in patients with AIDS/HIV. In addition, toxoplasmosis is also a clinically important opportunistic pathogen in cancer treatment, organ transplant and in newborns that are infected in utero. The life cycle of T. gondii is complex, with multiple differentiation steps that are critical to the survival of the parasite in human and feline hosts (Kim et al., 2004). Infection by oocysts containing sporozoites shed by cats or by bradyzoites contaminating ingested meat leads to differentiation into the rapidly growing tachyzoites that are responsible for the clinical manifestations in humans. The immune response, among other stresses to the parasite, induces the conversion of the tachyzoites into bradyzoites. These latent bradyzoites are responsible for chronic disease due to their ability to evade the immune system, to resist commonly used drug treatments and to reactivate into virulent tachyzoites. Therefore, the tachyzoite to bradyzoite interconversion is a critical step for the pathogenesis and survival of the parasite.
Transcriptome studies of the various life cycle stages of the parasite have revealed a complex pattern of expression associated with each form of the parasite. Sequential analysis of gene expression throughout development showed that 18% of genes were stage-specifically expressed, with co-regulated genes that are specific to each form of the life cycle (Radke et al., 2005). Similarly, the timing of expression for more than 2500 genes is dependent on the tachyzoite cell cycle (Behnke et al., 2010). Interestingly, some of the genes specifically regulated during the tachyzoite-bradyzoite transition are expressed throughout mitosis, cytokinesis and early G1 phases (Behnke et al., 2010), in concordance with the cell cycle arrest observed during bradyzoite differentiation (Radke et al., 2003). These co-regulated genes are scattered across the genome, indicating the importance of the promoter and trans-acting factors for the regulation of each particular gene (Radke et al., 2005; Behnke et al., 2010). This notion is strongly supported by the presence of most basal transcription machinery required for the transcriptional control of protein-coding gene expression in the T. gondii genome. Moreover, a number of DNA motifs were found to be active in the promoters of developmentally regulated genes (Behnke et al., 2008; Kibe et al., 2005; Gissot et al., 2009). Although tachyzoites and bradyzoites have fundamental differences in their gene expression profiles and cell proliferation, the pathways responsible for controlling the expression of these genetic traits remain unknown. DNA-sequence-specific transcription factors were initially regarded as poorly represented in apicomplexan genomes, leading to speculation that epigenetic or post-transcriptional mechanisms were the primary mode of gene regulation. The discovery of a specific pattern of chromatin markers in the promoters of T. gondii genes suggested an important role for chromatin structure in the parasite’s gene regulation (Saksouk et al., 2005; Gissot et al., 2007). In addition, the GCN5A histone acetyltransferase has been linked to gene activation during bradyzoite differentiation (Naguleswaran et al., 2010). Subsequent analysis of apicomplexan genomes led to the discovery of a plant-like family of transcription factors, each with an AP2 DNA binding domain (Balaji et al., 2005). Approximately fifty putative AP2 transcription factors were first reported in the T. gondii genome (Iyer et al., 2007), although 68 have since been annotated on the T. gondii genome database (www.toxodb.org) while only 27 ApiAP2 genes were found in Plasmodium (Painter et al.,2011).
In plants, protein members of the AP2/ethylene response factor family, which encompass one or two AP2 domains, specifically bind DNA and participate in developmental and stress responses (Riechmann et al., 1998). Recombinant proteins containing Plasmodium and Cryptosporidium AP2 DNA binding domains were reported to bind specific DNA motifs (De Silva et al., 2008; Campbell et al., 2010). Interestingly, gene disruptions of two AP2 transcription factor candidates performed in P. berghei resulted in alterations in the transcript expression profile and significant reductions in mosquito-invasive stages (Yuda et al., 2009) and sporozoites (Yuda et al., 2010). Altogether, these observations indicate that the ApiAP2 family of transcription factors may participate in the control of gene expression in apicomplexan parasites. In T. gondii, the steady-state levels of 24 transcripts encoding different AP2 proteins were shown to be dependent on the cell cycle, suggesting that these factors may play a role in controlling expression of sets of cell cycle regulated genes (Behnke et al., 2010). Five of those AP2 proteins were confirmed to have expression patterns similar to their cognate transcript (Behnke et al., 2010). While T. gondii AP2 proteins are hypothesized to act as transcription factors, their cognate DNA binding motifs and functions in gene modulation remain to be demonstrated. To our knowledge, there are no reports characterizing a DNA-sequence specific transcription factor involved in T. gondii bradyzoite differentiation, although the TgAP2XII-6 gene is disrupted in one of the differentiation mutants produced by random insertion (Lescault et al., 2010). In this paper, we show that T. gondii AP2XI-4 protein is a novel nuclear factor that binds a specific DNA motif and that ablation of the AP2XI-4 gene profoundly impairs the stress-induced bradyzoite gene expression program.
Results
TgAP2XI-4 expression is up-regulated in bradyzoites
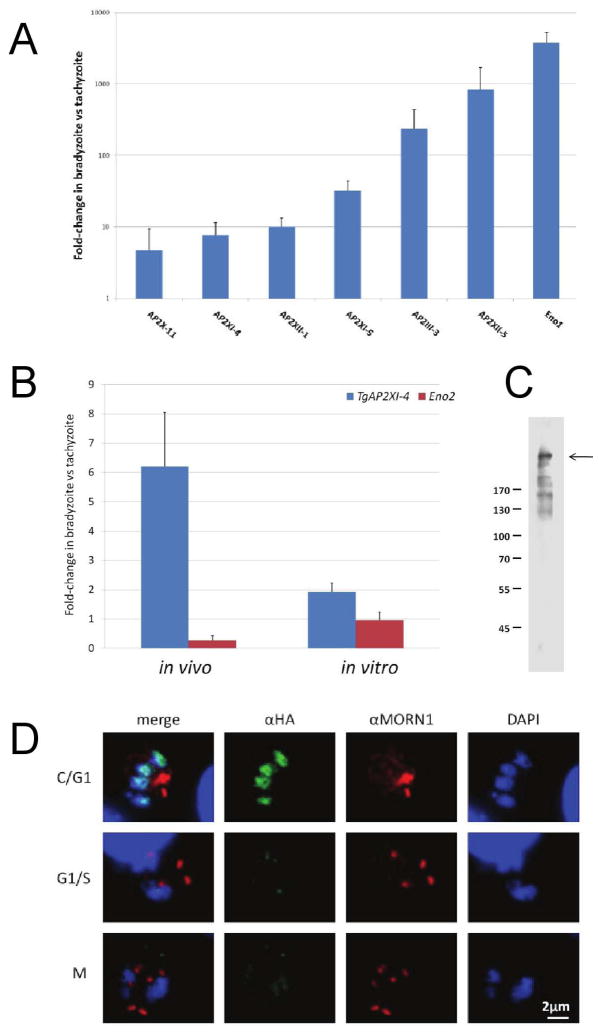
The conversion of T. gondii tachyzoites into bradyzoites is associated with changes in the transcriptional profiles of many genes involved in diverse biological functions. In order to identify potential regulators of this process, preliminary qRT-PCR was carried out on genes coding over twenty putative AP2-like transcription factors of T. gondii, comparing cDNA synthesized from purified T. gondii Type II bradyzoite cysts (produced in vivo) and tachyzoites (in vitro) (Fig. S1). The transcript levels of a number of AP2 genes were more abundant in bradyzoites than in tachyzoites (Fig. 1A). Among these ‘bradyzoite-specific AP2s’, we noticed TgAP2XI-4 was up-regulated 6-fold. This gene was chosen for further analysis because it encodes a protein that has a strong homology with the DNA-binding domain of one Plasmodium’s AP2 protein (PFD0985w). Enolase 1 (Eno1) was used as a control gene for bradyzoite-specific transcriptional up-regulation (Dziersizinski et al., 2001) and as expected, more Eno1 transcripts were detected in bradyzoites in-vivo than in tachyzoites (Fig. 1A). In addition, an in vitro model of bradyzoite differentiation using alkaline pH 8.2-stress to induce the expression of bradyzoite genes was used to verify that the transcription of TgAP2XI-4 is also up-regulated in the T. gondii ΔKu80 Type I strain. Quantitative RT-PCR analysis revealed that the abundance of TgAP2XI-4 transcripts was almost 2-fold higher in the pH 8.2-stressed parasites versus those cultured under conditions that favour tachyzoite growth (Fig 1B, in vitro). Enolase 2 (Eno2), a tachyzoite-specific gene that is not up-regulated in bradyzoites in-vivo (Dzierszinski et al., 2001), was used as a negative control. As expected, Eno2 mRNA levels were not increased in bradyzoites either in vivo or in in vitro.
Figure 1. TgAP2XI-4 expression is regulated during bradyzoite differentiation and throughout the tachyzoite cell-cycle.
(A) Wild type parasites from a T. gondii type II 76K strain were either used to produce bradyzoite tissue cysts (in vivo) or were cultured under normal conditions for tachyzoite growth (in vitro) (Dzierszinski et al., 2001). Total RNA was purified from all samples and analysed by quantitative RT-PCR to determine the relative levels of TgAP2 genes and Eno1 mRNA. The values are presented as the fold-changes in the bradyzoite samples relative to the corresponding tachyzoite samples.
(B) Wild type parasites from a T. gondii type II strain were used to produce bradyzoite tissue cysts (in vivo) while wild type parasites from a type I strain were cultured under alkaline (pH 8.2) stress (in vitro) to induce the expression of bradyzoite genes. For comparison, type I and type II tachyzoite strains were grown in vitro under control conditions (pH 7.0). Total RNA was purified from all samples and analysed by quantitative RT-PCR to determine the relative levels of TgAP2XI-4 and Eno2 mRNA. The values are presented as the fold-change in the bradyzoite samples relative to the corresponding tachyzoite samples.
(C) A whole-cell protein lysate from the TgAP2XI-4-HA mutant was fractionated on a 10% SDS-PAGE gel under reducing conditions. Western blot was carried out using a mouse monoclonal αHA antibody. An arrow indicates the band corresponding to TgAP2XI-4-HA. Molecular weight markers are in kDa.
(D) Immunofluorescence assays were conducted on TgAP2XI-4 parasites fixed 24-hours after infection of HFF cells. The mouse monoclonal αHA antibody was used in combination with a rabbit αMORN1 antibody and detected with anti-mouse Alexa488 (green) and anti-rabbit Alexa594 (red), respectively. Daughter cell formation and mitosis were effectively monitored with αMORN1 and DAPI counterstaining (blue), respectively. TgAP2XI-4 protein expression peaks during the cytokinesis and early G1 phase. C, cytokinesis; G1, gap phase; S, synthesis phase; and M, mitosis. It should be noted that MORN1 localizes at ring structures at the apical and posterior ends of the inner membrane complex and to the centrocone. The centrocone-associated MORN1 concentrates at a focal point during G1 or as two focal points during S/M phase and is absent during C phase.
Expression of nuclear factor TgAP2XI-4 is regulated throughout the tachyzoite cell cycle
Microarray data reveal that the level of TgAP2XI-4 transcript is also regulated throughout the in-vitro tachyzoite cell-cycle (Behnke et al., 2010; also see www.ToxoDB.org). To monitor the expression of TgAP2XI-4 in T. gondii Δku80 Type I strain, the TgAP2XI-4 protein was tagged at the C-terminal end with the HA epitope. Using anti-HA monoclonal antibody (αHA), Western blots of total protein extracts from this transgenic parasite revealed a large >300 kDa protein (Fig. 1C), consistent with the predicted molecular weight of 347 kDa of the TgAP2XI-4 protein. Other smaller molecular weight protein bands, possibly representing degradation products of TgAP2XI-4-HA, were also detected, albeit with less intensity compared to the main protein band. Immunofluorescence assays confirmed the nuclear localization of TgAP2XI-4 in T. gondii tachyzoites (Fig. 1D). Furthermore, expression of the TgAP2XI-4 protein was regulated throughout the tachyzoite cell-cycle as monitored by dual staining with polyclonal antibodies specific to MORN1, a marker of the centrocone, and the anti-HA antibody. We found that in contrast to the prediction of the microarray data, where TgAP2XI-4 transcription peaks during mitosis and cytokinesis (Behnke et al., 2010), the expression peak of TgAP2XI-4 protein was detected during the end of cytokinesis (C) and the beginning of the gap phase (G1), with minimal protein visualized during the synthesis phase (S) and mitosis (M). Additional observations of the varying expression of TgAP2XI-4 during the tachyzoite cell cycle are also provided in Figure S2.
TgAP2XI-4 binds to a sequence-specific DNA motif
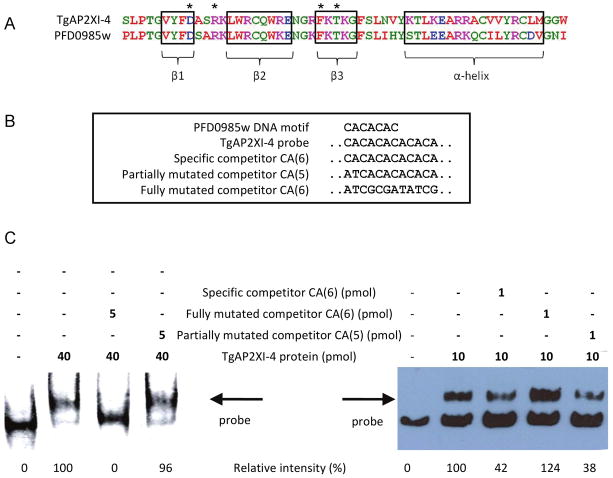
There are currently 68 different AP2-like genes predicted in the T. gondii database, compared with 27 homologues of AP2 in the related Apicomplexa, Plasmodium falciparum. A blastp query of the predicted protein database of P. falciparum using the entire TgAP2XI-4 protein sequence identified two potential homologues, including PFD0985w (1.2e−13) and PF14_0533 (1.0e−05), based primarily on the conservation of the AP2 domain. Amino acid conservation of the TgAP2XI-4 AP2 domain (residues 3137 3191) was estimated at 85% (47/55) similarity for PFD0985w but only 69% (36/55) for PF14_0533. An alignment of the conserved AP2 domains of TgAP2XI-4 and PFD0985w is represented in Fig. 2A. Three anti-parallel β-sheets (Fig. 2A, boxed regions), representing the AP2 regions that bind directly with the DNA target molecules (Lindner et al., 2010), are conserved between the two sequences as well as the four positions potentially involved in direct DNA-binding (Fig. 2A, amino acids indicated with a star). In a recent study, protein-binding microarrays were used to resolve the putative DNA regulatory elements targeted by many of the P. falciparum AP2s, including several TgAP2XI-4-homologues (Campbell et al., 2010). The DNA motif ‘CACACAC’ was identified as the putative regulatory element targeted by the first AP2 domain of PFD0985w (conserved with the AP2 domain of TgAP2XI-4) and the second AP2 domain of PF14_0533. To accommodate possible variations in the size of the DNA regulatory element recognized by TgAP2XI-4, an electrophoretic mobility shift assay (EMSA) was carried out using a 179 bp PCR-amplified region of DNA containing an extended ‘CACACACACACA’ motif (Fig. 2B). This 179-bp DNA was chosen from the sequence of one of the gene promoters identified by micro-array studies and listed in Table 1. A recombinant protein spanning the AP2 domain of TgAP2XI-4 was able to bind this DNA probe (Fig. 2C). The binding of the TgAP2XI-4 factor was inhibited by pre-incubation with 500-fold excess of a specific-competitor probe (a 40bp double-stranded oligonucleotide containing the motif, named “specific competitor CA(6)”). No inhibition of DNA-binding was observed using 500-fold excess of a non-specific competitor, where only the extended motif was mutated, demonstrating the sequence-specific DNA-binding of TgAP2XI-4 recombinant protein. To discover the minimal number of CA repeats needed for the specific TgAP2XI-4 recombinant protein binding, we competed the protein-probe interaction using an 100-fold excess probes containing different sizes of the CA repeats (specific competitor CA(6), partially mutated competitor CA(5) and partially mutated competitor CA(4)). To assess the degree of competition of the interaction by each competitor, we measured the intensity of the band representing the TgAP2XI-4 recombinant protein-probe binding. We show that an 100-fold excess of an oligonucleotide containing 5 CA repeats (partially mutated competitor CA(5)) was able to reduce by 48 % the interaction while the partially mutated competitor CA(4) yielded only 12 % reduction of the protein-probe interaction (Fig. S3B). The specific competitor CA(6) was the most efficient in inhibiting the TgAP2XI-4 recombinant protein-probe interaction with 70 % inhibition of the interaction. Using others concentrations of the specific CA(6) competitor, we were able to compete the interaction (Fig 2C, left panel and Fig S3) while the same concentrations of the fully mutated competitor did yield any significant reduction of the interaction.
Figure 2. Sequence-specific DNA-binding of TgAP2XI-4.
(A) The amino acid sequence of the TgAP2XI-4 AP2 region was aligned to the first of two AP2 domains in the P. falciparum homologue, PFD0985w. Three completely conserved β-sheets (β1, β2 and β3) are highlighted, along with a less conserved α-helix.
(B) The solved PFD0985w DNA motif (Campbell et al., 2010) was used to design a 179 bp TgAP2XI-4 probe containing an extended version of the DNA motif. Specific and non-specific competitor probes were designed with the extended motif or a mutated motif, respectively.
(C) An electrophoretic mobility shift assay was carried out using a recombinant protein spanning the TgAP2XI-4 AP2 domain. The shift caused by the binding of TgAP2XI-4 to the biotinylated probe is indicated by an arrow. TgAP2XI-4 binding was completely inhibited by a 500-fold excess of specific competitor but not by a non-specific competitor.
Table 1. Bradyzoite genes are most affected by the knock-out of TgAP2XI-4.
A complete list of the 22 T. gondii genes that are down-regulated in ΔTgAP2XI-4 following pH 8.2-stress reveals several well-characterized bradyzoite genes. The gene identification and biological or putative annotation is listed alongside the log fold-change (logFC) observed at pH 8.2 in ΔTgAP2XI-4 vs the wild type. Evidence of each bradyzoite-specific gene is provided with the logFC values in the wild type following pH 8.2-stress.
| Gene | Annotation | logFC of ΔTgAP2XI-4 vs WT (at pH8.2) | logFC of pH8.2 vs pH7.0 (in WT) |
|---|---|---|---|
| TGME49_091040 | lactate dehydrogenase (LDH2) | −4.115 | 5.445 |
| TGME49_068860 | enolase 1 | −4.024 | 6.844 |
| TGME49_002020 | hypothetical (DnaK-TPR) | −3.865 | 4.271 |
| TGME49_080570 | SAG4, bradyzoite surface antigen | −3.647 | 7.203 |
| TGME49_007210 | Hypothetical | −3.592 | 6.910 |
| TGME49_016140 | ankyrin repeat-containing domain protein | −3.383 | 4.348 |
| TGME49_078080 | Hypothetical | −3.367 | 3.830 |
| TGME49_021550 | CMGC kinase, MAPK family, MEK kinase-related | −3.279 | 4.241 |
| TGME49_007150 | SAG49C (=SAG2D) | −3.215 | 3.766 |
| TGME49_007130 | SAG49A (=SAG2Y) | −3.071 | 4.222 |
| TGME49_059020 | BAG1 | −2.974 | 5.636 |
| TGME49_120190 | SRS16B (= SRS9) | −2.780 | 0.491* |
| TGME49_040470 | Hypothetical | −2.609 | 3.770 |
| TGME49_024170 | SRS domain-containing protein | −2.600 | 4.181 |
| TGME49_025290 | nucleoside-triphosphatase (B-NTPase) | −2.508 | 4.168 |
| TGME49_080580 | P18 surface antigen | −2.499 | 3.039 |
| TGME49_061650 | Hypothetical | −2.442 | 3.872 |
| TGME49_009760 | Hypothetical | −2.338 | 3.641 |
| TGME49_111370 | methylmalonate-semialdehyde dehydrogenase, putative | −2.179 | 3.501 |
| TGME49_052640 | plasma-membrane proton-ATPase | −2.167 | 3.086 |
| TGME49_053330 | rhoptry kinase family protein, truncated | −2.137 | 5.634 |
| TGME49_053340 | Hypothetical | −2.063 | 4.578 |
TGME49_120190 (SRS9) is a known bradyzoite marker (Van, 2007), despite the lack of strong up-regulation in pH 8.2-stressed wild type parasites in this experiment.
Alkaline stress-induced activation of bradyzoite genes is impaired in a type I RH TgAP2XI-4 knock-out mutant
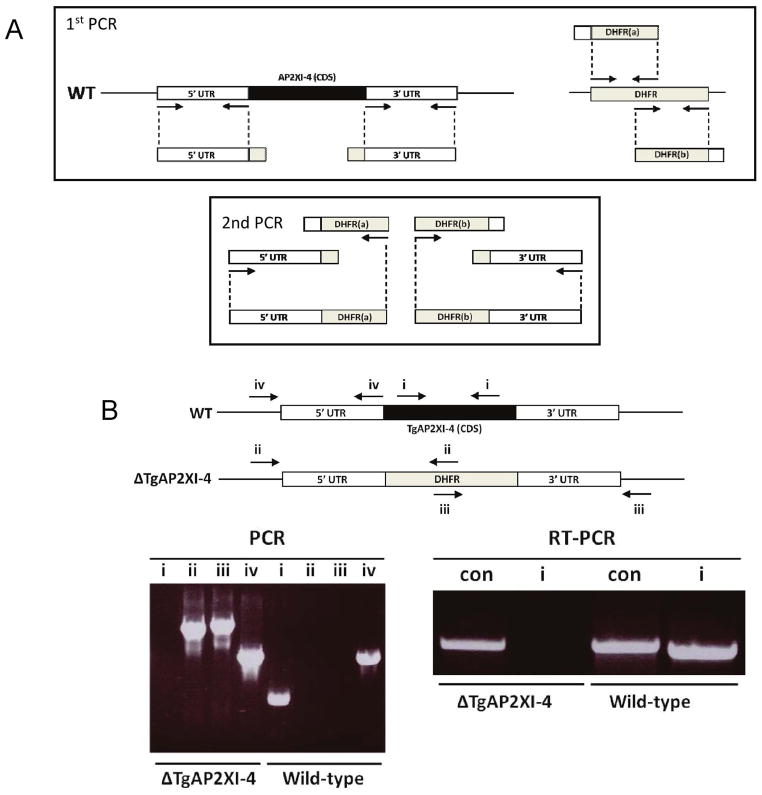
In order to assess the role of TgAP2XI-4 in T. gondii, a knock-out mutant was first produced in the ΔKu80 Type I strain of T. gondii using a modified version of the technique previously described (Upadhya et al., 2011). For the fusion PCR, the 5′ upstream and 3′ downstream flanking regions of the TgAP2XI-4 gene were amplified and then each was fused via a secondary PCR to either the first or second half of a DHFR cassette, respectively (Fig. 3A). These two constructs, each containing an incomplete pyrimethamine-resistance drug cassette, were transfected together into the ΔKu80 Type I strain. Both the successful integration of the two PCR products, representing the ΔTgAP2XI-4 knock-out construct, and the resulting knock-out of the TgAP2XI-4 gene were confirmed by PCR, while RT-PCR analysis confirmed the absence of TgAP2XI-4 transcripts (Fig. 3B).
Figure 3. Construction of RH ΔTgAP2XI-4 using fusion PCR.
(A) In the first PCR, the 5′ upstream and 3′ downstream flanking regions of the TgAP2XI-4 gene were amplified along with the first and second half of a pyrimethamine resistant DHFR cassette. Due to the introduction of complementary flanking regions, the TgAP2XI-4 5′ and 3′ products could be fused to the first and second halves of the DHFR products, respectively, in a second PCR. The two resulting PCR products were directly transfected into the RH ΔKu80 T. gondii type I strain.
(B) PCR was used to confirm the correct integration of the fused ΔTgAP2XI-4 construct (using primer pairs ii and iii) and deletion of the TgAP2XI-4 coding sequence (primer pair i). The absence of TgAP2XI-4 transcripts was confirmed by RT-PCR (primer pair i). Primers for TgAP2XI-5 (TGME49_016220) were used as a control (con).
In vitro proliferation assays carried out in conditions suitable for tachyzoite growth revealed no clear differences between the wild-type and ΔTgAP2XI-4 parasites (Fig. S4A). Likewise, parasites lacking the TgAP2XI-4 gene displayed the same level of virulence (in vivo) as the wild-type Type I strain (Fig. S4B). In addition, both wild type and ΔTgAP2XI-4 parasites have a similar growth rate under alkaline stress conditions (Fig. S5A and S5B). Microarray analysis was carried out to identify more subtle changes in gene expression for the ΔTgAP2XI-4 mutant. Total RNA was purified from ΔTgAP2XI-4 and wild type parasites grown under normal tachyzoite growth conditions and subjected to microarray analysis using the Affymetrix ToxoGeneChip microarray containing probe sets for approximately 8,000 T. gondii genes (Bahl et al., 2010). Analysis of the wild type (WT) and ΔTgAP2XI-4 microarray data revealed very few changes in the transcription profile of T. gondii (Fig. 4A). Among 39 genes that displayed significant regulation due to tgap2XI-4 knock-out, only 12 genes had a logFC of <−2 (Table S3). In contrast, a direct comparison of microarray data from ΔTgAP2XI-4 and WT parasites after 2 days of culturing under pH8.2-stress conditions revealed a significant shift in the transcription profile of T. gondii due to the knock-out of tgap2XI-4 (Fig. 4B). A total of 72 genes showed significant fold-changes between ΔTgAP2XI-4 and the wild type (Table S5), and 22 genes had log fold-changes of <−2.0 in the knock-out mutant (Table 1). Interestingly, many of these 22 genes code for previously characterized bradyzoite-specific proteins including LDH2, BAG1, ENO1, P18, B-NTPase and DnaK-TPR. In addition, a comparison of the effect of pH 8.2-stress on ΔTgAP2XI-4 and the WT in Fig. 4C revealed that the knock-out is relatively impaired in its ability to regulate the transcription of many genes (R2 = 0.179).
Figure 4. Microarray analysis of RH ΔKu80 ΔTgAP2XI-4 under control and pH 8.2-stress conditions.
Total RNA purified from ΔTgAP2XI-4 and wild type parasites cultured either in control (pH 7) or stress (pH 8.2) conditions was analysed by microarray. Scatter plots represent the log fold-change (logFC) of T. gondii gene hits (plotted against average expression) in ΔTgAP2XI-4 vs wild type, at either pH 7 (A) or pH 8.2 (B). A broken green line surrounds an apparent sub-population of genes that show down-regulation shifts in ΔTgAP2XI-4 at pH 8.2.
(C) A scatter plot compares the logFC due to pH 8.2 stress in the wild type and ΔTgAP2XI-4. Plotted points were limited to genes displaying logFC of >2.0 in the pH 8.2-stressed wild type (i.e. putative bradyzoite genes). The regression line is in green. All points below the broken black represent putative bradyzoite genes with reduced transcriptional activation in ΔTgAP2XI-4.
Next, we analysed the putative promoter regions (the 5′ region of 1500 bp located upstream of the start ATG site) of the genes listed in Table 1 for the putative TgAP2XI-4 DNA regulatory element ‘CACACAC’. We found that 10 out of 22 (45%) of these genes contained this DNA motif. This is a higher frequency than for the putative promoters (including the 1500 bp region directly 5′ of the start ATG site) of all 8102 T. gondii genes in the Type I strain GT1 (approximately 35%). However, we cannot rule out the possibility that TgAP2XI-4 protein might also bind to other cis-acting elements because only a truncated protein was used in these gel shift assays.
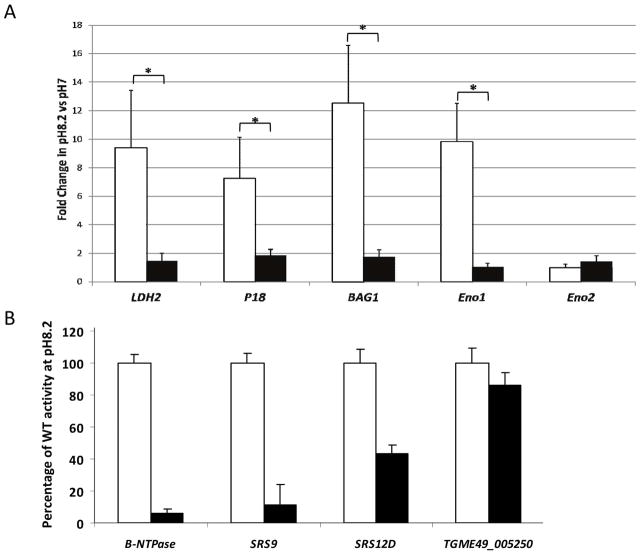
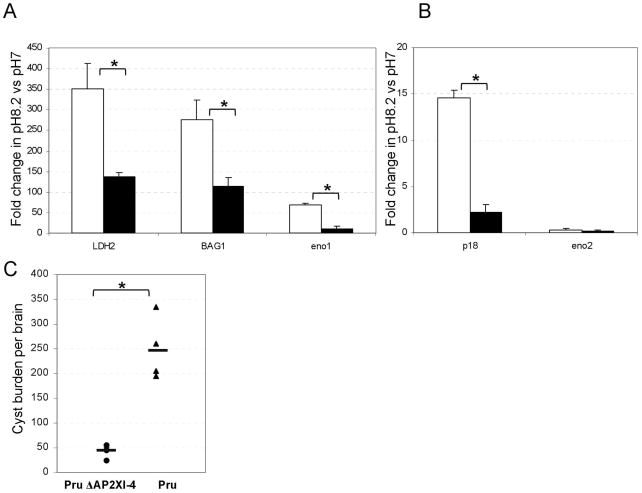
Validation of the microarray data was also carried out using qRT-PCR that compared transcripts from ΔTgAP2XI-4 and wild type samples. A number of well-characterised bradyzoite genes, including LDH2, P18, BAG1 and ENO1 were analysed, along with ENO2, which is not up-regulated during pH 8.2 stresses. Figure 5A clearly shows that pH 8.2-stress induces increased steady-state mRNA transcription of all four of the bradyzoite genes tested in the wild type, with a 9.4-fold increase for LDH2, 7.2-fold for P18, 12.5-fold for BAG1 and 9.8-fold ENO1. The degree to which these bradyzoite genes were up-regulated in ΔTgAP2XI-4 was estimated at 1.5-fold, 1.9-fold, 1.8-fold and 1.0-fold for LDH2, P18, BAG1 and ENO1, respectively. We also compared the promoter activities of selected bradyzoite genes in the ΔTgAP2XI-4 parasites versus the parental strain (Figure 5B). After shifting the parasite to alkaline pH, the promoter activity of the B-NTPase, SRS9 and SRS12D genes was measured in the parental and ΔTgAP2XI-4 strains using the luciferase assays. Figure 5B shows that the promoter activities of three bradyzoite specific-genes are significantly altered in the ΔTgAP2XI-4 strain while that of the unrelated gene TGME49_005250 remained unchanged.
Figure 5. (A) Quantitative RT-PCR of bradyzoite transcripts from RH ΔKu80 ΔTgAP2XI-4.
Total RNA purified from ΔTgAP2XI-4 and wild type parasites cultured either under control (pH 7) or stress (pH 8.2) conditions was analysed by quantitative RT-PCR. Genes coding for known bradyzoite proteins, including LDH2, P18, BAG1 and Eno1, were analysed alongside Eno2, which is not affected by alkaline stress. Values are presented as fold-change in pH8.2-stressed parasites relative to those under control conditions. An asterisk indicates a significant difference (P < 0.05) between the wild type and ΔTgAP2XI-4 mutant for individual genes.
(B) Promoter assay in the parental and ΔTgAP2XI-4 strains.B-NTPase, SRS9 and SRS12D bradyzoite gene promoters were assayed in a luciferase promoter assay as well as the promoter of the TGME49_005250 gene as a negative control. After shifting the culture to alkaline pH, the Firefly luciferase activity was measured and normalised to the Renilla luciferase activity of the co-transfected tubulin promoter in the parental strain (empty bars) and in the ΔTgAP2XI-4 strain (black bars). Luciferase activity was measured in duplicate and is represented as a percentage of the activity produced by a given promoter in the parental strain.
Bradyzoite markers and cyst burden are reduced in the type II Pru TgAP2XI-4 knock-out mutant
In order to assess the role of TgAP2XI-4 in T. gondii bradyzoite differentiation in chronically infected mice, a knock-out mutant was produced in the ΔKu80 Type II strain of T. gondii (Fox et al., 2011). As described in Figure 6, both the successful integration of the ΔTgAP2XI-4 knock-out construct and the resulting knock-out of the TgAP2XI-4 gene were confirmed by PCR (Fig. 6A). RT-PCR analysis also confirmed the absence of TgAP2XI-4 transcripts (Fig. 6B).
Figure 6. Construction of Pru ΔKu80 ΔTgAP2XI-4.
(A) Schematics representing the WT and recombinant TgAP2XI-4 locus. PCR was used to confirm the correct integration of the fused ΔTgAP2XI-4 construct (using primer pairs ii and iii) and deletion of the TgAP2XI-4 coding sequence (primer pair i).
(B) Absence of TgAP2XI-4 transcripts was confirmed by RT-PCR (primer pair i). Primers for TgAP2XI-5 (TGME49_016220) were used as a control (con).
In vitro proliferation assays carried out under conditions suitable for tachyzoite growth revealed no clear differences between the parental and Pru ΔTgAP2XI-4 parasites (Fig. S6A). Given that knock-out of tgap2XI-4 in the type I strain perturbed the activation of genes strongly associated with bradyzoite development, we assessed the ability of the type II Pru ΔTgAP2XI-4 parasites to grow and produce bradyzoite markers after 5 days of pH8.2 treatment, a potent inducer of bradyzoite markers in type II strains. As shown in Figure S6B, growth of the type II Pru ΔTgAP2XI-4 parasites were not significantly different from the parental parasites. However, expression of bradyzoites markers as measured by RT-qPCR was reduced when compared to the parental parasites (Fig. 7A and 7B). Finally, we measured the cyst burden in mouse brain 5 weeks post-infection. We found that the Pru ΔTgAP2XI-4 strain produced at least 5-fold fewer cysts per brain than the Pru parental strain, indicating a role for the TgAP2XI-4 gene in controlling bradyzoite differentiation and cyst formation (Fig. 7C). It should be noted that cysts from the Pru ΔTgAP2XI-4 strain were of a similar size to those of the parental strain, and were similarly able to differentiate back to tachyzoites after pepsin treatment (data not shown).
Figure 7. Phenotypic analysis of Pru ΔKu80 ΔTgAP2XI-4.
(A) Quantitative RT-PCR of bradyzoite transcripts from Pru ΔKu80 ΔTgAP2XI-4. Total RNA purified from ΔTgAP2XI-4 and parental parasites cultured under either control (pH 7) or stress (pH 8.2) conditions were analysed by quantitative RT-PCR. Genes coding for known bradyzoite proteins including LDH2, P18, BAG1 and Eno1 were analysed alongside Eno2, which is not affected by alkaline stress. Values are presented as fold-change in pH 8.2-stressed parasites relative to those in control conditions. An asterisk indicates a significant difference (P < 0.05) between the wild type and the ΔTgAP2XI-4 mutant for individual genes.
(B) Dolichos bifluorus lectin staining of parental and ΔTgAP2XI-4 strain after 2 days of treatment at pH 8.2. The percentage of vacuoles positive for lectin staining is represented. Data are means +/− SD. An asterisk indicates a significant difference (P < 0.05).
(C) Cyst burden in mouse brain. Cysts were enumerated after Dolichos bifluorus lectin staining of the cyst wall. A minimum of five mice was used per group. Brains of mice infected with the Pru ΔTgAP2XI-4 strain are represented with circles. Brains of mice infected with the Pru strain are represented with triangles. The mean cyst burden for each group is represented by a horizontal bar.
Discussion
As in other apicomplexan parasites, T. gondii gene expression is tightly regulated during stage conversion and differentiation. However, little is known about the actors coordinating the maintenance of and changes in this expression program. Although DNA motifs have been found in the promoters of genes activated during tachyzoite-to-bradyzoite differentiation, the specific transcription factors binding those elements are unknown. We examined the possibility that one of the putative ApiAP2 transcription factors might be involved in regulation of the specific stress-induced bradyzoite expression program. The results presented here demonstrate that TgAP2XI-4 is involved in regulating the induction of the bradyzoite expression program in vitro after alkaline stress. Most importantly, the ablation of TgAP2XI-4 has significantly impaired bradyzoite differentiation and cyst formation in the brains of chronically infected mice.
T. gondii encodes a large number of putative AP2 proteins and most of them have been shown to be expressed at the tachyzoite stage (Behnke et al., 2010). In contrast to Plasmodium, which encodes only 27 ApiAP2 genes, the expansion of the number of AP2s in T. gondii (68) increases the possibility of a large functional redundancy. In order to screen for genes potentially involved in bradyzoite differentiation, we identified those over-expressed in bradyzoites when compared to tachyzoites. TgAP2XI-4 was found to have strong transcript abundance in brain tissue cysts (in vivo) and to be moderately over-expressed after stress-induced differentiation in vitro. Most strikingly, TgAP4XI-4 transcript and protein expression are regulated according to the tachyzoite cell cycle, with maximal protein expression during the cytokinesis and early G1 phases. However, the tgap2XI-4 gene knock-out had no marked defects in normal tachyzoite growth conditions, exhibiting only modest changes in gene expression (Table S3). This raises into question the exact role of TgAP2XI-4 regulation during the tachyzoite cell cycle. In contrast, the TgAP2XI-4 knock-out mutant showed a marked perturbation in its ability to induce the bradyzoite expression program. Moreover, the presence of the protein during cytokinesis and the early G1 phase of the tachyzoite cell cycle matches the expression pattern of several cell cycle-regulated bradyzoite markers (Behnke, 2010); this may be crucial for explaining the rapid response to stress-induced bradyzoite differentiation. There is a close relationship between cell cycle and bradyzoite differentiation (Bohne et al., 1994; Radke et al., 2003) and differentiation cannot occur when cell cycle progression is blocked (Radke et al., 2001). In fact, tachyzoites which have stopped their proliferation undergo differentiation from a novel late S/G2 subpopulation (Radke et al., 2001). Because the type I strain of T. gondii is not the best model for studying bradyzoite differentiation in vivo, we performed a knock-out of this gene in the recently available Pru ΔKu80 type II strain (Fox et al., 2011). Both expression of bradyzoite markers in vitro and in vivo cyst burden were significantly altered in the type II strain tgap2XI-4 knock-out mutant, when compared to the parental strain. This strongly suggests that TgAP2XI-4 may play a role in the processes leading to bradyzoite differentiation and cyst formation.
The AP2 transcription factors of Plasmodium have been shown to bind double-stranded DNA in a sequence-specific manner (Campbell et al., 2010). In our study, the ‘CACACAC’ motif was investigated as a putative DNA regulatory target of TgAP2XI-4 based on its clear homology with PFD0985w, the P. falciparum AP2 for whose DNA binding preference was solved in silico. The ability of TgAP2XI-4 to specifically bind to an extended version of this motif in EMSA studies confirms its DNA-binding capacity. In vitro, our results show that 5 CA repeats must be present in the oligonucleotide probe for efficient competition between TgAP2XI-4 and the DNA motif. However, EMSA were performed using a recombinant protein containing only the AP2 domain, so this specific binding does not account for possible contributions by the remaining protein or for other interacting factors that may change its binding specificity. While the CACACAC motif can be found in 45% of the putative promoters of genes most clearly affected by the knock-out of tgap2XI-4 at pH 8.2 (Table 1), other cis or trans acting factors may also be involved in regulating those genes. One such example is the well-characterized DNA element (TACTGG) in the BAG1 gene promoter, which is essential for its expression in the bradyzoites (Behnke et al., 2008). TgAP2XI-4 may also be able to bind other DNA elements that were not found with our bioinformatics analyses, despite several attempts with different motif-search softwares. Importantly, another cis regulatory element has been characterized in the promoter of B-NTPase (Behnke et al., 2008; Nakaar et al., 1998), a bradyzoite-specific gene (Nakaar et al., 1998) whose transcription is also significantly affected by the knock-out of tgap2XI-4. The element contains not only the CACACAC motif but a slightly mutated form (2 base substitutions) of the extended motif used for the EMSA analysis of TgAP2XI-4. The B-NTPase element has been shown to drive gene expression during bradyzoite development. Furthermore, trans-activating regulators capable of binding this element were detected in nuclear extracts of in vitro bradyzoites, and this element is sufficient to confer bradyzoite-inducible expression to a tachyzoite gene (Behnke et al., 2008). Unfortunately, we were not able to show that TgAP2XI-4 can bind to this promoter in vivo because chromatin immunoprecipitation experiments (ChIP-on-chip and ChIP-Seq) carried out unfortunately failed, even when these experiments were run under both control and alkaline-stress conditions. This failure is possibly due to the relatively low level of protein expression. When we compared the expression profile of the ΔTgAP2XI-4 strain with other type I mutants defective for bradyzoite differentiation (Lescault et al., 2010) under alkaline pH stress, we discovered that about 60% of the genes differentially regulated in the ΔTgAP2XI-4 strain are common to previous studies. Importantly, mice infected with the Pru ΔTgAP2XI-4 strain show a 5-fold reduction of cyst burden, indicating that TgAP2XI-4 is involved in bradyzoite differentiation and not exclusively in the in-vitro alkaline stress response pathway. Therefore, it is possible that the knock-out of tgap2XI-4 could be causing broad downstream effects, particularly given that some of the genes differentially expressed in ΔTgAP2XI-4 may play a role in the regulation of bradyzoite-specific genes. As an example, two zinc finger proteins (TGME49_048330 and TGME49_062970) are down-regulated in the knock-out and may play a role as putative downstream effectors.
It is also worth noting that numerous attempts to complement the ΔTgAP2XI-4 mutant with an exogenous copy of the gene were unsuccessful, due to the unavailability of cosmid clones encompassing the large size TgAP2XI-4 gene (17 kb) or its corresponding cDNA (>10 kb). Nevertheless, together with TgAP2XII-6 (Lescault et al., 2010), TgAP2XI-4 are two known sequence specific transcription factors described so far to be involved in the regulation of the bradyzoite expression program and cyst production in vivo, despite numerous attempts by reverse genetics and other strategies (Singh et al., 2002; Matrajt et al., 2002). Further studies of the biological functions of TgAP2XI-4 will shed light on how this novel T. gondii nuclear factor that binds to a specific DNA motif regulates the bradyzoite expression program and differentiation, leading to cyst formation in vivo.
Experimental procedures
Parasite tissue culture and manipulation
T. gondii strain RH ΔKu80 (Huynh, 2009) tachyzoites were propagated in vitro in human foreskin fibroblasts (HFF) using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS (Fetal Calf Serum), 2mM glutamine and 1% penicillin-streptomycin. T. gondii tachyzoites were grown in ventilated tissue culture flasks at 37°C and 5% CO2. Transgenes were introduced by electroporation into tachyzoites of the T. gondii RH ΔKu80 or Pru ΔKu80 strain and stable transformants were selected in the presence of 2 μM pyrimethamine. Clonal lines were obtained by limiting dilution. Prior to RNA and protein purification, intracellular parasites were purified by sequential syringe passage through 17 gauge and 26 gauge needles and filtration through a 3 μm polycarbonate membrane filter.
In vivo parasite infections
For RNA harvest, the brains of mice chronically infected after ingestion of 20 cysts of the type II 76K strain of T. gondii were collected after 6 weeks to isolate cysts that were then purified using Percoll gradients, washed with PBS, and counted by inverted phase microscopy as previously described (Tomavo et al., 1991). Encysted bradyzoites were released using 0.05% pepsin/HCl, and mRNA was isolated and cDNA was synthesized as previously described (Dzierszinski et al., 2001). Purified tachyzoites from the parental and knock-out mutant were also inoculated into groups of four female 6–8-week-old Balb/c mice and monitored until death or survival for 1 month. C57b/6 mice (5) were intraperitoneally injected with 200 parasites. After 4 weeks, brains were collected and homogenised as described (Fox et al., 2011). Cyst counts were performed after Dolichol biflorus lectin labelling of the cyst wall for 30 min at room temperature in PBS. Ten slides corresponding to one fifth of each brain were scored for the presence of lectin-positive cysts.
In vitro stress conditions and parasite growth assays
To induce bradyzoite development in vitro, freshly lysed parasites were allowed to infect HFF cells for 4 hours. The normal DMEM culture medium described above was then replaced with RPMI 1640 medium supplemented with 20 mM HEPES, 1% penicillin-streptomycin and 1% FCS and adjusted to pH 8.2. For RNA purification and protein purification, parasites were then cultured at 37°C without CO2 for 2 days for the RH ΔKu80 strain and 5 days for the Pru ΔKu80 strain. Proliferation assays were carried out using 2.5×104 parasites per sample over 24 hours, and were cultured under either control or alkaline stress conditions in 24-well plates. For invasion assays, extracellular parasites were incubated in control or alkaline media for 2 hours at 37°C, after which 103 parasites were inoculated onto confluent HFF monolayers in 24-well plates. Plaques were left to grow over 5 days under normal culture conditions. Fixation and staining for proliferation and invasion assays were carried out using the RAL555 kit.
Quantitative real time PCR (qRT-PCR)
All primers were designed online using Primer2 v.0.4.0 (http://frodo.wi.mit.edu/primer3/) and are listed in Table S1. The cDNA samples were synthesized from total RNA samples using the Revert Aid™ First Strand cDNA Synthesis Kit (Fermentas). The qRT-PCR was carried out on an Mx3000P System (Agilent Technologies). Individual reactions were prepared with 0.5 μM of each primer, ~5 ng of cDNA and SYBR Green PCR Master Mix (Applied Biosystmes, CA) to a final volume of 20 μl. All experiments were performed twice with separate biological replicates. For each experiment, reactions were performed in triplicate and expression of individual genes was normalized to housekeeping tubulin gene Ct values.
DNA manipulation
TgAP2XI-4 (TGME49_115760) was amplified from genomic DNA of the parental strain and cloned into the pLIC.HA9.DHFR vector (Huynh et al., 2009) kindly provided by Vern Carruthers and Michael White. A region of the TgAP2XI-4 coding sequence corresponding to amino acids 2917 to 3229 was PCR-amplified and cloned into the pGex-6P3 vector using EcoRI and BamHI sites. Positive clones were selected using ampicillin. All PCR reactions for both the generation of ΔTgAP2XI-4 constructs and the screening of mutant clones were carried out using High-Fidelity PCR Enzyme Mix (Fermentas) as per manufacturer’s instructions.
The knock-out of TgAP2XI-4 in the RH ΔKu80 strain was carried out using a modified version of the protocol as described (Upadhya et al., 2011). TgAP2XI-4 5′ and 3′ flanking regions were amplified from type I ΔKu80 T. gondii genomic DNA and the two halves of the pyrimethamine-resistant DHFR cassette were amplified from pLIC.HA9.DHFR plasmid DNA. PCR products generated from the first PCR were used as templates for the second PCR (illustrated in Fig. 3a). The PCR products from the second PCR were transfected together in equimolar quantities (typically between 5 and 10 μM per transfection).
The knock-out of TgAP2XI-4 in the Pru ΔKu80 strain was performed after amplification of 2000 bp of the 5′ and 3′ flanking regions of the TgAP2XI-4 gene. The flanking regions were cloned into the pDHFR plasmid using ApaI and HindIII (5′ flanking) and SpeI and NotI (3′ flanking). The plasmid was linearised with BglI before transfection. A list of primers used is provided in supplementary Table 1.
GST fusion protein purification
The pGex-6P3 plasmid was used for the expression of a truncated TgAP2XI-4 protein with an N-terminal GST-tag. The plasmid was transformed into BL21 E. coli, grown to OD600 = 0.6 and induced by the addition of 1 mM IPTG. After incubation for 12 hours at 22°C, the bacteria were centrifuged at 180 rpm and the pellet was lysed by sonication in PBS supplemented with 1 mM PMSF. Following incubation with 1% Triton X-100 on ice for 30 min, the samples were centrifuged. The GST-tagged TgAP2XI-4 protein was then purified from the soluble fraction by binding to glutathione Sepharose 4B resin (GE Healthcare) and eluted with 20 mM reduced glutathione. The purified protein was washed with PBS using a 30 kDa cut-off centrifugal filter (Amicon® Ultra) and quantified using the Bio-Rad protein assay.
Electro-Mobility Shift Assays (EMSA)
A 179-bp region within the putative promoter of TGME49_017700 that contained the extended ‘CACACACACACA’ motif was amplified using the primers listed in Table S1. Biotinylation was carried out using the Biotin DecaLabel™ DNA Labelling Kit (Fermentas) according to manufacturer’s instructions. All other probes (listed in Table S2) were purchased from Sigma-Aldrich. The annealing of sense and anti-sense ssDNA probes was carried out by incubating complementary probes for 5 min at 95°C in 10 mM Tris, 1 mM EDTA, 50 mM NaCl (pH 8.0) and then cooling overnight at room temperature. EMSA were performed using the LightShift Chemiluminescent EMSA Kit (Pierce) as follows: 1) the provided 10X binding buffer was supplemented with 1 μg of Poly (dI·dC) and pre-incubation with 2.5 μg (40pmol) of recombinant protein and 1.0 pmol of appropriate competitor was carried out for 10 min at room temperature; 2) 10 fmol of biotinylated probe was added and the samples were incubated for a further 30 minutes at room temperature; 3) the mixture was then resolved on a pre-run 5% acrylamide gel prepared in 0.5X TBE. The DNA was then transferred onto a Nylon membrane and blotted with Steptavidin-HRP beads according to the manufacturer’s instructions.
Luciferase assay
Promoters of the B-NTPase, SRS9 and SRS12D genes were cloned to drive the expression of the firefly luciferase. Fifty μg of these plasmids were co-transfected together with 10 μg of the Tubulin promoter-Renilla Luciferase plasmid into the Pru ΔKu80 and the Pru ΔKu80 ΔTgAP2XI-4 strains. Luciferase activity was measured after switching the cultures for 5 days in alkaline pH media using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Purification of RNA and microarray
Total RNA was purified from the RH ΔKu80 strain after 2 days of in-vitro culture under either control (pH 7.0) or alkaline (pH 8.2) stress conditions. Total RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel) according to manufacturer’s instructions. At least two independent experiments were performed. Experiments were performed starting from 0.5 μg of total RNA. A one-cycle labelling procedure for the synthesis of fluorescent targets was followed. This protocol allows for signal amplification due to the synthesis of a cDNA intermediate including a T7-RNA polymerase cassette, which is used to obtain in vitro transcription (IVT). Biotinylated ribonucleotide analogues are incorporated in target RNA during IVT. This biotinylated RNA is then fragmented before being hybridized on an Affymetrix chip. After hybridization, the biotin is recognised by a streptavidin-phycoerythrin and the signal is amplified by the presence in the labelling reaction of biotinylated anti-streptavidin antibody. All hybridizations were performed on the Toxoplasma Geneship Affymertix microarray as described (Bahl, 2010).
After hybridizations, the raw data were analysed using the RMA (Robust Multi-array Average)(Irizarry et al., 2003)and LIMMA (Linear Models for Microarray Data) (Smyth et al., 2004) packages that were run under the statistical language R. A normalization protocol, consisting of a within-array loess normalization to correct for dye and spatial effects (Yang et al., 2002), was applied on the background corrected median or mean intensities. After normalization, identification of statistically significant regulation was performed using moderated t-statistic with empirical Bayes shrinkage of the standard errors.
Supplementary Material
Figure S1: Comparative expression of selected TgAP2 genes in bradyzoite tissue cysts and tachyzoites. The 76K type II strain was either used to produce bradyzoite tissue cysts (in vivo) or cultured under normal tachyzoite conditions (in vitro). Total RNA was purified from the two samples and analysed by quantitative RT-PCR to determine the relative levels of TgAP2 genes, Eno1 and Eno2 mRNA. The values are presented as the fold-change in the bradyzoite samples relative to the corresponding tachyzoite samples.
Figure S2: TgAP2XI-4 expression is regulated throughout the tachyzoite cell-cycle. Immunofluorescence assays were conducted on PAF-fixed intracellular TgAP2XI-4 parasites at 24-hours post-infection. A mouse monoclonal αHA antibody was used in combination with a rabbit αMORN1 antibody and detected with anti-mouse Alexa488 (green) and anti-rabbit Alexa594 (red), respectively. Daughter cell formation and mitosis were monitored with αMORN1 and DAPI staining (blue), respectively.
Figure S3: EMSA using different sizes of the CA competitor.
(A) An electrophoretic mobility shift assay was carried out using a recombinant protein spanning the TgAP2XI-4 AP2 domain. The shift caused by the binding of TgAP2XI-4 to the biotinylated probe is indicated by an arrow. TgAP2XI-4 binding was inhibited by a 250-fold or 500-fold excess of specific competitor CA(6) but not by a non-specific competitor.
(B) An electrophoretic mobility shift assay was carried out using a recombinant protein spanning the TgAP2XI-4 AP2 domain. The shift caused by the binding of TgAP2XI-4 to the biotinylated probe is indicated by an arrow. TgAP2XI-4 binding was inhibited by a 100-fold excess of specific competitor CA(6) and partially mutated CA(5) but not by a partially mutated competitor CA(4).
Figure S4: ΔTgAP2XI-4 proliferation and virulence assays. (A) Proliferation assays were carried out over 24 hours in vitro using ΔTgAP2XI-4 and wild type parasites under normal pH 7 conditions. Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites. (B) Virulence assays were carried out using Balb/c mice infected with ΔTgAP2XI-4 and wild type parasites, respectively. The percentage of survival in each group of infected mice was monitored over 10 days.
Figure S5: Effect of alkaline pH 8.2-stress on ΔTgAP2XI-4 proliferation and invasion.
(A) Proliferation assays were carried out for 24 hours in vitro using ΔTgAP2XI-4 and wild type parasites under alkaline pH 8.2-stress conditions. Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites. (B) Invasion assays were performed for 5 days in vitro using ΔTgAP2XI-4 and wild type parasites following an extracellular treatment for 2 hour under either control pH7 or alkaline pH8.2-stress conditions. Results are presented as the number of plaques counted at pH 8.2 relative to the number at pH 7.
Figure S6: Growth of the Pru ΔKu80 ΔTgAP2XI-4 mutant. (A) Proliferation assays were carried out over 48 hours in vitro using ΔTgAP2XI-4 and wild type parasites under normal conditions (pH 7). Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites. (B) Proliferation assays were carried out over 48 hours in vitro on ΔTgAP2XI-4 and wild type parasites under alkaline pH conditions (pH 8.2). Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites.
Table S1: Primers used in this study. The names and sequences of all primers used in this study are listed along with their associated gene targets and experimental applications. Underlined regions of primer sequences indicate additional, non gene-specific regions required for either cloning or fusion PCR. FWD = forward primer, REV = reverse primer.
Table S2: EMSA probes used in this study. Four single-stranded oligonucleotides were synthesized for the construction of two different EMSA probes specific to the putative promoter region of the gene TGME49_017700. The underlined regions indicate the putative cis-regulatory element of the specific competitor, while the regions in bold and italics indicate the mutated sequence of the non-specific competitor.
Table S3: Differentially regulated genes in ΔTgAP2XI-4 vs wild type at pH7. Microarray analysis of ΔTgAP2XI-4 grown under control pH 7 conditions reveals the differential regulation of 39 genes compared to wild type. Genes displayed were significantly regulated (adjusted P value < 0.05) by a log fold change of at least +/− 1.0.
Table S4: Genes up-regulated in wild type ΔKu80 Type I T. gondii following in vitro bradyzoite induction. Microarray analysis of T. gondii ΔKu80 type I (wild type) cultured under alkaline pH 8.2-stress conditions reveals increased transcription of 272 genes compared to control conditions. Genes that were significantly up-regulated (adjusted P value < 0.05) by a log fold change of at least 1.0 are shown.
Table S5: Differentially regulated genes in ΔTgAP2XI-4 vs wild type at pH 8.2.
Microarray analysis of ΔTgAP2XI-4 cultured under alkaline pH 8.2-stress conditions reveals the differential regulation of 73 genes compared to wild type. Genes that were significantly regulated (adjusted P value < 0.05) by a log fold change of at least +/− 1.0 are shown. Two unique microarray probes*, designed to what were originally thought to be two unique genes, actually match different regions of the same gene, TGME49_009750.
Acknowledgments
The authors are grateful to Thomas Mouveaux and Joelle Duflot for technical assistance. We also thank Vern Carruthers, Michael White, Marc–Jan Gubbels and Gary Ward for providing reagents. This work was supported by the Centre National de la Recherche Scientifique (CNRS), Institut National de la Sante and de la Recherche Medicale (INSERM) and a grant from the French National Research Agency (ANR) number ANR-09-MIEN-002-01 (BradyToxoFT). MMC was supported by the Training Program in Cellular and Molecular Biology and Genetics, funded by NIH T32 GM007491 awarded to the Albert Einstein College of Medicine. KK was supported by NIH grants R01 AI087625 and RC4 AI092801.
References
- Bahl A, Davis PH, Behnke M, Dzierszinski F, Jagalur M, Chen F, et al. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics. 2010;11:603. doi: 10.1186/1471-2164-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Radke JB, Smith AT, Sullivan WJ, White MW. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol. 2008;68:1502–1518. doi: 10.1111/j.1365-2958.2008.06249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, et al. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS ONE. 2010;5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne W, Heesemann J, Gross U. Reduced replication of Toxoplasma gondii is necessary for induction of bradyzoite-specific antigens: a possible role for nitric oxide in triggering stage conversion. Infect Immun. 1994;62:1761–1767. doi: 10.1128/iai.62.5.1761-1767.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinás M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierszinski F, Mortuaire M, Dendouga N, Popescu O, Tomavo S. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J Mol Biol. 2001;309:1017–1027. doi: 10.1006/jmbi.2001.4730. [DOI] [PubMed] [Google Scholar]
- Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, et al. Type II Toxoplasma gondii KU80 knock-out strains enable functional analysis of genes required for cyst development and latent infection. Eukaryotic Cell. 2011;10:1193–1206. doi: 10.1128/EC.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 2007;3:e77. doi: 10.1371/journal.ppat.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissot M, Kim K, Schaap D, Ajioka JW. New eukaryotic systematics: a phylogenetic perspective of developmental gene expression in the Apicomplexa. Int J Parasitol. 2009;39:145–151. doi: 10.1016/j.ijpara.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryotic Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2007 doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Kibe MK, Coppin A, Dendouga N, Oria G, Meurice E, Mortuaire M, et al. Transcriptional regulation of two stage-specifically expressed genes in the protozoan parasite Toxoplasma gondii. Nucleic Acids Res. 2005;33:1722–1736. doi: 10.1093/nar/gki314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Weiss LM. Toxoplasma gondii: the model apicomplexan. Int J Parasitol. 2004;34:423–32. doi: 10.1016/j.ijpara.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescault PJ, Thompson AB, Patil V, Lirussi D, Burton A, Margarit J, et al. Genomic data reveal Toxoplasma gondii differentiation mutants are also impaired with respect to switching into a novel extracellular tachyzoite state. PLoS ONE. 2010;5:e14463. doi: 10.1371/journal.pone.0014463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, De Silva EK, Keck JL, Llinas M. Structural determinants of DNA binding by a P. falciparum ApiAP2 transcriptional regulator. J Mol Biol. 2010;395:558–567. doi: 10.1016/j.jmb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrajt M, Donald RGK, Singh U, Roos DS. Identification and characterization of differentiation mutants in the protozoan parasite Toxoplasma gondii. Mol Microbiol. 2002;44:735–747. doi: 10.1046/j.1365-2958.2002.02904.x. [DOI] [PubMed] [Google Scholar]
- Naguleswaran A, Elias EV, McClintick J, Edenberg HJ, Sullivan WJ., Jr Toxoplasma gondii lysine acetyltransferase GCN5-A functions in the cellular response to alkaline stress and expression of cyst genes. PLoS Pathog. 2010;6:e1001232. doi: 10.1371/journal.ppat.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaar V, Bermudes D, Peck KR, Joiner KA. Upstream elements required for expression of nucleoside triphosphate hydrolase genes of Toxoplasma gondii. Mol Biochem Parasitol. 1998;92:229–239. doi: 10.1016/s0166-6851(97)00220-x. [DOI] [PubMed] [Google Scholar]
- Painter HJ, Campbell TL, Llinás M. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol Biochem Parasitol. 2011;176:1–7. doi: 10.1016/j.molbiopara.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke JR, Striepen B, Guerini MN, Jerome ME, Roos DS, White MW. Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:165–175. doi: 10.1016/s0166-6851(01)00284-5. [DOI] [PubMed] [Google Scholar]
- Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW. The transcriptome of Toxoplasma gondii. BMC Biol. 2005;3:26. doi: 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke JR, Guerini MN, Jerome M, White MW. A change in the premitotic period of the cell cycle is associated with bradyzoite differentiation in Toxoplasma gondii. Mol Biochem Parasitol. 2003;131:119–127. doi: 10.1016/s0166-6851(03)00198-1. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25:10301–14. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva EK, Gehrke AR, Olszewski K, León I, Chahal JS, Bulyk ML, et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci USA. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U, Brewer JL, Boothroyd JC. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol Microbiol. 2002;44:721–733. doi: 10.1046/j.1365-2958.2002.02903.x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Tomavo S, Fortier B, Soete M, Ansel C, Camus D, Dubremetz JF. Characterization of bradyzoite-specific antigens of Toxoplasma gondii. Infect Immun. 1991;59:3750–3753. doi: 10.1128/iai.59.10.3750-3753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Kim K, Hogue-Angeletti R, Weiss LM. Improved techniques for endogenous epitope tagging and gene deletion in Toxoplasma gondii. J Microbiol Methods. 2011;85:103–113. doi: 10.1016/j.mimet.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van TT, Kim SK, Camps M, Boothroyd JC, Knoll LJ. The BSR4 protein is up-regulated in Toxoplasma gondii bradyzoites, however the dominant surface antigen recognised by the P36 monoclonal antibody is SRS9. Int J Parasitol. 2007;37:877–885. doi: 10.1016/j.ijpara.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Vanchinathan P, Brewer JL, Harb OS, Boothroyd JC, Singh U. Disruption of a locus encoding a nucleolar zinc finger protein decreases tachyzoite-to-bradyzoite differentiation in Toxoplasma gondii. Infect Immun. 2005;73:6680–6688. doi: 10.1128/IAI.73.10.6680-6688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuda M, Iwanaga S, Shigenobu S, Kato T, Kaneko I. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol Microbiol. 2010;75:854–863. doi: 10.1111/j.1365-2958.2009.07005.x. [DOI] [PubMed] [Google Scholar]
- Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, Waters AP, et al. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol Microbiol. 2009;71:1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Comparative expression of selected TgAP2 genes in bradyzoite tissue cysts and tachyzoites. The 76K type II strain was either used to produce bradyzoite tissue cysts (in vivo) or cultured under normal tachyzoite conditions (in vitro). Total RNA was purified from the two samples and analysed by quantitative RT-PCR to determine the relative levels of TgAP2 genes, Eno1 and Eno2 mRNA. The values are presented as the fold-change in the bradyzoite samples relative to the corresponding tachyzoite samples.
Figure S2: TgAP2XI-4 expression is regulated throughout the tachyzoite cell-cycle. Immunofluorescence assays were conducted on PAF-fixed intracellular TgAP2XI-4 parasites at 24-hours post-infection. A mouse monoclonal αHA antibody was used in combination with a rabbit αMORN1 antibody and detected with anti-mouse Alexa488 (green) and anti-rabbit Alexa594 (red), respectively. Daughter cell formation and mitosis were monitored with αMORN1 and DAPI staining (blue), respectively.
Figure S3: EMSA using different sizes of the CA competitor.
(A) An electrophoretic mobility shift assay was carried out using a recombinant protein spanning the TgAP2XI-4 AP2 domain. The shift caused by the binding of TgAP2XI-4 to the biotinylated probe is indicated by an arrow. TgAP2XI-4 binding was inhibited by a 250-fold or 500-fold excess of specific competitor CA(6) but not by a non-specific competitor.
(B) An electrophoretic mobility shift assay was carried out using a recombinant protein spanning the TgAP2XI-4 AP2 domain. The shift caused by the binding of TgAP2XI-4 to the biotinylated probe is indicated by an arrow. TgAP2XI-4 binding was inhibited by a 100-fold excess of specific competitor CA(6) and partially mutated CA(5) but not by a partially mutated competitor CA(4).
Figure S4: ΔTgAP2XI-4 proliferation and virulence assays. (A) Proliferation assays were carried out over 24 hours in vitro using ΔTgAP2XI-4 and wild type parasites under normal pH 7 conditions. Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites. (B) Virulence assays were carried out using Balb/c mice infected with ΔTgAP2XI-4 and wild type parasites, respectively. The percentage of survival in each group of infected mice was monitored over 10 days.
Figure S5: Effect of alkaline pH 8.2-stress on ΔTgAP2XI-4 proliferation and invasion.
(A) Proliferation assays were carried out for 24 hours in vitro using ΔTgAP2XI-4 and wild type parasites under alkaline pH 8.2-stress conditions. Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites. (B) Invasion assays were performed for 5 days in vitro using ΔTgAP2XI-4 and wild type parasites following an extracellular treatment for 2 hour under either control pH7 or alkaline pH8.2-stress conditions. Results are presented as the number of plaques counted at pH 8.2 relative to the number at pH 7.
Figure S6: Growth of the Pru ΔKu80 ΔTgAP2XI-4 mutant. (A) Proliferation assays were carried out over 48 hours in vitro using ΔTgAP2XI-4 and wild type parasites under normal conditions (pH 7). Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites. (B) Proliferation assays were carried out over 48 hours in vitro on ΔTgAP2XI-4 and wild type parasites under alkaline pH conditions (pH 8.2). Counts are presented as the percentage of vacuoles containing 2, 4, 8 or 16 parasites.
Table S1: Primers used in this study. The names and sequences of all primers used in this study are listed along with their associated gene targets and experimental applications. Underlined regions of primer sequences indicate additional, non gene-specific regions required for either cloning or fusion PCR. FWD = forward primer, REV = reverse primer.
Table S2: EMSA probes used in this study. Four single-stranded oligonucleotides were synthesized for the construction of two different EMSA probes specific to the putative promoter region of the gene TGME49_017700. The underlined regions indicate the putative cis-regulatory element of the specific competitor, while the regions in bold and italics indicate the mutated sequence of the non-specific competitor.
Table S3: Differentially regulated genes in ΔTgAP2XI-4 vs wild type at pH7. Microarray analysis of ΔTgAP2XI-4 grown under control pH 7 conditions reveals the differential regulation of 39 genes compared to wild type. Genes displayed were significantly regulated (adjusted P value < 0.05) by a log fold change of at least +/− 1.0.
Table S4: Genes up-regulated in wild type ΔKu80 Type I T. gondii following in vitro bradyzoite induction. Microarray analysis of T. gondii ΔKu80 type I (wild type) cultured under alkaline pH 8.2-stress conditions reveals increased transcription of 272 genes compared to control conditions. Genes that were significantly up-regulated (adjusted P value < 0.05) by a log fold change of at least 1.0 are shown.
Table S5: Differentially regulated genes in ΔTgAP2XI-4 vs wild type at pH 8.2.
Microarray analysis of ΔTgAP2XI-4 cultured under alkaline pH 8.2-stress conditions reveals the differential regulation of 73 genes compared to wild type. Genes that were significantly regulated (adjusted P value < 0.05) by a log fold change of at least +/− 1.0 are shown. Two unique microarray probes*, designed to what were originally thought to be two unique genes, actually match different regions of the same gene, TGME49_009750.