Abstract
The roots of Salvia miltiorrhiza (“Danshen”) are used in traditional Chinese medicine for the treatment of numerous ailments including cardiovascular disease, hypertension, and ischemic stroke. Extracts of S. miltiorrhiza roots in formulation “Compound Danshen Dripping Pill” is undergoing clinical trials in the U.S. To date, the active components of this material have not been conclusively identified. We have determined that S. miltiorrhiza roots contain potent human carboxylesterase (CE) inhibitors, due to the presence of tanshinones. Ki values in the nM range were determined for inhibition of both the liver and intestinal CEs. As CEs hydrolyze clinically used drugs, the ability of tanshinones and S. miltiorrhiza root extracts to modulate the metabolism of the anticancer prodrug irinotecan (CPT-11) was assessed. Our results indicate that marked inhibition of human CEs occurs following incubation with both pure compounds and crude material, and that drug hydrolysis is significantly reduced. Consequently, a reduction in the cytotoxicity of irinotecan is observed following dosing with either purified tanshinones or S. miltiorrhiza root extracts. It is concluded that remedies containing tanshinones should be avoided when individuals are taking esterified agents, and that patients should be warned of the potential drug-drug interaction that may occur with this material.
In the past, drug discovery approaches traditionally used natural products as templates for the design of novel therapeutic agents. However recently, with the advent of alternative screening and chemical synthesis strategies, a considerable number of newly approved drugs have been developed using totally synthetic scaffolds. However, the majority of these molecules tend to be insoluble in aqueous media and have relatively poor bioavailability. To overcome these liabilities, medicinal chemists typically add chemotypes such as esters, sulfonamides and amides. Therefore many new clinically used agents (e.g. Lunesta® (eszopiclone), Ritalin® (methylphenidate), Altace® (ramipril)) contain ester moieties and are, as a consequence, substrates for carboxylesterase enzymes (CE).1, 2 These proteins hydrolyze these small molecules into the corresponding carboxylic acid and the alcohol,3, 4 a process that can either activate, or inactivate the drug.
In humans, two CE isoforms have been characterized 5–8 that demonstrate markedly different substrate specificities and patterns of expression. hCE1 (CES1) is primarily expressed in the liver and hydrolyzes small, relatively compact molecules. This includes drugs such as Plavix® (clopidogrel),9 methylphenidate,1 and Tamiflu® (oseltamivir).10 In contrast, hiCE (CES2; hCE2) is localized to the epithelia of the gut, the liver, and the kidney, and demonstrates considerably more plasticity with respect to compounds that can be hydrolyzed.11 This includes the anticancer agent irinotecan, (CPT-11, 7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin)5, 8 and the alkaloid cocaine.12 The difference in substrate specificity of these proteins is likely due to the regulated access of the ester to the catalytic residues that are buried deep within the protein, at the bottom of a long hydrophobic gorge.13–15
Recently, we identified several different classes of selective CE inhibitors and demonstrated that some of these molecules are cell permeable.16–20 These compounds resulted in the modulation of drug metabolism, and with irinotecan, led to reduced cytotoxicity in cell culture models 18. These results confirmed that inhibition of CEs can be deleterious with respect to prodrug hydrolysis, and that such activities may compromise the therapeutic efficacy of clinically used agents. While a few reports have appeared on the occurrence of CE inhibitors of plant-origin, the majority of these compounds demonstrate only modest activity, with Ki or IC50 values in the low micromolar range.21, 22 These molecules were identified by screening different extracts against purified pig liver using a colorimetric substrate. In the present investigation a different approach was adopted, using a defined pharmacophore model to identify structural analogues and then assaying these purified compounds. Using this methodology, the tanshinones have been identified as potent CE inhibitors, and it has been demonstrated that these molecules are present within the Chinese herbal medicine Danshen. Extracts of the latter, and the tanshinones, modulate the efficacy of irinotecan-induced cytotoxicity by inhibiting human CEs.
RESULTS AND DISCUSSION
Identification of Tanshinones as Potential Inhibitors of Human CEs
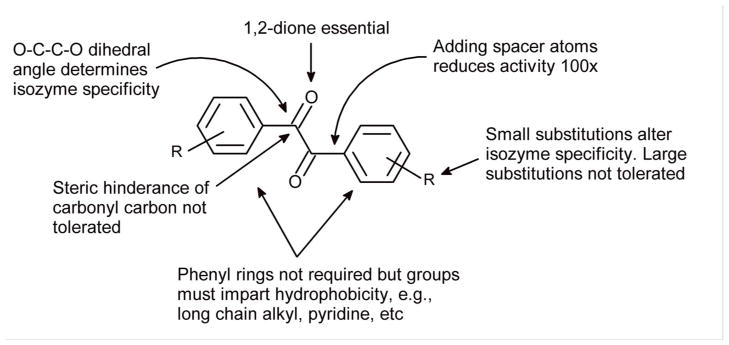
Based on our previous observations using benzils, isatins, 1,2-quinones, alkyl-1,2-diones and 1-phenylalkyl-1,2-diones,16, 19, 23–25 our group has identified the essential components of small molecules that account for CE inhibition. These include the necessity for the 1,2-dione chemotype that is not sterically hindered, the presence of relatively small hydrophobic domains, and the absence of a heteroatom in the α-position with respect to the carbonyl carbon atom (Figure 1). Using this information, a pharmacophore model was generated based on ten previously identified potent CE inhibitors using MOE 2011.10 software. These molecules, and their associated Ki values, are presented in Table 1. The development of the pharmacophore was driven principally by observations that efficient CE inhibition was only observed by molecules containing a 1,2-dione chemotype that had a relatively high clogP value (typically >3; Figure 119, 23–25). Therefore, two molecules each, of five different chemical classes were selected that met these criteria (Table 1), and a suitable model was generated that could be used for database searching. Alignment of these molecules using MOE software (Figure 2A) demonstrated that the 1,2-dione moiety demonstrated excellent overlap, with the corresponding pharmacophore model (Figure 2B) consisting of essentially six different elements (the electrophilic oxygen atoms, the carbonyl carbon atoms, and two hydrophobic centers derived from the adjacent domains of the compounds).
Figure 1.
QSAR parameters for CE inhibition by benzil.
Table 1.
Ki Values for Human CEs with 1,2-Diones used for the Development of the Pharmacophore Model.
| name | structure | hCE1 Ki (nM ± SE) | hiCE Ki (nM ± SE) | reference |
|---|---|---|---|---|
| Bbenzil (1,2-diphenylethane-1,2-dione) |
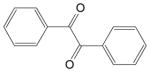
|
45 ± 3 | 15 ± 2 | 19 |
| 1,2-bis(3,5-difluorophenyl)-ethane-1,2-dione |
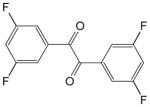
|
74 ± 8 | 23 ± 4 | 19 |
| acenapthoquinone |
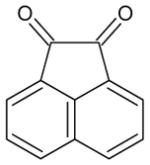
|
31 ± 4 | 170 ± 22 | 24 |
| phenanthrene-9,10-dione |
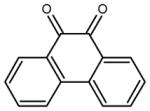
|
2.5 ± 0.2 | 52 ± 3 | 24 |
| phenyl isatin |
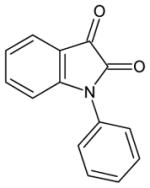
|
23 ± 2 | 950 ± 120 | 23 |
| 1-(4-chlorobenzyl)isatin) |
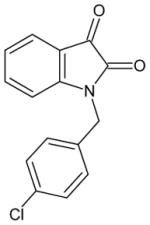
|
25 ± 3 | 32 ± 3 | 23 |
| tetradecane-7,8-dione |
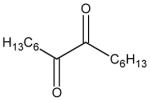
|
4.8 ± 0.3 | 77 ± 9 | 25 |
| hexadecane-8,9-dione |
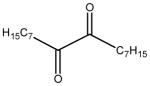
|
7.6 ± 1.6 | 26 ± 2 | 25 |
| 1-phenyl-1,2-hexanedione |
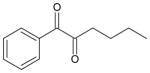
|
26 ± 2 | 73 ± 5 | 25 |
| 1-phenyl-1,2-heptanedione |
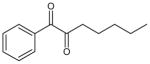
|
6.7 ± 0.2 | 46 ± 6 | 25 |
Figure 2.
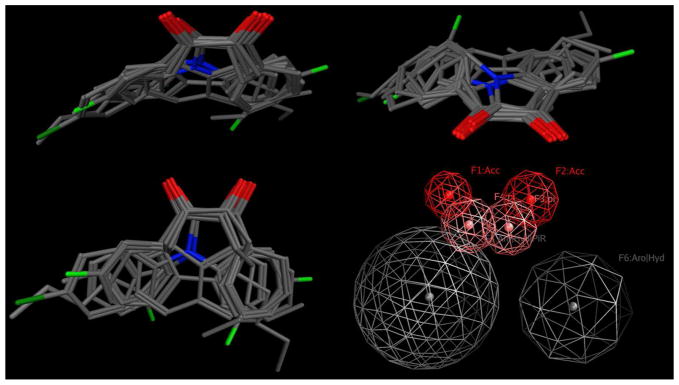
Overlays (top 2 and lower left hand panel) of the ten 1,2-diones used for the generation of the pharmacophore model. The lower right hand panel indicates the descriptors for this model that were employed for searching the MDPI natural product database.
This pharmacophore was then used to search a subset of natural products present in the ZINC database (the Molecular Diversity Preservation International (MDPI) collection), resulting in 4512 hits. To eliminate molecules that did not contain the ethane-1,2-dione chemotype, filtering was undertaken using the C(=O)C(=O) motif (the SMILES notation for the 1,2-dione). This resulted in 265 candidate compounds, and Table 2 displays the structures and root mean squared deviation (RMSD) values for the top 20 scoring compounds. Subsequent filtering was then employed to remove amides since it has been demonstrated previously that the inclusion of the nitrogen atom deactivates the ketone, making these compounds inactive towards CEs.25 This eliminated compounds 5, 6, 8–11, 13, 16, 18, and 19. It should be noted that due to the symmetry of the pharmacophore, duplicates were frequently obtained since the model could align with the target compounds in at least two different conformations. Examination of the compounds that met all of the search criteria (1–4, 7, 12, 14, 15, 17, and 20) indicated that in general, these molecules were polycyclic and contained at least one aromatic ring, with the 1,2-dione usually present within a cyclohexyl moiety. Commercial sources for these compounds were determined using the ZINC database and at the time of these searches (March 2012), compounds 1–4 were unable to be obtained. However, compound 7 (tanshinone IIA) was readily available and was used to assess inhibition of the human CE and cholinesterases (ChEs).
Table 2.
Top 20 Compounds Obtained from Database Searches Using the Pharmacophore Model Based on the 1,2-Dione-containing CE Inhibitors.a
| Code | name | structure | total number conformations (positions) | RMSD |
|---|---|---|---|---|
| 1 | 1-phenyl-1,3a,10,10a-tetrahydropyrazolo[3,4-b][1] benzazepine-4,5-dione |
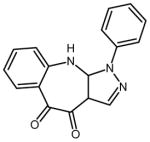
|
8 (1–13,15–18) | 0.1612 |
| 2 | 2,5,6,8-tetrahydroxy-7-(1-hydroxypropan-2-yl)-1,1,4a-trimethyl-1,2,3,4,4a,10a-hexahydrophenanthrene-9,10-dione |
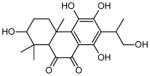
|
8 (22, 23 48, 49, 106–109) | 0.1666 0.1667 0.1668 |
| 3 | 2-(1,3,4,7-tetrahydroxy-4b,8,8-trimethyl-9,10-dioxo-4b,5,6,7,8,8a,9,10-octahydrophenanthren-2-yl) propane-1,3-diyl diacetate |
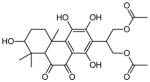
|
2 (34, 50) | 0.1666 0.1667 |
| 4 | 2-benzyl-1H-β-carboline-1,3,4(2H,9H)-trione |
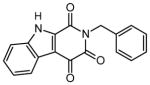
|
2 (41,45) | 0.1667 |
| 5 | (N′1E,N′2E)-N′1,N′2-bis(1-(2-oxo-2H-chromen-3-yl) ethylidene)oxalohydrazide |
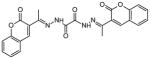
|
2 (42, 43) | 0.1667 |
| 6 | N-(1H-tetrazol-5-ylmethyl)ethanediamide |
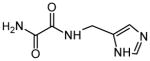
|
2 (44, 76) | 0.1667 0.1668 |
| 7 | 1,6,6-trimethyl-6,7,8,9-tetrahydrophenanthro[1,2-b] furan-10,11-dione (tanshinone IIA) |
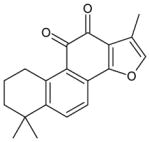
|
1 (56) | 0.1667 |
| 8 | N′1-[(1E)-1-(4-ethyl-3-hydroxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-yl) ethylidene]ethane dihydrazide |
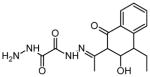
|
1 (73) | 0.1668 |
| 9 | N′1-[(1E)-1-(2-oxo-2H-chromen-3-yl) ethylidene]ethane dihydrazide |
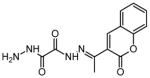
|
1 (75) | 0.1668 |
| 10 | 4-(6-chloro-2,3-dioxo-3,4-dihydroquinoxalin-1(2H)-yl) butanenitrile |
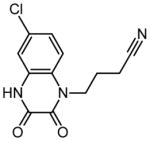
|
2 (82, 135) | 0.1668 |
| 11 | N′1-[(1E)-1-(2-hydroxy-4-oxo-1-phenyl-1,4-dihydroquinolin-3-yl)ethylidene] ethanedihydrazide |
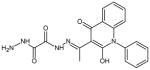
|
1 (83) | 0.1668 |
| 12 | 5,6,8-trihydroxy-7-isopropyl-1,1,4a-trimethyl-1,2,3,4,4a,10a-hexahydrophenanthrene-9,10-dione |
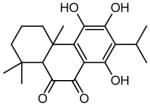
|
2 (84,85) | 0.1668 |
| 13 | 1,4-dihydroquinoxaline-2,3-dione |
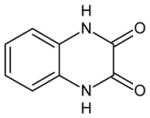
|
1 (98) | 0.1668 |
| 14 | N-(4-methylphenyl)-2-oxopropanamide |
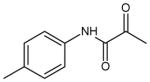
|
1 (115) | 0.1668 |
| 15 | 7-(1,3-dihydroxypropan-2-yl)-2,5,6,8-tetrahydroxy-1,1,4a-trimethyl-1,2,3,4,4a,10a-hexahydrophenanthrene-9,10-dione |
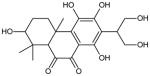
|
2 (119,120) | 0.1668 |
| 16 | N-[(6-methylpyridin-2-yl)methyl]ethanediamide |
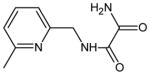
|
1 (136) | 0.1668 |
| 17 | 5,6-dihydroxycyclohex-5-ene-1,2,3,4-tetrone |
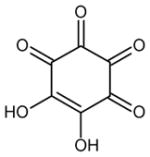
|
4 (149, 150, 159, 160) | 0.1668 |
| 18 | (N′1E,N′2E)-N′1,N′2-bis(4-butoxybenzylidene)oxalo hydrazide |
|
2 (163, 164) | 0.1668 |
| 19 | diethyl 2,2′-(2,3-dioxo-2,3-dihydroquinoxaline-1,4-diyl) diacetate |
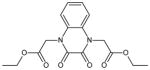
|
2 (169, 170) | 0.1668 |
| 20 | phenanthrene-9,10-dione |
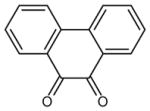
|
2 (171, 172) | 0.1668 |
Scores are ranked based upon RMSD.
Inhibition of Human Esterases by Tanshinones
Inhibition of hiCE, hCE1, hAChE, hBChE was assessed using tanshinone IIA (7) and while modest activity was seen with the CEs when using the generic esterase substrate o-nitrophenyl acetate (o-NPA; 2.4 μM and 6.9 μM, for hiCE and hCE1, respectively; Table 3), 7 was much more potent when irinotecan was used (69 nM with hiCE). Since irinotecan is a very poor substrate for hCE1, inhibition of hydrolysis of this compound was not determined. Interestingly however, 7 did not inhibit either human AChE or BChE. Since several analogues of tanshinone IIA are commercially available, these compounds were obtained and assessed for enzyme inhibition. As indicated in Table 3, different levels of activity were observed, with the most potent compounds being miltirone (23) and cryptotanshinone (24). Both of these molecules demonstrated mid-nanomolar Ki values for hiCE with both o-NPA and irinotecan, and modest activity towards hCE1. Indeed the lowest inhibition constant observed with this series of compounds (40 nM for 23 with hiCE and o-NPA) was comparable with that seen with the benzil analogues.
Table 3.
Inhibition Constants for Tanshinone IIA (7) and its Analogues with Human Carboxylesterases and Cholinesterases.
| compound | code | structure | name | Ki ± SE (nM) | ||||
|---|---|---|---|---|---|---|---|---|
| hiCE o-NPA | hiCE CPT-11 | hCE1 o-NPA | hAChE | hBChE | ||||
| tanshinone IIA | 7 |
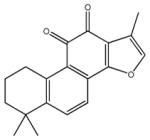
|
1,6,6-trimethyl-6,7,8,9-tetrahydrophenanthro [1,2-b] furan-10,11-dione | 2,450 ± 369 | 69.4 ± 6.1 | 6,890 ± 424 | >100,000 | >100,000 |
| tanshinone I | 21 |
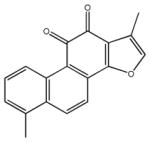
|
1,6-dimethyl-phenanthro [1,2-b] furan-10,11-dione | 14,550 ± 1,560 | 1,410 ± 420 | 26,250 ± 1,730 | >100,000 | >100,000 |
| dihydrotanshinone | 22 |
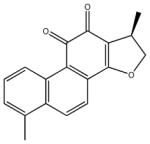
|
1,2-dihydro-1,6-dimethyl-(1R)-phenanthro [1,2-b] uran-10,11-dione | 118 ± 21 | 1,830 ± 294 | 398 ± 66 | 1,950 ± 381 | >100,000 |
| miltirone | 23 |
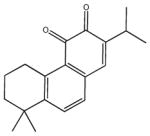
|
5,6,7,8-tetrahydro-8,8-dimethyl-2-(1-methylethyl)-3,4-phenanthrenedione | 40.0 ± 3.4 | 83.70 ± 10.0 | 2,530 ± 1,040 | >100,000 | >100,000 |
| cryptotanshinone | 24 |
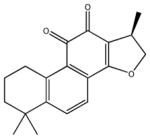
|
1,2,6,7,8,9-hexahydro-1,6,6-trimethyl-(1R)-phenanthro [1,2-b] furan-10,11-dione | 141 ± 23 | 290 ± 110 | 544 ± 64 | 6,410 ± 1,160 | >100,000 |
| tanshinone IIA sulfonate | 25 |
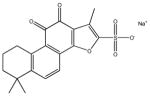
|
6,7,8,9,10,11-hexahydro-1,6,6-trimethyl-10,11-dioxo-phenanthro[ 1,2-b] furan-2-sulfonic acid, sodium salt | 3,900 ± 254 | 28,790 ± 12,790 | >100,000 | >100,000 | >100,000 |
While most compounds were more potent towards hCE1 than hiCE when using o-NPA as a substrate, the most active molecule, miltirone (23), generated Ki values similar to those observed for compounds used in the development of the pharmacophore. Furthermore, when using irinotecan as the substrate, molecules 7, 23, and 24 yielded inhibition constants that were similar to that seen for benzil.12 Since our group has demonstrated previously that benzil can reduce the toxicity of irinotecan in cell culture assays (by limiting intracellular drug hydrolysis and therefore concentrations of the active metabolite SN-38), it was considered likely that the tanshinone derivatives would exhibit similar activity.
It was also observed that dihydrotanshinone (22) and cryptotanshinone (24) demonstrated modest activity towards hAChE, but not BChE. Since both of these small molecules have increased saturation of the furan ring, and a chiral carbon in the β-position with respect to the cyclic oxygen (components not seen in the other analogues), this suggest that these domains are important for AChE inhibition. Clearly however, this only applies for hAChE since no inhibition of hBChE was observed for any of the tanshinone analogues.
Kinetic analyses for all enzyme/substrate/inhibitor combinations indicated that the mode of enzyme inhibition was partially competitive. This indicates that the inhibitor demonstrates structural similarity to an esterified substrate molecule, but is unable to completely inhibit product formation. This is consistent with all of the previous results obtained for the 1,2-dione-containing inhibitors.18, 19, 25, 26 In addition, the results presented here provide further support for the hypothesis that the catalytic serine Oγ atom within the enzyme attacks the carbonyl carbon atom of the inhibitor, but fails to cleave the carbon-carbon bond within the dione.19, 25 This is likely due to the increased strength and decreased polarity of the latter, as compared to the weaker, more polar bond present within an ester.
Isolation of Tanshinones from Salvia miltiorrhiza roots
Since tanshinones are abundant in the roots of Salvia miltiorrhiza Bunge (Lamiaceae; red sage) and this material is used in Chinese herbal medicine under the name “Danshen”, a sample of the latter was obtained and the ability of extracts of this material to inhibit CEs was assessed. Three different extracts were generated using hot water, acetone or 56% ethanol. The latter was used since in Chinese medicine, “Danshen” is frequently steeped in “Baijui”, a distilled sorghum liquor, that contains high concentrations of ethanol. As indicated in Table 4, organic solvent extracts of S. miltiorrhiza roots contained large amounts of compounds that inhibited both hiCE and hCE1. Indeed, IC50 values as low as 15 ng/mg were observed for this material. Due to the likely presence of numerous tanshinones in these extracts it is not possible to generate an accurate Ki value for enzyme inhibition. However, if we assume that the active components present within the sample have a molecular weight of ~300 Da (the molecular weights of tanshinone I and IIA are 276.2 and 294.3, respectively), for the most potent extracts, this would equate to a Ki value of ~60 nM. Since this is similar to the values seen for the benzil and benzene sulfonamide-based hiCE inhibitors,17, 19, 20 this indicates that very potent compounds are present within the S. miltiorrhiza root extracts.
Table 4.
Inhibition of hiCE by Different Salvia miltiorrhiza Root Extracts.
| extract/fraction | concentration (mg/mL) | IC50 ± SE (μg/ml) | IC50 ± SE (μg/mg) | ||
|---|---|---|---|---|---|
| o-NPA | CPT-11 | o-NPA | CPT-11 | ||
| water | 41.8 | 4,080 ± 990 | ND | 97.6 ± 23 | ND |
| acetone | 1.13 | 0.16 ± 0.05 | 0.51 ± 0.12 | 0.14 ± 0.04 | 0.45 ± 0.10 |
| ethanol (56%) | 37.1 | 179 ± 16 | 41.4 ± 9.3 | 4.82 ± 0.43 | 1.12 ± 0.25 |
| acetone fraction 16 | 1a | 0.27 ± 0.02 | 0.52 ± 0.04 | 0.27 ± 0.02 | 0.52 ± 0.04 |
The dried chromatography fraction was dissolved in DMSO at 1 mg/mL.
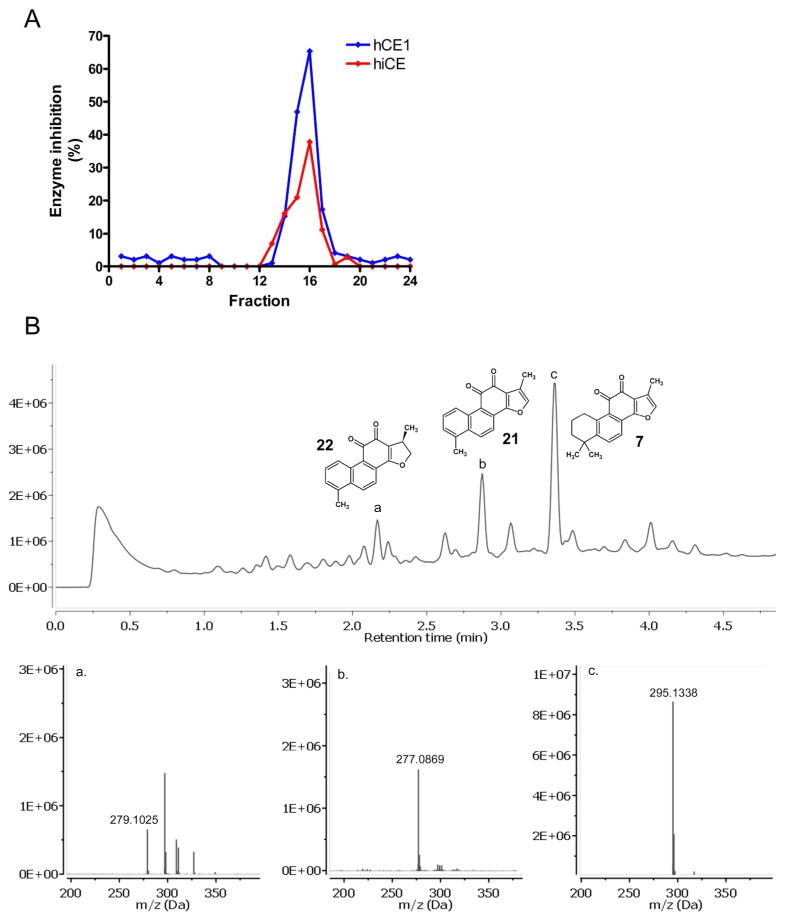
Having established that CE inhibitors were present within S. miltiorrhiza root extracts, chromatographic separation of the acetone sample was undertaken and the fractions obtained were assessed for inhibition of hiCE and hCE1 using o-NPA as substrate. As depicted in Figure 3A, significant enzyme inhibition was observed in fractions 13.–17, with peak activity occurring in fraction 16. To evaluate the molecule(s) responsible for this inhibition, this fraction was subjected to UPLC/HRMS and the masses of the major components in the sample were determined. As indicated in Figure 3B, tanshinone I (21), tanshinone IIA (7), and dihydrotanshinone (22) were detected in this material, confirming that CE inhibition is likely due to these compounds. While we do not know the relative contribution toward enzyme inhibition and the exact amounts of all of the inhibitory compounds in these samples, our preliminary semi-quantitative MS analyses indicate that up to 10 mg of tanshinone IIA is present in acetone extracts of 5g of S. miltiorrhiza root. Hence, this herbal material may represent an important source of tanshinones that might mediate modulation of esterified drug metabolism.
Figure 3.
A. CE inhibition profile of fractions obtained following chromatographic separation of an acetone extract of S. miltiorrhiza root. Fractions were assayed with either hCE1 (blue) or hiCE (red) at a final concentration of 100 ng/mL. B. UPLC/HRMS analysis of chromatographic fraction 16. High-resolution spectra are included for the major peaks identified in the sample. (a) Dihydrotanshinone (22; HREIMS m/z 279.10210, calcd for C18H14O3, 279.10250); (b) Tanshinone I (21; HREIMS m/z 277.086445, calcd for C18H12O3, 277.08690); (c) Tanshinone IIA (7; HREIMS m/z 295.13340, calcd for C19H18O3, 295.13380).
Intracellular Inhibition of hiCE by Tanshinones and S. miltiorrhiza root extracts
Having demonstrated that the tanshinones are potent CE inhibitors toward purified enzymes in vitro, their ability to effect intracellular inhibition of hiCE was evaluated using 4-methylumbelifferone acetate (4-MUA) as a substrate. In these studies, a fluorescence-based assay was used, as previously developed,18 which monitors the in situ hydrolysis of 4-MUA in U373MG cells. If reduced substrate metabolism is observed in these assays, this indicates that the inhibitor is cell permeable, and can inhibit hiCE intracellularly. As shown in Table 5, all tanshinones except tanshinone sulfonate (25) were able to inhibit hiCE, indicating that the molecules can cross cell membranes and reduce substrate hydrolysis by up to 90%, as compared to the control. These values are similar to that seen for benzil (Table 518). Furthermore, both acetone and ethanolic (not shown) extracts of S. miltiorrhiza root, as well as fraction 16 of the former, also exhibited intracellular inhibition of hiCE in U373MG cells (Table 5). It is not surprising that the sodium salt of tanshinone sulfonate (25) failed to inhibit in these assays since this molecule is charged and its cell permeability is likely to be very poor. In addition, since this was one of the least potent compounds evaluated, it is unlikely that even if the compound were able to enter the cell that it would effectively inhibit hiCE.
Table 5.
Intracellular CE inhibition and Reduction of Sensitization of Cells to CPT-11 by Tanshinones and S. miltiorrhiza root (Danshen) extracts.
| Expt numbera | Sample | Intracellular CE inhibitionb (% ± SD) | CPT-11 GI50 (μM) | Fold change in GI50 (as compared to DMSO control for respective experiment) |
|---|---|---|---|---|
| 1 | None (DMSO) | - | 1.1 | - |
| Benzil | 83.8 ± 4.7 | 6.0 | 5.4 | |
| Miltirone (23) | 83.0 ± 3.3 | 8.5 | 7.7 | |
| 2 | None (DMSO) | - | 0.9 | - |
| Tanshinone I (21) | 68.7 ± 1.1 | 3.3 | 3.9 | |
| Dihydrotanshinone (22) | 80.8 ± 2.6 | 8.9 | 9.9 | |
| 3 | None (DMSO) | - | 0.44 | - |
| Tanshinone IIA (7) | 80.8 ± 2.7 | 2.2 | 5 | |
| Cryptotanshinone (24) | 81.0 ± 7.7 | 2.0 | 4.5 | |
| 4 | None (DMSO) | - | 1.3 | - |
| Danshen acetone extract (2.5μg/ml) | 88.1 ± 0.7 | 5.1 | 3.9 | |
| 5 | None (DMSO) | - | 2.0 | - |
| Fraction 16 (2.5μg/ml) | 89.4 ± 4.7 | 7.4 | 3.7 | |
| Tanshinone sulfonate (25) | 14.9 ± 6.7 | ND | - |
Five independent experiments were conducted for growth inhibition studies due to the large number of data points generated by the cell-based assays. The differences in the GI50 values observed are dependent upon the levels of expression of hiCE. Since this varies, each independent experiment has its own DMSO control, and fold changes are calculated from this value.
Data represent percentage reduction in 4-MUA metabolism 30s after addition of substrate. Results represent the data obtained from 3 individual experiments
Based upon these data, it was assessed as to whether the pure tanshinones and S. miltiorrhiza root extracts were capable of modulating irinotecan hydrolysis in vivo. We have previously demonstrated that benzil can reduce SN-38 hydrolysis in cells expressing hiCE, and this alters the sensitivity of the cells to the drug.18 Hence, it is likely that the natural product compounds evaluated here would have similar properties. To assess whether the reduction in hiCE activity generated by these compounds would reduce the sensitivity of cells to CPT-11, growth inhibition assays were undertaken. In these studies, cells expressing hiCE were preincubated with either purified tanshinones or S. miltiorrhiza root extracts for 1 h and the GI50 values for irinotecan were determined. As indicated in Table 5, changes in cellular sensitivity to the drug were observed for all tanshinones and extracts, due to reduced production of SN-38. Indeed, with some compounds such as dihydrotanshinone (22), the GI50 value for CPT-11 increased as much as ten-fold as compared to cells treated with DMSO (Table 5). Hence, all of the tanshinones (with the exception of 25), as well as the acetone root extract of S. miltiorrhiza, were able to reduce the sensitivity of U373G cells expressing hiCE to irinotecan. These data further confirm that these molecules are cell permeable and can modulate SN-38 production in vivo.
The metabolism of esterified drugs by CEs represents a key process in the either activation or inactivation of numerous clinically used agents.1, 5, 8–10 Hence compounds that modulate the activity of these enzymes will likely affect drug efficacy. We have identified a class of agents (tanshinones), present within the Chinese herbal medicine “Danshen” (S. miltiorrhiza roots), that are polycyclic compounds and contain a 1,2-dione moiety that are potent human CEs inhibitors.
“Danshen” has been recommended for use in Chinese medicine for numerous conditions including cardiovascular and cerebrovascular disease. However, it has recently been approved by the FDA for clinical trials in the United States under the moniker “Compound Danshen Dripping Pills”. According to the clinicaltrials.gov website (http://www.clinicaltrials.gov), four clinical trials are being conducted, or have been completed, in the U.S. using this material. The specific conditions that are being targeted in these studies are hypertension, coronary heart problems and polycystic ovary disease. To date, the active components in “Compound Danshen Dripping Pills” have not been identified, although in vitro and in vivo experiments have ascribed several different properties to S. miltiorrhiza root. These include the cytotoxicity of tanshinones towards tumor cells,27–29 the modulation of levels of oxidative species in cells treated with these agents,30–32 as well as specific effects on cellular biochemistry.33–35 Clearly therefore, S. miltiorrhiza root and the compounds present within the extracts generated have considerable biological activity. Interestingly however, in our cell culture-based assays (either the 4-MUA or the growth inhibition experiments with irinotecan), no adverse effects were observed at 1 μM. However, at these concentrations, significant intracellular inhibition of hiCE and modulation of drug sensitivity to irinotecan were demonstrated (Table 5). This would suggest that the levels of tanshinones present that are required to affect cell survival are considerably greater than that necessary to inhibit CEs.
These results portend that individuals who may take extracts containing S. miltiorrhiza roots (“Danshen”) would likely demonstrate reduced hydrolysis of esterified drugs. Since numerous clinically used agents contain this chemotype, potentially, significant drug-drug interactions might be observed when these compounds are combined. Clearly, changes in drug hydrolysis would depend upon the pharmacokinetics of the tanshinones, the formulation and amounts of S. miltiorrhiza roots used (e.g., organic tinctures vs. herbal teas), the source of the material (e.g., crude roots vs. finely ground powders) etc. However, due to the potency of the inhibition that we have observed and the ubiquitous nature of the ester chemotype in many drugs, we believe that further investigation of the likelihood for drug-drug interactions are warranted. Both in vitro and in vivo studies to assess this hypothesis are currently underway.
EXPERIMENTAL SECTION
General Experimental Procedures
The glioblastoma line (U373MG), that has been engineered to overexpress hiCE, has been described previously.18 Tanshinone I, tanshinone IIA, tanshinone IIA sulfonate, miltirone, dihydrotanshinone and cryptotanshinone were obtained from LKT Labs (St. Paul, MN), ChromaDex (Irvine, CA), Carbocore (The Woodlands, TX) or Bosche Scientific (New Brunswick, NJ). Benzil, o-NPA, and 4-MUA were obtained from Sigma Chemicals (St. Louis, MO). CPT-11 was a kind gift from Dr. J.P. McGovren (Pfizer, NY).
Human CEs (hiCE and hCE1) were purified from baculovirus-infected Spodoptera frugiperda cells as previously described.12, 36 Human acetyl- and butyrylcholinesterase (hAChE; hBChE) were purchased from Sigma Chemicals.
For identification of components within extracts using HRMS, samples were separated and analyzed on an Acquity UPLC coupled to a Xevo G2 QToF mass spectrometer (Waters Technology Co., Milford, MA). Identity was validated by comparison to chromatographic retention times obtained for commercially available tanshinone standards and by HRMS.
Plant Material
Dried roots of S. miltiorrhiza Bunge (lot number 6069902) were purchased from a local Chinese grocery in Memphis, originally provided by South Project Ltd. (Hong Kong). A voucher specimen (#P0001) was deposited in the Department of Chemical Biology and Therapeutics, St. Jude Children’s Research Hospital.
Extraction and Isolation
Extracts of S. miltiorrhiza root (“Danshen”) were generated by incubating 5 g of plant material with 100 mL of solvent (water, acetone, or 56% ethanol/44% water). Different approaches (refluxing, microwave extraction), times (up to 18 h) and temperatures (ambient to solvent boiling point) were used. Solutions were clarified by filtration, solvents removed under reduced pressure, and resulting solids were then weighed and dissolved in methanol. Extracts that demonstrated the highest potency towards the inhibition of CEs were then subjected to chromatographic separation using a previously published procedure.37 Briefly, following removal of polyphenols, compounds were separated by reversed-phase semi-preparative HPLC on a C18 column (30 × 50 mm, 110 Å, 5 μM) using a water/methanol gradient. This resulted in ~25 fractions that were dried, weighed, and dissolved in DMSO.
Pharmacophore Development and Library Searches
Alignment of molecules and development of pharmacophore models was achieved using MOE 2011.10 software (Chemical Computing Group, Montreal, Canada). Briefly, candidate compounds were minimized and the Flexible Alignment module was used to overlay compounds with the default settings, except that the logP option was selected. This was chosen since an inverse correlation between clogP and inhibitor potency has been demonstrated previously.23, 25 The top scoring alignment for these studies was then used for the development of the pharmacophore.
Pharmacophore models were calculated using the Pharmacophore option in MOE with constraints developed for the C=O bonds and the carbon atoms within the 1,2-dione group. Derived models were then used to search the Molecular Diversity Preservation International database (ca. 22,000 unique molecules). The results were sorted using the mean RMSD distances, and compounds lacking the 1,2-dione moiety were discarded.
Enzyme Inhibition
Inhibition of CE was determined using a spectrophotometric assay with o-NPA as a substrate. Typically at least 8 concentrations of inhibitor were used, and data were evaluated using the following multifactorial equation:
where i is the fractional inhibition, [I] is the inhibitor concentration, [s] is the substrate concentration, α is the change in affinity of the substrate for enzyme, β is the change in the rate of enzyme substrate complex decomposition, Ks is the dissociation constant for the enzyme substrate complex, and Ki is the inhibitor constant.12, 19, 20, 38, 39 Examination of the curve fits, where α ranged from 0 to ∞ and β ranged from 0 to 1, was performed using GraphPad Prism software and Perl data language.19, 20 Curves were generated and analyzed using Akaike’s information criteria.40, 41 Finally, using the equation generated by Prism considered to be the best fit for the datasets, Ki values were then calculated.19, 20 For crude extracts, data are presented as IC50 values (amount of sample required to reduce enzymatic activity by 50%) since the amounts and molecular weights of the components present within the material are unknown.
Inhibition of hAChE and hBChE were assessed using acetylthiocholine or butyrylthiocholine as substrates, respectively.42–44
Intracellular Inhibition of hiCE
To assess intracellular inhibition of CEs, U373MG cells expressing hiCE were exposed to 1 μM inhibitor for 1 h, and, after addition of 4-MUA to a final concentration of 750 μM, the reduction of the production of 4-methylumbelliferone was determined by fluorescence spectrometry.18 Data were recorded continuously for 2 min and the percentage of enzyme inhibition 30 s following 4-MUA addition was calculated by comparison to untreated (DMSO only) cells.18
Inhibition of CPT-11 Hydrolysis
In vitro assays to assess inhibition of CPT-11 metabolism were performed as previously described.20 CPT-11 and SN-38 were detected and quantitated using HPLC with fluorescence detection.
Growth Inhibition Assays
Growth inhibition assays were undertaken using previously described methods.18 In combination studies, inhibitors were added to cells for 1 h before application of CPT-11. Two h later, the medium containing drug and inhibitor was removed and replaced, and cells were then allowed to grow for a further 72 h. Typically, data points were determined in triplicate using at least eight different drug concentrations. After cell counting, concentrations of CPT-11 that resulted in 50% growth inhibition (GI50) were calculated using Prism software (GraphPad, San Diego, CA).
Acknowledgments
This work was supported in part by NIH grants CA108775, a Cancer Center Core grant CA21765, and by the American Lebanese Syrian Associated Charities (ALSAC) and St. Jude Children’s Research Hospital (SJCRH).
References
- 1.Sun Z, Murry DJ, Sanghani SP, Davis WI, Kedishvili NY, Zou Q, Hurley TD, Bosron WF. J Pharmacol Exp Ther. 2004;310:469–76. doi: 10.1124/jpet.104.067116. [DOI] [PubMed] [Google Scholar]
- 2.Grima M, Welsch C, Michel B, Barthelmebs M, Imbs JL. Hypertension. 1991;17:492–496. doi: 10.1161/01.hyp.17.4.492. [DOI] [PubMed] [Google Scholar]
- 3.Potter PM, Wadkins RM. Curr Med Chem. 2006;13:1045–1054. doi: 10.2174/092986706776360969. [DOI] [PubMed] [Google Scholar]
- 4.Satoh T, Hosokawa M. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 5.Humerickhouse R, Lohrbach K, Li L, Bosron W, Dolan M. Cancer Res. 2000;60:1189–1192. [PubMed] [Google Scholar]
- 6.Brzezinski MR, Spink BJ, Dean RA, Berkman CE, Cashman JR, Bosron WF. Drug Metab Dispos. 1997;25:1089–96. [PubMed] [Google Scholar]
- 7.Schwer H, Langmann T, Daig R, Becker A, Aslanidis C, Schmitz G. Biochem Biophys Res Comm. 1997;233:117–120. doi: 10.1006/bbrc.1997.6413. [DOI] [PubMed] [Google Scholar]
- 8.Khanna R, Morton CL, Danks MK, Potter PM. Cancer Res. 2000;60:4725–4728. [PubMed] [Google Scholar]
- 9.Tang M, Mukundan M, Yang J, Charpentier N, LeCluyse EL, Black C, Yang D, Shi D, Yan B. J Pharmacol Exp Ther. 2006;319:1467–76. doi: 10.1124/jpet.106.110577. [DOI] [PubMed] [Google Scholar]
- 10.Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, Akhlaghi F, Yan B. J Pharmacol Exp Ther. 2006;319:1477–84. doi: 10.1124/jpet.106.111807. [DOI] [PubMed] [Google Scholar]
- 11.Hatfield MJ, Tsurkan L, Garrett M, Shaver T, Edwards CC, Hyatt JL, Hicks LD, Potter PM. Biochem Pharmacol. 2011;81:24–31. doi: 10.1016/j.bcp.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatfield MJ, Tsurkan L, Hyatt JL, Yu X, Edwards CC, Hicks LD, Wadkins RM, Potter PM. Br J Pharmacol. 2010;160:1916–28. doi: 10.1111/j.1476-5381.2010.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bencharit S, Morton CL, Howard-Williams EL, Danks MK, Potter PM, Redinbo MR. Nat Struct Biol. 2002;9:337–342. doi: 10.1038/nsb790. [DOI] [PubMed] [Google Scholar]
- 14.Bencharit S, Morton CL, Xue Y, Potter PM, Redinbo MR. Nat Struct Biol. 2003;10:349–356. doi: 10.1038/nsb919. [DOI] [PubMed] [Google Scholar]
- 15.Wierdl M, Tsurkan L, Hyatt JL, Edwards CC, Hatfield MJ, Morton CL, Houghton PJ, Danks MK, Redinbo MR, Potter PM. Cancer Gene Ther. 2008;15:183–92. doi: 10.1038/sj.cgt.7701112. [DOI] [PubMed] [Google Scholar]
- 16.Hicks LD, Hyatt JL, Moak T, Edwards CC, Tsurkan L, Wierdl M, Ferreira AM, Wadkins RM, Potter PM. Bioorg Med Chem. 2007;15:3801–3817. doi: 10.1016/j.bmc.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hicks LD, Hyatt JL, Stoddard S, Tsurkan L, Edwards CC, Wadkins RM, Potter PM. J Med Chem. 2009;52:3742–52. doi: 10.1021/jm9001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyatt JL, Tsurkan L, Wierdl M, Edwards CC, Danks MK, Potter PM. Mol Cancer Ther. 2006;5:2281–2288. doi: 10.1158/1535-7163.MCT-06-0160. [DOI] [PubMed] [Google Scholar]
- 19.Wadkins RM, Hyatt JL, Wei X, Yoon KJ, Wierdl M, Edwards CC, Morton CL, Obenauer JC, Damodaran K, Beroza P, Danks MK, Potter PM. J Med Chem. 2005;48:2905–2915. doi: 10.1021/jm049011j. [DOI] [PubMed] [Google Scholar]
- 20.Wadkins RM, Hyatt JL, Yoon KJ, Morton CL, Lee RE, Damodaran K, Beroza P, Danks MK, Potter PM. Mol Pharmacol. 2004;65:1336–1343. doi: 10.1124/mol.65.6.1336. [DOI] [PubMed] [Google Scholar]
- 21.Djeridane A, Brunel JM, Vidal N, Yousfi M, Ajandouz EH, Stocker P. Chem Biol Interact. 2008;172:22–6. doi: 10.1016/j.cbi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Djeridane A, Yousfi M, Nadjemi B, Maamri S, Djireb F, Stocker P. J Enzyme Inhib Med Chem. 2006;21:719–26. doi: 10.1080/14756360600810399. [DOI] [PubMed] [Google Scholar]
- 23.Hyatt JL, Moak T, Hatfield JM, Tsurkan L, Edwards CC, Wierdl M, Danks MK, Wadkins RM, Potter PM. J Med Chem. 2007;50:1876–1885. doi: 10.1021/jm061471k. [DOI] [PubMed] [Google Scholar]
- 24.Hyatt JL, Wadkins RM, Tsurkan L, Hicks LD, Hatfield MJ, Edwards CC, Ii CR, Cantalupo SA, Crundwell G, Danks MK, Guy RK, Potter PM. J Med Chem. 2007;50:5727–5734. doi: 10.1021/jm0706867. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson EI, Jason Hatfield M, Tsurkan L, Hyatt JL, Edwards CC, Hicks LD, Yan B, Potter PM. Bioorg Med Chem. 2011;19:4635–43. doi: 10.1016/j.bmc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyatt JL, Stacy V, Wadkins RM, Yoon KJ, Wierdl M, Edwards CC, Zeller M, Hunter AD, Danks MK, Crundwell G, Potter PM. J Med Chem. 2005;48:5543–5550. doi: 10.1021/jm0504196. [DOI] [PubMed] [Google Scholar]
- 27.Yan MY, Chien SY, Kuo SJ, Chen DR, Su CC. Int J Mol Med. 2012;29:855–63. doi: 10.3892/ijmm.2012.908. [DOI] [PubMed] [Google Scholar]
- 28.Lee WY, Chiu LC, Yeung JH. Food Chem Toxicol. 2008;46:328–38. doi: 10.1016/j.fct.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Wu WL, Chang WL, Chen CF. Am J Chin Med. 1991;19:207–16. doi: 10.1142/S0192415X91000284. [DOI] [PubMed] [Google Scholar]
- 30.Fu J, Huang H, Liu J, Pi R, Chen J, Liu P. Eur J Pharmacol. 2007;568:213–21. doi: 10.1016/j.ejphar.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Zhang HA, Gao M, Zhang L, Zhao Y, Shi LL, Chen BN, Wang YH, Wang SB, Du GH. Biochem Biophys Res Commun. 2012;421:479–83. doi: 10.1016/j.bbrc.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 32.Qiao Z, Ma J, Liu H. Molecules. 2011;16:10002–12. doi: 10.3390/molecules161210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S, Little PJ, Lan T, Huang Y, Le K, Wu X, Shen X, Huang H, Cai Y, Tang F, Wang H, Liu P. Arch Biochem Biophys. 2011;515:72–9. doi: 10.1016/j.abb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Koon CM, Woo KS, Leung PC, Fung KP. J Ethnopharmacol. 2011;138:175–83. doi: 10.1016/j.jep.2011.08.073. [DOI] [PubMed] [Google Scholar]
- 35.Steinkamp-Fenske K, Bollinger L, Voller N, Xu H, Yao Y, Bauer R, Forstermann U, Li H. Atherosclerosis. 2007;195:e104–11. doi: 10.1016/j.atherosclerosis.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 36.Morton CL, Potter PM. Mol Biotechnol. 2000;16:193–202. doi: 10.1385/MB:16:3:193. [DOI] [PubMed] [Google Scholar]
- 37.Tu Y, Jeffries C, Ruan H, Nelson C, Smithson D, Shelat AA, Brown KM, Li XC, Hester JP, Smillie T, Khan IA, Walker L, Guy K, Yan B. J Nat Prod. 2010;73:751–754. doi: 10.1021/np9007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadkins RM, Hyatt JL, Edwards CC, Tsurkan L, Redinbo MR, Wheelock CE, Jones PD, Hammock BD, Potter PM. Mol Pharmacol. 2007;71:713–723. doi: 10.1124/mol.105.021683. [DOI] [PubMed] [Google Scholar]
- 39.Webb JL. Enzyme and Metabolic Inhibitors. Volume 1 General Principles of Inhibition. Academic Press, Inc; New York: 1963. [Google Scholar]
- 40.Akaike H. In: Petrov BN, Csaki F, editors. Information theory and an extension of the maximum likelihood principle; Second International Symposium on Information Theory; Budapest. 1973; Budapest: Akademiai Kiado; 1973. pp. 267–281. [Google Scholar]
- 41.Akaike H. IEEE Trans Automatic Control. 1974;AC-19:716–723. [Google Scholar]
- 42.Doctor BP, Toker L, Roth E, Silman I. Anal Biochem. 1987;166:399–403. doi: 10.1016/0003-2697(87)90590-2. [DOI] [PubMed] [Google Scholar]
- 43.Ellman GL, Courtney KD, Anders V, Featherstone RM. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 44.Morton CL, Wadkins RM, Danks MK, Potter PM. Cancer Res. 1999;59:1458–1463. [PubMed] [Google Scholar]