Abstract
Programmed cell death (PCD) has well-established roles in the development and physiology of animals, plants, and fungi. Although aspects of PCD control appear evolutionarily conserved between these organisms, the extent of conservation remains controversial. Recently, a putative bacterial PCD protein homolog in plants was found to play a significant role in cell death control, indicating a conservation of function between these highly divergent organisms. Interestingly, these bacterial proteins are thought to be evolutionarily linked to the Bcl-2 family of proteins. In this Opinion article, we propose a new unifying model to describe the relationship between bacterial and plant PCD systems and propose that the underlying control of PCD is conserved across at least three Kingdoms of life.
An emerging hypothesis
Programmed cell death (PCD) is a genetically regulated process of cellular suicide and is well known to play a fundamental role in a wide variety of developmental and physiological functions in animals, plants, and fungi [1–3]. Over the past two decades, our knowledge regarding the role and regulation of animal PCD, in particular apoptosis, has increased at a rapid pace, providing important insight into the progression of a variety of diseases and resistance of cancers to therapeutic agents [4, 5]. By contrast, our understanding of cell death mechanisms in other organisms is less well understood. For example, studies suggest that a regulatory process similar to the intrinsic pathway of apoptosis exists in plants [6]; however, the core components of this pathway appear to be absent in plant genomes. In bacteria, controlled cell death has only been revealed relatively recently and an understanding of the regulatory components of this process has yet to be fully elucidated. In this Opinion article, we describe recent findings that suggest the presence of previously unrecognized similarities between cell death systems of animals, plants, and bacteria, and hypothesize that the regulatory control of these systems is evolutionarily related. Furthermore, we propose that these systems likely originated as a consequence of the endosymbiotic acquisition of bacteria by eukaryotic cells, which resulted in the formation of mitochondria and chloroplasts [7, 8].
The case for bacterial PCD
The existence of bacterial PCD was not considered, historically, primarily due to the characterization of these organisms as “unicellular”. After all, what benefit could there be to a single-celled organism to harbor genes that lead to its own demise? Although they obviously can exist quite well in a unicellular (or planktonic) state, it wasn’t until we appreciated the ability of bacteria to live in a multicellular (or sessile) state, within the context of a biofilm, that the concept of bacterial PCD began to make sense. Indeed, there have now been several systems proposed to control bacterial PCD [9] and even recent studies describing apoptosis-like properties in bacteria [10, 11]. Of interest here are the Staphylococcus aureus cid and lrg operons that control cell death and lysis and are important for biofilm development [12–16]. These paralogous operons encode similar proteins with opposing effects on cell death [13, 14]. Subsequent analysis has revealed that both operons are conserved in approximately 50% of bacterial species in which the genomes have been sequenced, as well as in several Archaeal species [12].
When originally identified, it was unclear what biological role controlled cell death and lysis might have in bacteria. However, studies of Pseudomonas aeruginosa biofilm provided the first clue to explain this phenomenon. In a study using purified DNase I, extracellular DNA (termed eDNA) was shown to be a major component of the matrix holding the biofilm together [15]. Although the mechanism by which this DNA was released by the bacterial cells was not described, the fact that the system under study was a mono-species biofilm strongly indicated that this was the result of an autolytic mechanism. Subsequent work in S. aureus revealed that biofilm produced by this organism also contained eDNA and that its release was modulated by the cid and lrg genes controlling death and lysis [15, 17], as well as by the LytSR two-component regulatory system that regulates lrgAB expression [18]. Although it is counterintuitive to think of a role for PCD in a “unicellular” organism, the biofilm context of this regulated cell death, in which subpopulations of cells exist within an organized multicellular cluster of bacteria, was essential for developing the concept of bacterial PCD.
Although the molecular mechanism by which the proteins encoded by the cid and lrg operons function to control death and lysis remain to be defined, the similarities of CidA and LrgA to the bacteriophage holin/antiholin family of proteins at the structural, biochemical, and functional levels [19] suggest a common strategy. Holins and antiholins are well known in the bacteriophage world as the proteins that control lysis of an infected host cell to release the newly formed viral particles into the extracellular environment [20]. They are small membrane proteins that, despite differing by as few as one amino acid at their N-termini, have opposing functions in cell lysis [21]. Like many proteins involved in bacterial cell lysis, the functional form of holins and antiholins are the oligomers they produce within the cytoplasmic membrane of the bacteria [22]. Once produced, these oligomers coalesce into channels, resulting in the depolarization of the membrane and cell death [23]. Lysis is a secondary response that is induced by these channels, which either allows for the release of autolytic enzymes that degrade the cell wall, or causes membrane depolarization that activates pre-secreted autolytic enzymes already associated with the cell wall [9]. Interestingly, the timing of the lytic event is dictated in large part by a balance between the holins and antiholins [20], and is suggested to have been fine-tuned by evolution to obtain the optimal bacteriophage burst size [24].
Unlike holins, which are commonly co-transcribed with a gene encoding a peptidoglycan hydrolase [23], the genes encoding cidA and lrgA are invariably co-transcribed with genes (designated cidB and lrgB, respectively) encoding other putative membrane proteins. The functions of these genes have not been defined but based on the observation that their expression is translationally coupled to their cidA and lrgA counterparts [13, 25], it seems likely that the functions of these proteins are linked, possibly in a stoichiometric manner. Current studies are focused on the possibility that the B components are involved in the control of cell death, while the A components are required for lysis (using a holin-/antiholin-like strategy) once the death program has been initiated.
A different twist for PCD in plants
Unlike apoptosis in animal cells, relatively little is known about PCD in plants, even though its role in plant development and physiology is well established [26–30]. Although the function of the mitochondria in plant PCD has been demonstrated [31, 32], it appears that chloroplasts also have a prominent role as illustrated by the following examples. First, reactive oxygen species (ROS) are involved in various types of PCD, and chloroplasts are the major site of ROS production in plants [33, 34]. Certain lesion mimic mutants (LMM), having defects in nucleus-encoded chloroplast proteins, exhibit necrotic or chlorotic lesions and misregulated cell death in leaves [35, 36]. Also, two Accelerated Cell Death genes, ACD1 and ACD2, encode pheophorbide a oxygenase (PAO) and red chlorophyll catabolite reductase (RCCR), respectively, for degradation of chlorophyll in chloroplasts [37–39]. Interestingly, during pathogen infection, ACD2 localizes dynamically between the chloroplast and mitochondria, apparently as a protective response against chloroplast-derived ACD2 substrate molecules that can target mitochondria and induce death [38], thus, illustrating the involvement of both organelles in PCD. The Staygreen (SGR)/Non-Yellowing/Mendel’s I Locus gene encodes a chloroplast-targeted protein, which functions in an early step of chlorophyll degradation via disruption of light-harvesting complexes [40]. Overexpression or mutation of SGR promotes or inhibits, respectively, the hypersensitive response and disease symptom development in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) [40–42]. Finally, chloroplastic FtsH proteases, involved in the catabolic processes of the photosystem II-associated light-harvesting complex (LHCII), appear to be regulators of plant PCD [43, 44].
Although plant mitochondria are important in the control of plant PCD, a prominent class of PCD proteins that has not been identified in plants is the mitochondria-associated Bcl-2 family of proteins (Box 1). Despite the absence of Bcl-2 paralogs, several studies have demonstrated that the expression of animal Bcl-2 family of proteins in plant cells can both promote and inhibit PCD. For example, overexpression of Bcl-xL and the Caenorhabditis elegans homolog Ced-9, which are well-characterized inhibitors of apoptosis in animal cells, suppresses plant cell death induced by UV-B irradiation, paraquat treatment, or virus infection [45], whereas overexpression of animal Bax promotes rapid cell death, resembling the hypersensitive response induced by virus infection [46]. In addition, plants expressing animal Bcl-2, Bcl-xL, Ced-9, or baculovirus Op-IAP transgenes conferred heritable resistance to necrotrophic fungal pathogens [47]. Finally, animal Bax-induced plant cell death can be down-regulated by overexpression of the Arabidopsis AtBI-1 gene [48]. Interestingly, when members of the mitochondria-associated, Bcl-2 family of proteins (see below), which are found in animals, were ectopically expressed in transgenic tobacco (Nicotiana tabacum), the Bcl-2 proteins were found to be localized in the chloroplasts as well as in the mitochondria, and to regulate PCD induced by chloroplast-targeted herbicides [6]. Thus, although Bcl-2-family proteins have not been identified in plants, these studies suggest that proteins with functions analogous to those of the Bcl-2 family exist in plants. Consistent with this is the identification of molecules, such as Bax Inhibitor-1 [49–53] and caspase-like proteases including the vacuolar processing enzyme (VPE) [54], plant subtilisin-like protease [55, 56], plant proteasome PBA1 subunit [57], and metacaspase [58], that are known in animals to interact with or respond to the functions of Bcl-2 proteins.
Box 1. The Bcl-2 family of proteins.
At the heart of PCD regulatory control in animals are the mitochondria-associated, Bcl-2 family of proteins, which are the primary components of what is often referred to as the control elements of the intrinsic pathway of apoptosis [4, 5, 66]. Upon stimulation, these proteins target the mitochondria and induce mitochondrial outer membrane permeabilization (MOMP), leading to the release of cytochrome c. Once in the cytoplasm, cytochrome c functions as a trigger for the caspase cascade that is important in cellular disassembly, and the phenotypic characteristics of apoptosis [5]. The caspase cascade can also be triggered by the extrinsic pathway of apoptosis, which is engaged on the plasma membrane by ligation of members of the tumor necrosis factor super-family to the death receptors, via the signaling adapter proteins [2, 5].
The Bcl-2 protein family has been classified into three functional subgroups based on their Bcl-2 homology (BH) motifs [5, 66]: (i) Bcl-2 and its closest homologs, which contain four conserved sequence motifs (BH1–BH4), all promote cell survival; (ii) the multidomain pro-apoptotic proteins, Bax and Bak; and (iii) the BH3-only proteins, which are direct or indirect activators of Bax and Bak. Despite the central role of the Bcl-2 family of proteins in apoptosis, the specific functions of these proteins are incompletely understood. It has been generally recognized that, upon apoptosis induction, the proapoptotic member Bax translocates from the cytosol to mitochondria, where it oligomerizes and permeabilizes the mitochondrial outer membrane. By contrast, the prosurvival Bcl-2 family of proteins such as Bcl-xL can restrain Bax by heterodimerization with Bax on the mitochondrial outer membrane [2, 5]. Recently, it has been suggested that Bcl-xL inhibits apoptosis by constantly retrotranslocating Bax from the mitochondria into the cytosol [76]. Bcl-xL may also regulate metabolic efficiency, for example, in neurons, through interaction with the mitochondrial F1F0 ATP synthase in the inner membrane [77]. Finally, Bcl-2 inhibits autophagy through a direct interaction with Beclin 1/ATG6 at the endoplasmic reticulum [78], whereas ATG12 promotes apoptosis through the binding and inactivation of Bcl-2 [79].
A unifying molecular design
In 2003, it was first proposed that the Bcl-2 and holin family of proteins are evolutionarily related [59], an idea that was based on the striking molecular and functional similarities shared by these proteins [12, 59]. Both families include small, membrane-associated proteins that spontaneously oligomerize to form channels, and both include structurally similar proteins with opposing functions. As with many features associated with mitochondria, it was hypothesized that the Bcl-2 family of proteins originated in bacteria [12, 59] and were transferred to eukaryotic cells as a result of an endosymbiotic relationship more than a billion years ago that resulted in the emergence of mitochondria [7, 8]. Indeed, we could even consider the steps occurring after activation of the Bcl-2 family as being analogous to bacterial lysis, with cytochrome c release and signaling simply being a hallmark of mitochondrial lysis [12]. Remarkably, evidence in support of this hypothesis was recently generated by demonstrating that the Bcl-2 system is able to functionally replace holins to promote bacterial lysis [60]. These studies not only revealed the oligomerization-dependent lytic activity of the two known direct effectors of apoptosis (Bax and Bak), but also exhibited positive and negative modulation of this activity by Bcl-xL and tBid, respectively, demonstrating that the Bcl-2 family of proteins are functional holins and that many of the molecular components of the apoptotic regulatory machinery can be functionally recapitulated in bacteria.
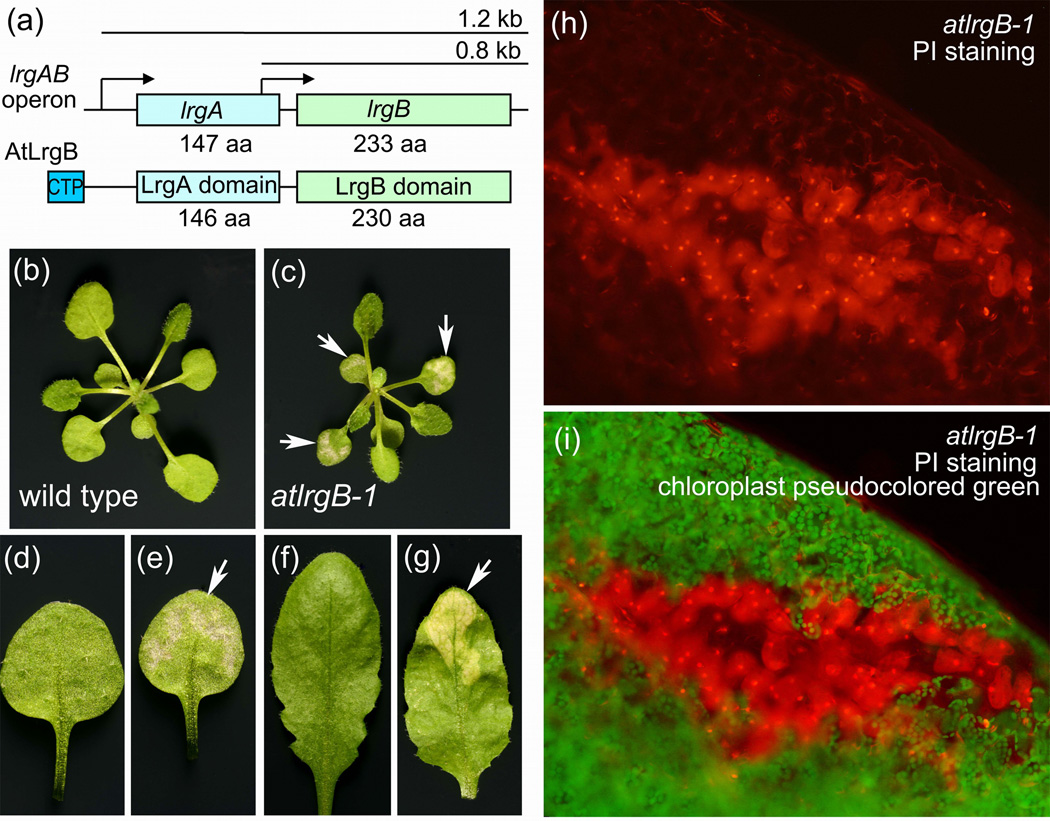
The recent demonstration that a Cid/Lrg ortholog exists in plants suggests that the holing model may apply to these organisms as well. As described above, the existence of plant proteins analogous to the Bcl-2 family have not been conclusively demonstrated. However, proteomic studies of Arabidopsis chloroplast envelope membranes [61–63], which identified the presence of putative bacterial Cid and Lrg orthologs (Figure 1a) suggested that these novel proteins may have a role in plant PCD [64]. In a recent study, the protein, designated AtLrgB, was predicted to have a chloroplast transit peptide of 13 amino-acids, and 12 transmembrane helices, five in the N-terminal region and seven in the C-terminal LrgB domain [64]. Bioinformatics analysis has suggested that the plant LrgB gene may have evolved from a gene fusion of lrgA and lrgB [64]. To determine the subcellular localization of the AtLrgB protein, a C-terminal translational fusion between the full-length AtLrgB and the EGFP protein was generated. The location of the fusion protein in the chloroplast inner envelope membrane was confirmed by confocal microscopy and protease sensitivity assays [64].
Figure 1.
Domain structures of AtLrgB protein and its role in plant cell death control. (a) Organization of the lrgAB operon in Staphylococcus aureus and the AtLrgB protein in Arabidopsis thaliana. Abbreviation: CTP, chloroplast transit peptide. (b–g) Phenotypic characterization of wild-type Arabidopsis (b,d,f) and atlrgB-1 mutants (c,e,g). Leaf sections of three-week-old seedlings (c,e) and six-week-old adult plants (g) undergoing cell death are indicated by white arrows. (h,i) Dead cells in the leaves of atlrgB-1 three-week-old seedlings are permeable to propidium iodide (PI), and fluoresce red. Chloroplasts are pseudocolored green.
To understand the function of AtLrgB, a mutant was obtained in which a T-DNA element was inserted in the first intron of the At1g32080 gene [64]. Interestingly, mutant plants produced interveinal chlorotic and premature necrotic leaves, consistent with a role for this gene in plant PCD (Figure 1b–g), as well as changes in carbon partitioning. These leaves also contained large regions of dead cells that were detectable by staining with propidium iodide (Figure 1h,i). Furthermore, overexpression of full-length AtLrgB (or its LrgA and LrgB domains, separately), under the control of CaMV 35S promoter, produced plants exhibiting veinal chlorosis and delayed greening [64]. Also, consistent with the putative membrane-damaging function of the Cid/Lrg protein family as holins/antiholins, AtLrgB could augment nystatin-induced membrane permeability in yeast cells [64]. The association of this gene with the induction of PCD was later confirmed in an independent study using transposon tagged mutants of AtLrgB [65]. At this point, however, we cannot exclude the possibility that the effect of the AtLrgB mutation on PCD is indirect, for example, as a consequence of its potential role in carbohydrate metabolism.
Although atLrgB is a single copy gene in Arabidopsis, in moss and higher plants there are typically two sets of lrgB paralogs just as there are in bacteria (Table 1). Interestingly, in most of these cases, one paralog has a predictable chloroplast transit peptide, whereas the other does not (Table 1), suggesting that only one of these proteins localizes to the chloroplast. In the Eukaryota, three kingdoms have LrgB genes: Plantae, Fungi, and Stramenopila [64]. Perhaps not coincidentally, these three kingdoms all have cell walls, and their LrgB genes are usually found as a pair of fused genes. In stramenopiles, some LrgB proteins even have predictable secretory signal peptides, suggesting that they are transmembrane proteins in the plasma membrane.
Table 1.
LrgB proteins in Plantae and Stramenopila
| Kingdom | Species | Gene namea | Amino acids |
Predictable CTPb |
GenBank ID | Refs |
|---|---|---|---|---|---|---|
| Plantae | Arabidopsis thaliana | AtLrgB | 512 | Yes | GI:18398254 | [64] |
| Oryza stativa, Japonica | OsLrgB1 | 519 | Yes | GI:222618542 | [67] | |
| OsLrgB2 | 458 | No | GI:115483664 | |||
| Glycine max | GmLrgB1 | 501 | Yes | GI:356559047 | [68] | |
| GmLrgB2 | 456 | No | GI:356495970 | |||
| Physcomitrella | PpLrgB1 | 529 | Yes | GI:168038766 | [69] | |
| Patens | PpLrgB2 | 447 | No | GI:168022538 | ||
| Zea mays | ZmLrgB1 | 544 | Yes | GI:226501414 | [70] | |
| ZmLrgB2 | 542 | Yes | GI:293334459 | |||
| Vitis vinifera | VvLrgB1 | 450 | No | GI:296085918 | [71] | |
| VvLrgB2 | 434 | No | GI:225439243 | |||
| Chlamydomons reinhardtii | CrLrgB | 455 | No | GI:159472707 | [72] | |
| Stramenopila | Phaeodactylum | PtLrgB1 | 524 | Yes | GI:219118995 | [73] |
| tricorntum | PtLrgB2 | 408 | No | GI:219118464 | ||
| Thalassiosira | TpLrgB1 | 525 | Yes | GI:224008725 | [74] | |
| pseudonana | TpLrgB2 | 413 | No | GI:224005114 |
LrgB homologues in Plantae and Stramenopila were collected from the Genbank resource of NCBI (http://www.ncbi.nlm.gov/). Amino acid sequence alignment of LrgB domains were conducted as described by Yang et al. [64].
CTP (chloroplast transit peptides) were predicted using the program ChloroP [75].
Conclusion
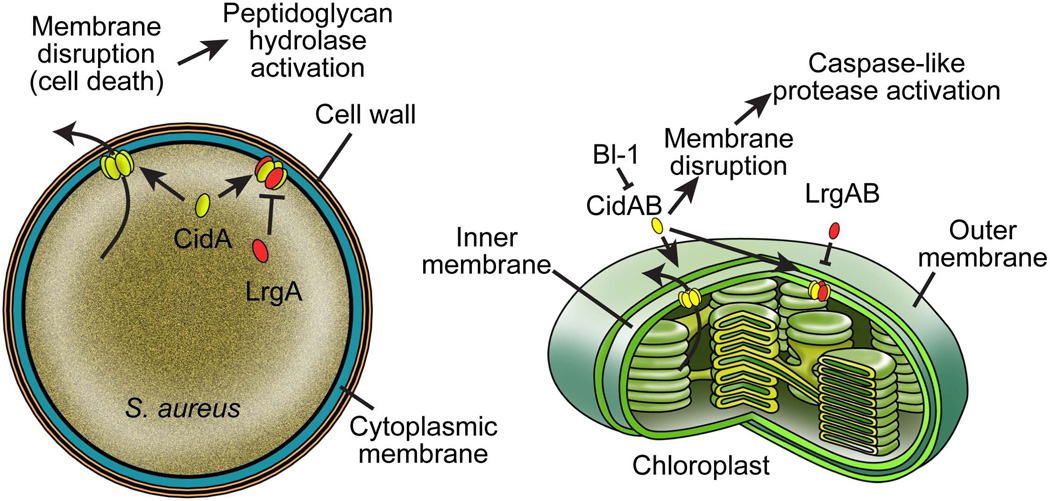
The identification of Cid/Lrg orthologs in plants and the demonstration that the Arabidopsis AtLrgB gene affects cell death in plants provides important insight, not only regarding the control of this cell death process, but also about the potential conservation of this mechanism in animals, plants, and bacteria. As described here, recent work has provided support for the model that the Cid/Lrg and Bcl-2 protein families are functional holins [19, 60]. As new members of this group, we propose that the LrgB orthologs in plants also function as holins. As an important component of plant PCD control, the chloroplast seems a logical target for these proteins. Thus, similar to the model describing the evolutionary relationship between bacteria and mitochondria and the control of cell death, we hypothesize that a comparable relationship exists between bacteria and chloroplasts (Figure 2). Indeed, this relationship between bacteria and plants is, in several ways, more conspicuous than the relationship between animals and bacteria, given that amino acid sequence similarity exists between these proteins and that the LrgB homolog localizes to the inner membrane of the chloroplasts, unlike the Bcl-2 proteins of animal cells. As depicted in Figure 2, we speculate that the plant LrgB orthologs oligomerize in the inner chloroplast membrane in a way similar to S. aureus CidA/LrgA holin-like proteins in the cytoplasmic membrane (and presumably the cytoplasmic membranes of other bacteria), as well as the mammalian Bcl-2 proteins in the mitochondrial outer membrane. As a consequence of this oligomerization, we anticipate that the energized state of the chloroplast inner membrane will be dissipated and that this represents an early event in the induction of plant cell death, much like the dissipation of the mitochondrial inner membrane is an early event in the induction of apoptosis.
Figure 2.
The molecular conservation of cell death mechanisms between chloroplasts and bacteria. Similar to previous hypotheses suggesting the relationship between bacterial and mitochondrial cell death pathways [9, 12, 14], it is envisioned that an important figure in plant cell death, the chloroplast, shares common cell death regulatory mechanisms with bacteria. Plant CidAB proteins are predicted to function as effectors of cell death and are counteracted by interactions with paralogous proteins similar to AtLrgB. As with the Bcl-2 family [60], plant CidAB and LrgAB proteins are hypothesized to function as holins and antiholins, respectively. Also, shown are the hypothetical interactions and inhibitory effects of plant BI-1-like proteins with CidAB. Finally, the ultimate consequences of Cid and Lrg function in both plants and bacteria are to activate cellular destruction, in bacteria by the stimulation of peptidoglycan hydrolases that cause lysis, and in plants by inducing protease activity analogous to the caspase cascade.
Arabidopsis may be unusual among plants in that it only has one orthologous LrgAB-encoding gene (Table 1). Most plants have two LrgAB orthologs, one of which is likely to be an LrgAB-like cell death inhibitor, whereas the other is likely to be an effector of cell death analogous to the CidAB proteins of S. aureus. We speculate that the N-terminal chloroplast-targeting domains found in one of the paralogs of each plant species may indicate an inhibitory (antiholin) function of these proteins. This is consistent with the observation that disruption of the AtLrgB gene results in plants exhibiting premature cell death. By contrast, those paralogs that do not contain this domain may indicate an effector (holin) function. In some respects, this would parallel the model for apoptosis control in that Bax, an effector of cell death, resides in the cytoplasm until a death-inducing signal is elicited, resulting in the localization of Bax to the mitochondria, where it oligomerizes and causes apoptosis [2, 5, 66]. Furthermore, evidence suggests that Bcl-2, the inhibitor of apoptosis, is more permanently localized to the mitochondrial membrane, functioning to suppress apoptosis until Bax activity is stimulated. However, many questions about the function of CidAB and LrgAB proteins remain (see Box 2).
Box 2. Outstanding questions.
What are the functions of the CidAB and LrgAB proteins in other plant species? Is CidAB an effector of cell death?
Do plant CidAB and LrgAB proteins result in chloroplast depolarization?
Are plant CidAB and LrgAB targets of ectopically expressed animal Bcl-2 family proteins?
Do CidAB and/or LrgAB crosstalk with other plant PCD regulators, such as BI-1 and ATG6 in the ER? If so, how?
Finally, the results of the studies described suggest that the control of cell death is a much more broadly conserved process than was previously appreciated. Indeed the existence of PCD in bacteria has only recently been seriously considered and the discovery of a protein family with conserved cell death functions in plants and bacteria provides strong support for the functions of these proteins in bacterial PCD. Similar to the model described previously suggesting a bacterial origin for the Bcl-2 family [12], we build on the widely held view of a bacterial origin of chloroplasts and propose that the mechanisms controlling cell death in plants, likewise, have a bacterial origin. Importantly, as with the control of apoptosis in animal cells, valuable lessons are likely to be learned about the control of plant cell death, by achieving a thorough understanding of the biochemical and physiological processes of cell death control in bacteria.
Acknowledgements
We thank Dr Muyuan Zhu for insightful discussions regarding the role of AtLrgB in plant cell death, as well as Xu Luo and Kari Nelson for the critical evaluation of this manuscript. This work was supported by the National Natural Science Foundation of China grant 31170211 (J.W.) and the National Institutes of Health grants P01-AI83211 (K.W.B.) and R01-AI038901 (K.W.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
The authors declare no conflict of interest.
References
- 1.Bozhkov PV, Lam E. Green death: revealing programmed cell death in plants. Cell Death Differ. 2011;18:1239–1240. doi: 10.1038/cdd.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teng X, et al. Gene-dependent cell death in yeast. Cell Death Dis. 2011;2:e188. doi: 10.1038/cddis.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- 5.Strasser A, et al. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Dickman MB. Bcl-2 family members localize to tobacco chloroplasts and inhibit programmed cell death induced by chloroplast-targeted herbicides. J Exp Bot. 2004;55:2617–2623. doi: 10.1093/jxb/erh275. [DOI] [PubMed] [Google Scholar]
- 7.Dyall SD, et al. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 8.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 9.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwyer DJ, et al. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012;46:561–572. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakansson AP, et al. Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLoS One. 2011;6:e17717. doi: 10.1371/journal.pone.0017717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 13.Groicher KH, et al. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice KC, et al. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol. 2003;185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice KC, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadykov MR, Bayles KW. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol. 2012 doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann EE, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma-Kuinkel BK, et al. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol. 2009;191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjit DK, et al. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol. 2011;193:2468–2476. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang IN, et al. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 21.Blasi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 22.Zagotta MT, Wilson DB. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J Bacteriol. 1990;172:912–921. doi: 10.1128/jb.172.2.912-921.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang IN. Lysis timing and bacteriophage fitness. Genetics. 2006;172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice KC, Bayles KW. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol. 2003;50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- 26.Beers EP, McDowell JM. Regulation and execution of programmed cell death in response to pathogens, stress and developmental cues. Curr Opin Plant Biol. 2001;4:561–567. doi: 10.1016/s1369-5266(00)00216-8. [DOI] [PubMed] [Google Scholar]
- 27.Jones AM. Programmed cell death in development and defense. Plant Physiol. 2001;125:94–97. doi: 10.1104/pp.125.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam E. Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol. 2004;5:305–315. doi: 10.1038/nrm1358. [DOI] [PubMed] [Google Scholar]
- 29.Love AJ, et al. Timing is everything: regulatory overlap in plant cell death. Trends Plant Sci. 2008;13:589–595. doi: 10.1016/j.tplants.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Williams B, Dickman M. Plant programmed cell death: can't live with it; can't live without it. Mol Plant Pathol. 2008;9:531–544. doi: 10.1111/j.1364-3703.2008.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond MaM, P F. Mitochondrial regulation of plant programmed cell death. In: Kempken F, editor. Plant Mitochondria, Advances in Plant Biology 1. Springer; 2011. pp. 439–465. [Google Scholar]
- 32.Lam E, et al. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- 33.De Pinto MC, et al. Redox regulation in plant programmed cell death. Plant Cell Environ. 2012;35:234–244. doi: 10.1111/j.1365-3040.2011.02387.x. [DOI] [PubMed] [Google Scholar]
- 34.Doyle SM, et al. Chloroplast and reactive oxygen species involvement in apoptotic-like programmed cell death in Arabidopsis suspension cultures. J Exp Bot. 2010;61:473–482. doi: 10.1093/jxb/erp320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorrain S, et al. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 36.Sun C, et al. RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics. 2011;38:29–37. doi: 10.1016/j.jcg.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Mach JM, et al. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci U S A. 2001;98:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattanayak GK, et al. Accelerated cell death 2 suppresses mitochondrial oxidative bursts and modulates cell death in Arabidopsis. Plant J. 2012;69:589–600. doi: 10.1111/j.1365-313X.2011.04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y, et al. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J Plant Physiol. 2011;168:1952–1959. doi: 10.1016/j.jplph.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 40.Mur LA, et al. Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 2010;188:161–174. doi: 10.1111/j.1469-8137.2010.03377.x. [DOI] [PubMed] [Google Scholar]
- 41.Jiang H, et al. Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J Integr Plant Biol. 2011;53:375–387. doi: 10.1111/j.1744-7909.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- 42.Mecey C, et al. A critical role of STAYGREEN/Mendel's I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol. 2011;157:1965–1974. doi: 10.1104/pp.111.181826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coll NS, et al. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo S, et al. Reduced levels of chloroplast FtsH protein in tobacco mosaic virus-infected tobacco leaves accelerate the hypersensitive reaction. Plant Cell. 2000;12:917–932. doi: 10.1105/tpc.12.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsuhara I, et al. Animal cell-death suppressors Bcl-x(L) and Ced-9 inhibit cell death in tobacco plants. Curr Biol. 1999;9:775–778. doi: 10.1016/s0960-9822(99)80341-8. [DOI] [PubMed] [Google Scholar]
- 46.Lacomme C, Santa Cruz S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc Natl Acad Sci U S A. 1999;96:7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dickman MB, et al. Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc Natl Acad Sci U S A. 2001;98:6957–6962. doi: 10.1073/pnas.091108998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawai-Yamada M, et al. Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1) Proc Natl Acad Sci U S A. 2001;98:12295–12300. doi: 10.1073/pnas.211423998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huckelhoven R, et al. Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis. Proc Natl Acad Sci U S A. 2003;100:5555–5560. doi: 10.1073/pnas.0931464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishikawa T, et al. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 2011;18:1271–1278. doi: 10.1038/cdd.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawai-Yamada M, et al. Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell. 2004;16:21–32. doi: 10.1105/tpc.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumura H, et al. Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L) cells. Plant J. 2003;33:425–434. doi: 10.1046/j.1365-313x.2003.01639.x. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe N, Lam E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006;45:884–894. doi: 10.1111/j.1365-313X.2006.02654.x. [DOI] [PubMed] [Google Scholar]
- 54.Hara-Nishimura I, et al. Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol. 2005;8:404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Chichkova NV, et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 2010;29:1149–1161. doi: 10.1038/emboj.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vartapetian AB, et al. A plant alternative to animal caspases: subtilisin-like proteases. Cell Death Differ. 2011;18:1289–1297. doi: 10.1038/cdd.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hatsugai N, et al. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009;23:2496–2506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsiatsiani L, et al. Metacaspases. Cell Death Differ. 2011;18:1279–1288. doi: 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayles KW. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 2003;11:306–311. doi: 10.1016/s0966-842x(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 60.Pang X, et al. Active Bax and Bak are functional holins. Genes Dev. 2011;25:2278–2290. doi: 10.1101/gad.171645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferro M, et al. Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci U S A. 2002;99:11487–11492. doi: 10.1073/pnas.172390399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Froehlich JE, et al. Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res. 2003;2:413–425. doi: 10.1021/pr034025j. [DOI] [PubMed] [Google Scholar]
- 63.Kleffmann T, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, et al. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 2012;193:81–95. doi: 10.1111/j.1469-8137.2011.03867.x. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi M, et al. Loss of the plastid envelope protein AtLrgB causes spontaneous chlorotic cell death in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:125–134. doi: 10.1093/pcp/pcr180. [DOI] [PubMed] [Google Scholar]
- 66.Reed JC. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 67.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 68.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 69.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 70.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 71.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 72.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 74.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 75.Emanuelsson O, et al. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein science : a publication of the Protein Society. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edlich F, et al. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alavian KN, et al. Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase. Nat Cell Biol. 2011;13:1224–1233. doi: 10.1038/ncb2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubinstein AD, et al. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]