Summary
Ferric enterobactin (FeEnt) acquisition is a highly efficient and conserved iron scavenging system in Gram-negative bacteria. Recently, we have characterized two FeEnt receptors (CfrA and CfrB) in Campylobacter jejuni and C. coli, the enteric human pathogens that do not produce any siderophores. In this study, whole genome sequencing and comparative genomic analysis identified a unique Ent trilactone esterase Cee (Cj1376) in C. jejuni. Genomic analysis and biochemical assay strongly suggested that Cee is the sole trilactone esterase in C. jejuni. Thin layer chromatography and HPLC analyses showed high efficiency of the purified Cee to hydrolyze Ent. Three Cee homologs previously characterized from other bacteria (IroE, IroD, and Fes) were also purified and analyzed together with Cee, indicating that Cee, Fes, and IroD displayed similar hydrolysis dynamics for both apo and ferric forms of Ent while IroE catalyzed Ent inefficiently. Unlike cytoplasmic Fes and IroD, Cee is localized in the periplasm as demonstrated by immunoblotting using Cee-specific antibodies. Genetic manipulation of diverse Campylobacter strains demonstrated that Cee is not only essential for CfrB-dependent FeEnt acquisition but also involved in CfrA-dependent pathway. Together, this study identified and characterized a novel periplasmic trilactone esterase and suggested a new model of FeEnt acquisition in Campylobacter.
Introduction
Iron is an essential nutrient required for all Gram-negative bacteria to survive. However, the levels of free iron in vivo are well below the levels required for the growth of Gram-negative pathogens. Thus, to obtain sufficient iron for survival and multiplication in the host, Gram-negative bacteria have evolved aggressive systems for iron uptake. Of these systems, high-affinity iron acquisition mediated by siderophore, a small iron chelator, is the most efficient iron scavenging mechanism in Gram-negative bacteria (Miethke & Marahiel, 2007). Enterobactin (Ent), a triscatecholate siderophore that has exceptionally high affinity for ferric iron, is an archetype for iron acquisition (Raymond et al., 2003). The active uptake of ferric Ent (FeEnt) is likely the best studied siderophore-mediated iron acquisition in bacteria.
As a highly conserved iron scavenging in Gram-negative bacteria, FeEnt acquisition requires coordination of multiple components from outer membrane to cytoplasm, such as specific receptor, TonB-ExbB-ExbD energy transduction system, ABC transporter system, and cytoplasmic trilactone esterase (Raymond et al., 2003, Miethke & Marahiel, 2007). It has been widely accepted that an efficient cytoplasmic trilactone esterase is required for intracellular hydrolytic iron release, the last and essential step in Ent-mediated iron acquisition. However, for most siderophores that have higher redox potential than Ent, direct reduction of ferric-siderophore complex by reductase is a common mechanism for iron release and utilization. Intracellular reduction of the FeEnt with extremely low redox potential has not been clearly demonstrated until recent characterization of the siderophore-interacting protein YqjH in Escherichia coli by Miethke et al. (2011).
Campylobacter species, including C. jejuni and C. coli, are the most common bacterial causes of human gastroenteritis in the United States and industrialized countries (Olson, 2008). Campylobacter is also associated with Guillain-Barré syndrome, an acute flaccid paralysis that may lead to respiratory muscle compromise and death (Rees et al., 1995). Several iron-uptake systems have been identified in C. jejuni and the Ent-mediated iron scavenging is tightly linked to the ability of in vivo survival and colonization of this organism (Palyada et al., 2004, Zeng et al., 2009, Xu et al., 2010). Notably, although Campylobacter does not produce any siderophores, Campylobacter could utilize catechol siderophore Ent and hydroxamate siderophore ferrichrome produced by other microorganisms (Field et al., 1986). Recently, rhodotorulic acid, another hydroxamate siderophore, was observed to be utilized by C. jejuni (Stintzi et al., 2008). Since both ferrichrome and rhodotorulic acid are primarily produced by certain soil fungi, Ent is the only known siderophore produced by enteric microorganisms for Campylobacter iron acquisition during colonization.
Recently, we characterized two FeEnt receptors, CfrA and CfrB, in Campylobacter and firmly established that Campylobacter utilize CfrA- and CfrB-dependent systems for efficient utilization of FeEnt during in vivo colonization (Zeng et al., 2009, Xu et al., 2010). Both CfrA and CfrB, which share approximately 34% amino acid identity, are dramatically induced under iron-restricted conditions. The CfrA and CfrB differ significantly in their prevalence and relative contribution to FeEnt acquisition in C. jejuni and C. coli, the two closely related Campylobacter species. Specifically, CfrA is widespread and produced in C. jejuni strains from various sources and plays a critical role in FeEnt acquisition in C. jejuni (Zeng et al., 2009). CfrB was produced in the majority of C. coli (41 out of 45) and in some C. jejuni (8 out of 32) primary strains (Xu et al., 2010). All of the CfrB-producing C. coli strains also produced CfrA, which was rarely observed in the tested C. jejuni strains. Genetic work further demonstrated that CfrB has a major role in FeEnt acquisition in C. coli (Xu et al., 2010). Interestingly, our work (Xu et al., 2010) strongly suggested that C. jejuni NCTC 11168 and JL 11 (ATCC 33560) contain an unidentified component critical for FeEnt acquisition with this component missing in C. jejuni 81–176, which prompted us to hunt for this new component by taking advantage of the finished genomes of NCTC 11168 and 81–176 (Parkhill et al., 2000, Hofreuter et al., 2006)
In this study, whole genome sequencing of ATCC 33560 and subsequent comparative genomic analysis identified a periplasmic Ent trilactone esterase, designated Cee (for Campylobacter enterobactin esterase). Biochemical and genetic characterization of Cee suggested a new pathway for CfrB-dependent FeEnt acquisition in which hydrolysis of FeEnt occurs in periplasm. In typical CfrA-dependent pathway, FeEnt is likely subjected to intracellular reduction for iron release and utilization. Genetic study further indicated that Cee plays a unique role in the two intersecting Ent utilization systems. These findings lead to a new model of FeEnt acquisition in Campylobacter.
Results
Identification of Cj1376 as a new component involved in FeEnt acquisition in Campylobacter
The whole genome of C. jejuni ATCC 33560 was sequenced using a 454 GS FLX sequencer for comparative genomic analysis (detailed in SI Results; Table S1, Fig. S1). Comparison of annotated genome of ATCC 33560 to 81–176 genome confirmed our previous finding that 81–176 and ATCC 33560 share identical major known genes that are involved in FeEnt utilization (Table S2). Comparative genomic analysis identified 10 genes that are absent in 81–176 but present in ATCC 33560 and 11168 (Table S3). Because the genes involved in iron metabolism are usually differentially expressed in response to iron, we then examined if any of these 10 genes are subjected to iron regulation by taking advantage of the published transcriptome profiling of C. jejuni (Palyada et al., 2004, Holmes et al., 2005). The Cj1376 is the only gene potentially induced under iron-limited condition because its upstream gene, Cj1375 (cmeG, encoding a multidrug efflux transporter) (Jeon et al., 2011), was induced under iron-limited conditions. In particular, the Cj1375 and Cj1376 appear to form an operon because stop codon (TGA) of Cj1375 overlaps the start codon (ATG) of Cj1376 by 4-bp nucleotides; co-transcription of Cj1375 and Cj1376 has been confirmed by recent characterization of Cj1375 (Jeon et al., 2011). In 81–176, the Cj1376 homolog is completely deleted (Fig. S2A) while the upstream CJJ81176_1378 shares almost identical sequence to its corresponding homolog (Cj1375) in NCTC 11168 except the last 12 nucleotides (data not shown). Interestingly, in C. coli RM2228 (Fouts et al., 2005), the Cj1376 homolog (CCO1491) together with its downstream 5 genes were in an opposite orientation of the corresponding genetic loci in C. jejuni (Fig. S2A).
Complementation of 81–176 with the Cj1375/1376 operon from NCTC 11168 successfully restored its ability to utilize FeEnt as a sole iron source for growth (Fig. 1A). The plasmid containing Cj1376 only, whose expression is driven by the promoter of Cj1375, still efficiently rescued 81–176 (Fig. 1A), indicating Cj1376 is the missing component essential for 81–176 to utilize FeEnt. The Cj1376 homolog from C. coli also fully rescued 81–176 for FeEnt utilization despite its low homology to the Cj1376 from NCTC 11168 (54% aa identity) (81–176/pCc1376 in Fig. 1A).
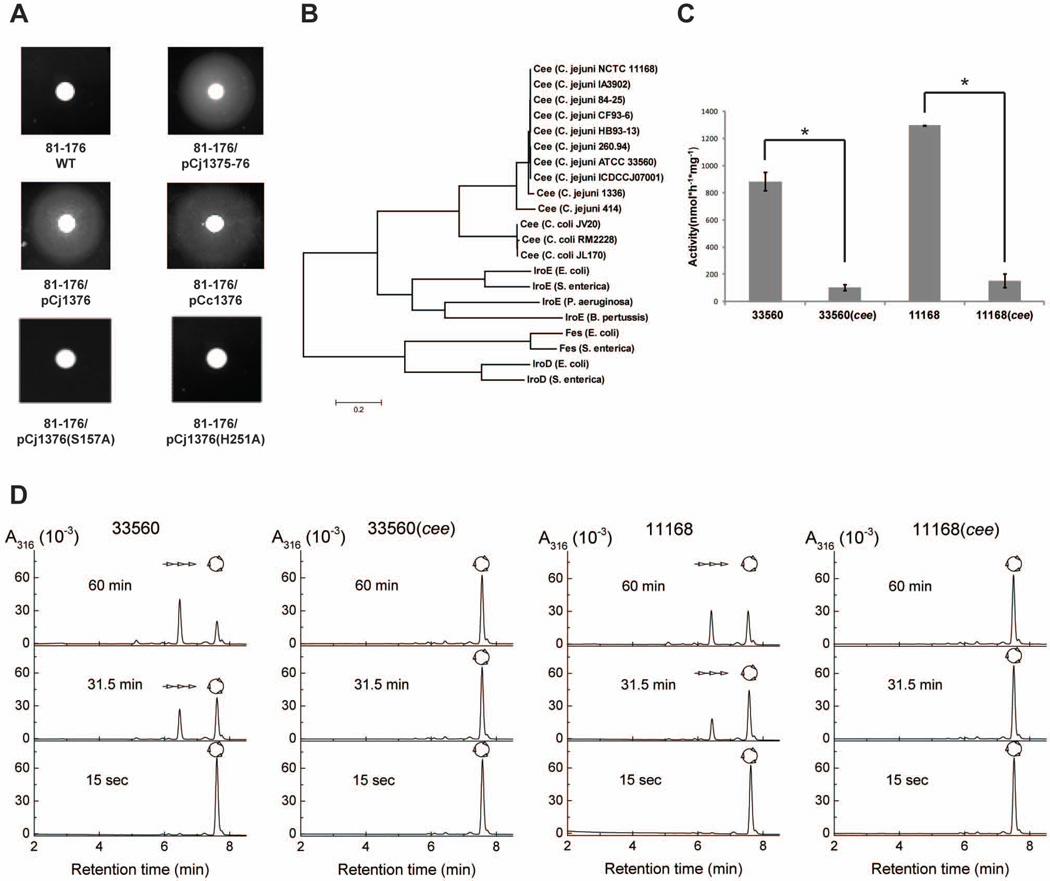
Figure 1.
Cee (Cj1376) is a putative Ent esterase required for FeEnt acquisition in Campylobacter. (A) Complementation of C. jejuni 81–176 with Cj1376 rescued its ability to utilize FeEnt as a sole iron source for growth. The standard Ent growth promotion assay was performed for wild-type (WT) 81–176 and its derivatives containing specific plasmids. (B) Phylogenetic relationship of Cee and the Ent esterases from different bacterial species. Both traditional PBS counter-extraction method (C) and HPLC (D) were performed to analyze hydrolysis of Ent by whole cell extracts from wild-type C. jejuni NCTC 11168, ATCC 33560, and their isogenic cee mutants. In panel C, each bar represents the mean ± SD from three independent measurements. The asterisk denotes statistically significant difference (P < 0.05). In panel D, reaction aliquots were quenched at different time points and analyzed by HPLC. The schematic representatives of cyclic Ent ( ) and the hydrolysis product (
) and the hydrolysis product ( linear trimer of DHBS) are shown.
linear trimer of DHBS) are shown.
Cj1376 is an enterobactin esterase
Sequence analysis indicated that Cj1376 (269 aa) is a putative periplasmic protein that displays low but significant homology to IroE of E. coli CFT073 (30% identity and 45% similarity in 164 aa overlap). IroE is a trilactone esterase with low efficiency for hydrolysis of Ent as well as its analog salmochelin in some E. coli and S. enterica strains (Lin et al., 2005a, Zhu et al., 2005, Baumler et al., 1996). Thus, we designated Cj1376 as Cee (Campylobacter Enterobactin Esterase). The structural modeling of Cee by using the crystal structure of IroE as the template revealed structural similarity between Cee and IroE (Fig. S2B), particularly with respect to the atypical dyad composed of two highly conserved and essential residues (Ser 189 and His 287 in IroE versus Ser 157 and His 251 in Cee) (Lin et al., 2005a). The Ser 157 and His 251 residues are highly conserved in the Cee of different Campylobacter strains (Fig. S2C). Alanine substitution of Ser 157 or His 251 in Cee abolished its ability to rescue 81–176 for utilization of FeEnt (Fig. 1A), further suggesting that Cee is an esterase for hydrolyzing trilactone backbone of Ent. Comparison of Cee to the trilactone esterase identified in other bacteria showed that Cee was phylogenetically closer to IroE than Fes and IroD, the two efficient Ent esterases localized in cytoplasm (Fig. 1B).
The extracts from ATCC 33560 and NCTC 11168 effectively catalyzed the hydrolysis of Ent with activity of 883 ± 66 and 1296 ± 2 nmol of Ent·h−1·mg−1, respectively (Fig. 1C). However, the extracts from corresponding isogenic cee mutants only displayed marginal hydrolysis activity. A high resolution HPLC further demonstrated that the stracts from both ATCC 33560 and NCTC 11168 hydrolyzed Ent, generating Ent breakdown products of 2,3-dihydroxybenzoylserine (DHBS) trimers; however, the cell extracts from isogenic cee mutants failed to hydrolyze Ent at any time points (Fig 1D). These findings strongly suggest that Cee is the sole Ent esterase in C. jejuni. Extensive genome analysis also indicated that no similar esterase homolog other than Cee was observed in C. jejuni (SI Results; Figure S3).
Cee is localized in the periplasm of C. jejuni
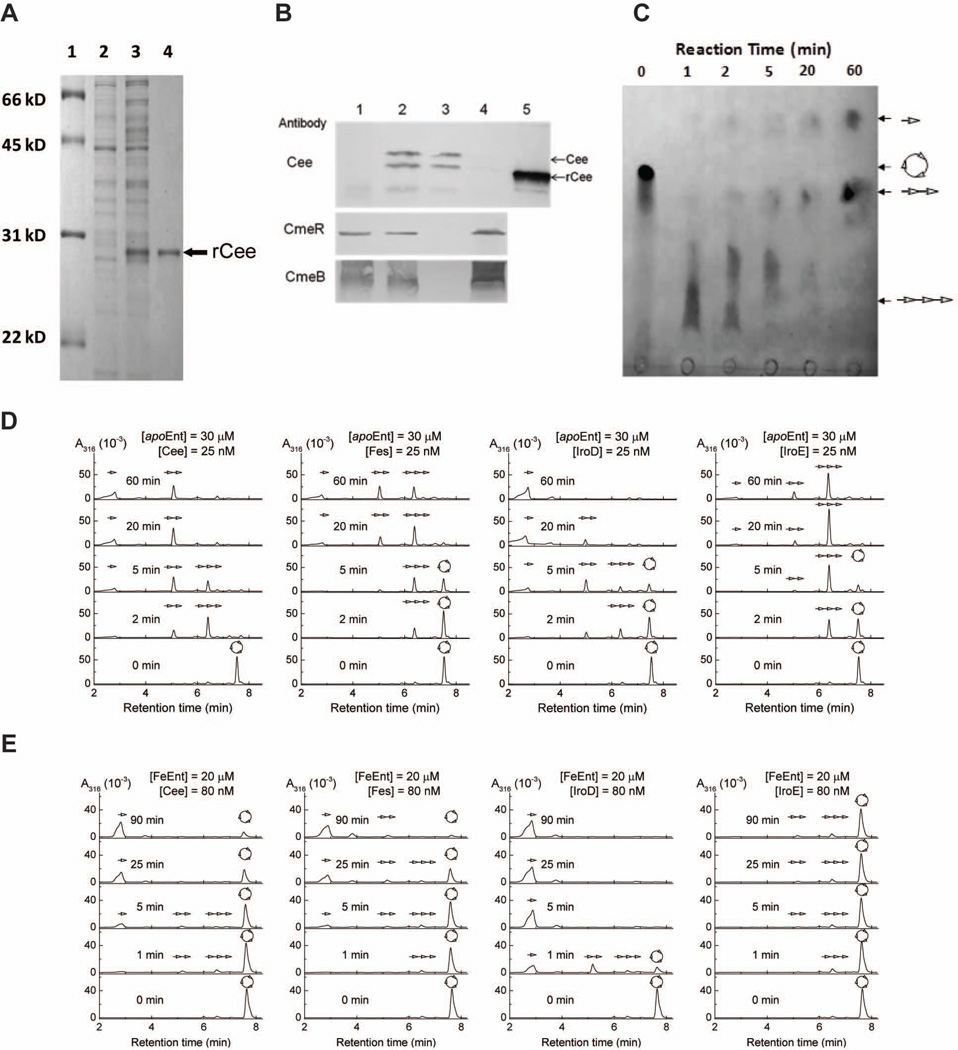
A His-tagged recombinant Cee (rCee) was produced with high purity (Fig. 2A) for raising specific antiserum and for in vitro enzyme activity assay in this study. Using Cee-specific antibody, immunoblotting was performed to determine the presence of Cee in different cell fractions. Because the Cee-specific antibody failed to detect the production of Cee in wild-type C. jejuni, likely due to the low titer of Cee antibody and/or low production level of Cee, we created a strain JL870 (Table S4) by complementing 81–176 with a vector in which production of Cee was driven by a strong promoter. As shown in Fig. 2B, Cee specific antibody did not react with any proteins in the whole cell lysate of 81–176 containing the control vector (lane 1). However, Cee antibody detected two vivid bands in the whole cell lysate of JL870 with approximate molecular weight of 29 kDa and 31 kDa, respectively (lane 2). The 31-kDa band may represent original precursor without removal of signal peptide or posttranslationally modified Cee. When using fractionated samples from JL870, Cee antibody only detected the two bands in periplasmic fraction (lane 3) but failed to react with any proteins in spheroplastic fraction (lane 4). To rule out the possibility of contamination of periplasmic fraction by the proteins from cytoplasm or inner membrane, the antibodies directed against CmeR (a 23-kDa regulator in cytoplasm) (Lin et al., 2005b) and CmeB (a 120-kDa transporter in inner membrane) (Lin et al., 2002) were also used for immunoblotting. Both CmeR and CmeB antibodies detected the corresponding proteins only in the spheroplastic fraction (lane 4) but failed to react with any proteins in periplasmic fraction (lane 3), indicating that high quality periplasmic fraction was used in this study.
Figure 2.
Cee is an enterobactin esterase located in periplasm of C. jejuni. (A) SDS-PAGE analysis of rCee production and purification. Lane 1, molecular weight marker (Bio-Rad); lane 2, whole cell lysate of noninduced E. coli; lane 3, whole-cell lysate of E. coli induced for 3h with 1 mM IPTG; lane 4, purified rCee purified. (B) Localization of Cee. The proteins samples were subjected to SDS-PAGE and immunoblotting analysis using CmeR as a cytoplasmic protein control and CmeB as an inner membrane control. Lane 1, whole cell lysate of C. jejuni 81–176 containing control vector pRY111 (JL229); lane 2, whole cell lysate of the C. jejuni 81–176 containing a vector that overproduces Cee (JL870); lane 3, periplasmic fraction of JL870; lane 4, spheroplastic fraction of JL870; and lane 5, the purified rCee. (C) Thin layer chromatography (TLC) analysis of the hydrolysis of Ent by rCee. The reaction time was indicated above the picture.  , Ent, or cyclic trimer of DHBS;
, Ent, or cyclic trimer of DHBS;  , linear trimer of DHBS;
, linear trimer of DHBS;  , dimer of DHBS;
, dimer of DHBS;  , DHBS monomer. (D) HPLC analysis of hydrolysis of apo Ent by Cee, Fes, IroD, and IroE. (E) HPLC analysis of hydrolysis of FeEnt by Cee, Fes, IroD, and IroE.
, DHBS monomer. (D) HPLC analysis of hydrolysis of apo Ent by Cee, Fes, IroD, and IroE. (E) HPLC analysis of hydrolysis of FeEnt by Cee, Fes, IroD, and IroE.
Comparative enzyme activity assay of Cee, Fes, IroD, and IroE with apo Ent and FeEnt
TLC analysis indicated that the rCee hydrolyzed Ent efficiently (Fig. 2C). Surprisingly, unlike its closest homolog IroE that tends to hydrolyze the trilactone of Ent only once to produce linear trimers (Lin et al., 2005a), the rCee hydrolyzed Ent to generate dimer and monomer products as early as 2 min (Fig. 2C); by 20 min of reaction, little linearized trimer was observed. To better analyze and compare the activity of Cee with those of previously characterized trilactone esterases from other bacteria, three Cee homologs (Fes, IroD, and IroE) were purified and analyzed together with the rCee using the same assay and HPLC conditions.
When using apo Ent as a substrate, the time course analysis indicated that Cee displayed high efficiency to hydrolyze Ent with similar pattern as Fes and IroD (Fig. 2D). In particular, hydrolysis of apo Ent with Cee and IroD for 20 min led to dimers and monomers only. Reaction of Fes with apo Ent for 20 min resulted in the formation of linear trimers, dimers, and monomers. In contrast, IroE hydrolyzed apo Ent slower than other esterases and displayed different catalytic pattern (Fig. 2D). Notably, the initial linear trimer products are not efficient substrates for further hydrolysis by IroE, making the reaction almost stop at linear trimer stage.
HPLC analysis also demonstrated that Cee could effectively hydrolyze FeEnt with similar dynamics as Fes and IroD (Fig. 2E). Notably, the predominant products of FeEnt catalyzed by Cee, Fes, or IroD are DHBS monomers. In contrast, IroE hydrolyzed FeEnt very inefficiently; hydrolysis of FeEnt with IroE for 90 min only generated trace amount of linear trimers and dimers with majority of FeEnt still not catalyzed (Fig. 2E).
Cee is essential for the CfrB-dependent FeEnt acquisition
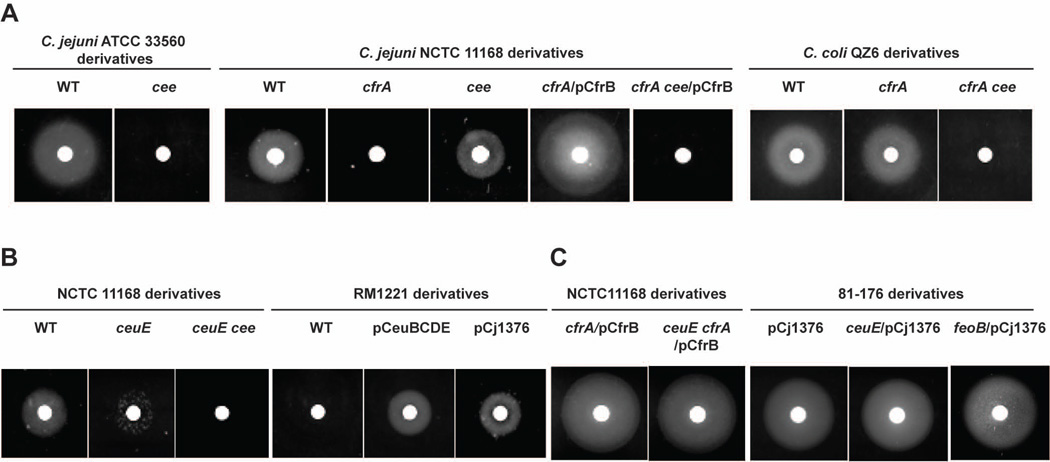
Inactivation of cee in ATCC 33560, a strain containing CfrB-dependent pathway only, completely abolished its ability to utilize FeEnt (Fig. 3A). For NCTC 11168, a strain containing functional CfrA-dependent FeEnt pathway as well as an intact Cee (Table S2), inactivation of cee had little effect on its ability to utilize Ent, indicating that Cee is dispensable for CfrA-dependent pathway (Fig. 3A). Complementation of the cfrA mutant of NCTC 11168 with wild-type cfrB fully rescued FeEnt utilization; inactivation of cee in the cfrA/pCfrB, a strain with genetically repaired CfrB-dependent pathway, abolished its ability to use FeEnt (Fig. 3A).
Figure 3.
Role and molecular interaction of Cee in CfrA-and CfrB-dependent FeEnt acquisition in C. jejuni. Standard Ent growth promotion assay was performed for wild-type strain and its mutant derivatives in different strain background. (A) Cee is essential for CfrB-dependent FeEnt utilization. (B) Cee could utilize CfrA-transported FeEnt. (C) CeuBCDE ABC transporter system and FeoB are dispensable for CfrB/Cee-dependent FeEnt acquisition pathway.
Most C. coli strains possess both CfrA- and CfrB-dependent systems but CfrB has a major role in FeEnt iron acquisition (Xu et al., 2010). Thus, inactivation of cfrA alone in QZ6, a swine C. coli isolate, did not significantly affect its ability to utilize FeEnt (Fig. 3A). However, cfrA cee double mutant failed to utilize Ent, demonstrating the essential role of Cee in CfrB-dependent pathway in C. coli. This observation was also confirmed in two different C. coli strains isolated from swine (QZ7 and QZ8) (data not shown).
Cee could utilize CfrA-transported FeEnt
In CfrA-dependent FeEnt acquisition, the CeuBCDE ABC transporter system is important (Miller et al., 2009, Richardson & Park, 1995). However, inactivation of ceuE in NCTC 11168 impaired but did not abolish its ability to utilize FeEnt (Palyada et al., 2004), which is also confirmed in this study (Fig. 3B). Further inactivation of cee in the ceuE mutant completely abolished its ability to utilize FeEnt (Fig. 3B), indicating Cee could utilize the CfrA-transported FeEnt for iron utilization. This observation is also supported by the findings from C. jejuni RM1221 (Fig. 3B). C. jejuni RM1221 cannot utilize FeEnt due to natural mutations in some genes, such as cfrB, and ceuE (Table S2). As expected, complementation of RM1221 with ceuBCDE operon restored its ability to utilize FeEnt (Fig. 3B) due to the presence of functional CfrA and TonB-ExbB-ExbD system (Table S2). Complementation of RM1221 with wild-type cee only (pCj1376) also restored its ability to utilize FeEnt (Fig. 3B), further demonstrating the interaction of Cee with CfrA-transported FeEnt.
CeuBCDE ABC transporter system and FeoB are dispensable for CfrB/Cee-dependent FeEnt acquisition pathway
To determine if the CeuBCDE ABC transporter system is involved in CfrB/Cee-dependent pathway, ceuE mutation was introduced into two strains that have functional CfrB/Cee-dependent pathway only: NCTC 11168 cfrA/pCfrB and 81-176/pCj1376. As shown in Fig. 3C, inactivation of ceuE in these two strains did not affect CfrB/Cee-dependent FeEnt utilization.
The in vitro enzyme activity assays (Fig. 2) suggest that FeEnt may be hydrolyzed by Cee efficiently for iron release in periplasm. One possible iron release mechanism is that the degraded Fe3+-DHBS products are subjected to ferric reduction. If this is the case, the released ferrous iron (Fe2+) should be assimilated via FeoB, the inner membrane ferrous transporter identified in C. jejuni (Naikare et al., 2006). However, insertional mutation of feoB in 81-176/pCj1376 did not affect its ability to utilize FeEnt, indicating feoB is not required for CfrB/Cee-dependent FeEnt utilization (Fig. 3B).
Role of Cee in C. jejuni colonization
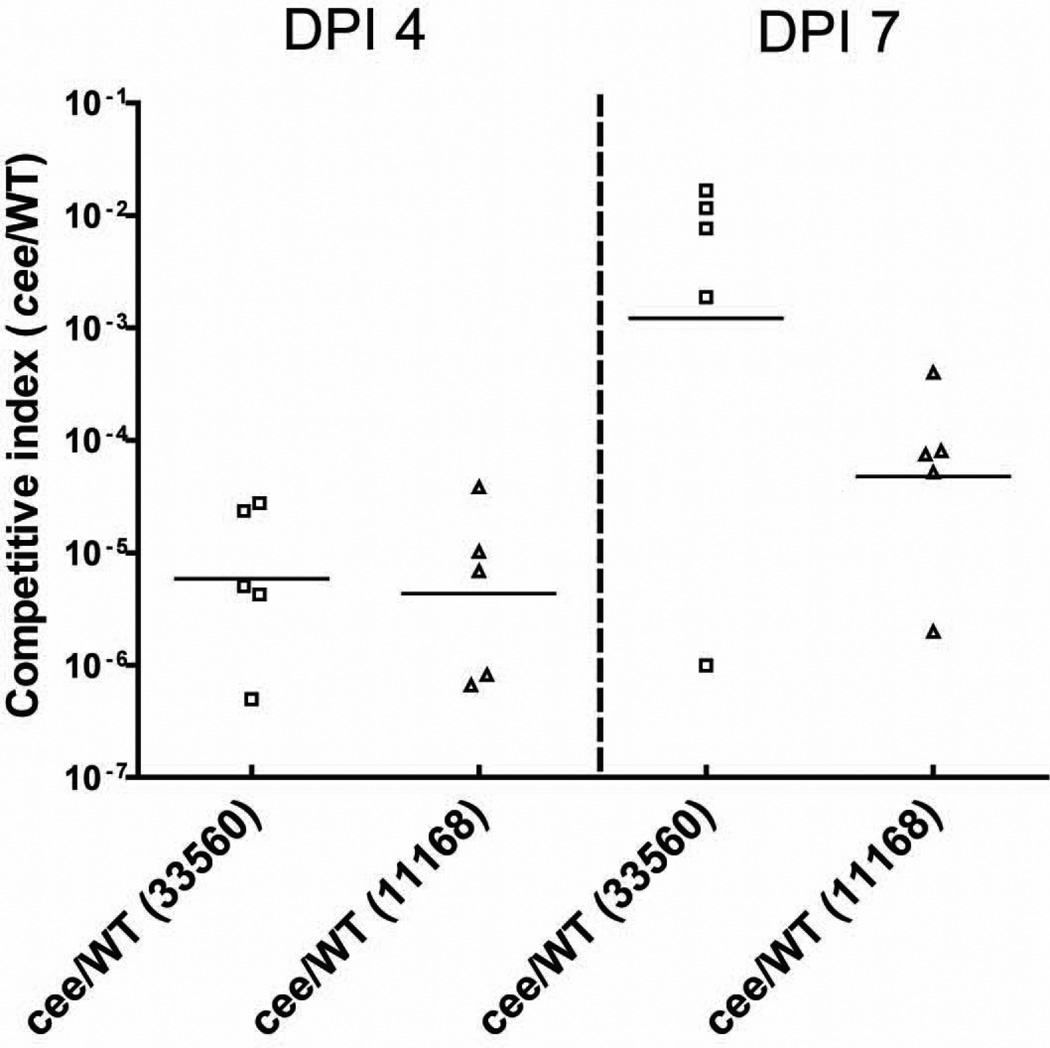
Since FeEnt acquisition is important for in vivo survival and colonization of C. jejuni (Palyada et al., 2004, Zeng et al., 2009, Xu et al., 2010), competitive C. jejuni colonization was performed in chickens. At 4 days postinoculation (DPI), colonization of both cee mutants was significantly outcompeted by their corresponding wild-type strains; the average competitive index is less than 10−5 (Fig. 4). By DPI 7, the cee mutants were still greatly outcompeted by their parent strains; there are no any cee mutants of JL11 detected in majority of chickens at DPI 7 (4 out of 5). Notably, an in vitro competitive assay was also performed in parallel with the in vivo assay; the competition index in MH broth for both pairs is around 1 at 4 days and 7 days post co-culturing, suggesting that inactivation of cee did not cause any growth defect in vitro (data not shown).
Figure 4.
Competitive C. jejuni colonization experiment in chickens. The chickens in each group (10 birds) were inoculated with a 1:1 mixture of C. jejuni ATCC 33560 or NCTC 11168, and its isogenic cee mutant. Each symbol represents the ratio of cee mutant/wild-type (competitive index) recovered from a single chicken. The bar represents the mean log transformed in vivo competitive index of each pair of strains at the indicated DPI (days postinfection).
Prevalence of cee in Campylobacter
Examination of available Campylobacter genomes showed cee is present in more than half of C. jejuni strains (11 out of 20) and in all C. coli strains (Table 1). By taking advantage of the sequence data from a recent Campylobacter pan-genome project (Lefebure et al., 2010), we also observed that cee was present in all 42 examined C. coli strains (data not shown).
Table 1.
Presence of cfrA, cfrB and cee in currently available Campylobacter genomes
| Campylobacter strainsa | cfrA | cfrB | cee |
|---|---|---|---|
| Campylobacter jejuni jejuni NCTC 11168 | + | pseudogeneb | + |
| Campylobacter jejuni jejuni 81–176 | − | + | − |
| Campylobacter jejuni jeuni ATCC 33560 (JL11) | − | + | + |
| Campylobacter jejuni RM1221 | + | pseudogene | − |
| Campylobacter jejuni doylei 269.97 | + | + | − |
| Campylobacter jejuni jejuni 1336 | − | + | + |
| Campylobacter jejuni jejuni 260.94 | + | + | + |
| Campylobacter jejuni jejuni 414 | NDc | + | + |
| Campylobacter jejuni jejuni 81116 | − | + | − |
| Campylobacter jejuni jejuni 84-25 | + | pseudogene | + |
| Campylobacter jejuni jejuni CF93-6 | + | pseudogene | + |
| Campylobacter jejuni jejuni CG8421 | + | pseudogene | − |
| Campylobacter jejuni jejuni CG8486 | + | pseudogene | − |
| Campylobacter jejuni jejuni HB93-13 | ND | + | + |
| Campylobacter jejuni jejuni IA3902 | + | pseudogene | + |
| Campylobacter jejuni jejuni M1 | ND | + | − |
| Campylobacter jejuni jeuni ICDCCJ07001 | + | + | + |
| Campylobacter jejuni jeuni S3 | + | + | − |
| Campylobacter jejuni subsp. jejuni 305 | + | pseudogene | +d |
| Campylobacter jejuni subsp. jejuni 327 | ND | + | − |
| Campylobcter coli RM2228 | + | + | + |
| Campylobacter coli JV20 | + | + | + |
Genome sequences were retrieved from Xbase (http://www.xbase.ac.uk/campydb/).
pseudogene, the gene with the frameshift mutation.
ND, not detected in corresponding draft genomes deposited in Xbase, normally due to the absence of the gene or the gene located in the gap between contigs of the draft genome.
cee is located in the boundary of the corresponding contig, thus incomplete sequence (553 bp).
Discussion
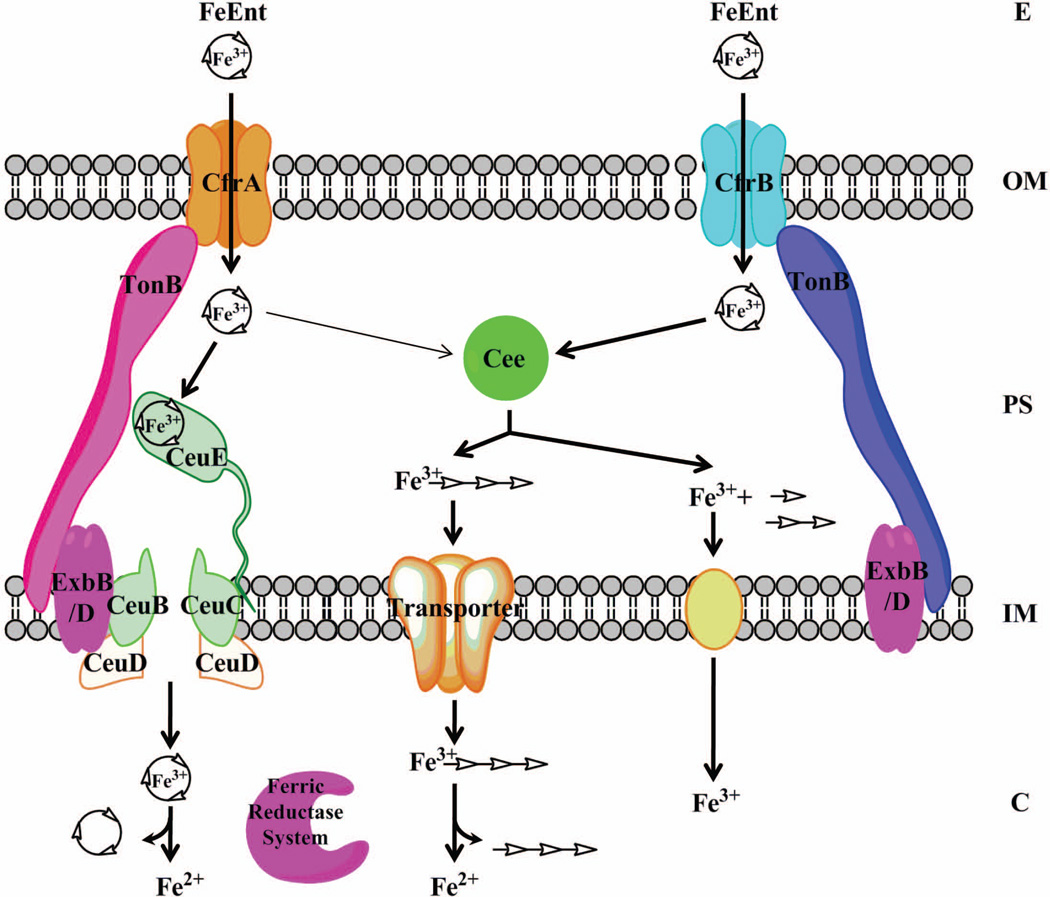
In this study, we identified a periplasmic Ent esterase Cee that plays an important and unique role in Ent-mediated iron acquisition in Campylobacter. In terms of sequence and structure, Cee shares the highest homology to IroE, a periplasmic trilactone hydrolase recently identified in E. coli and Salmonella (Larsen et al., 2006, Zhu et al., 2005). However, Cee displayed faster hydrolysis rate and higher efficiency than IroE for both apo and ferric forms of Ent (Fig. 2). Consistent with previous observation (Lin et al., 2005a), IroE tends to hydrolyze Ent just once to produce linearized trimers that still display high affinity to ferric iron. Thus, Lin et al. (2005a) proposed that IroE is likely involved in export rather than assimilation of triscatecholate siderophores. Clearly, genetic evidence in this study demonstrated that Cee is involved in Ent-mediated iron uptake in Campylobacter, which is further supported by the evidence that Cee could hydrolyze both apo and ferric forms of Ent effectively (Fig. 2). Notably, comparative enzyme activity assays showed that Cee is functionally similar to Fes and IroD, the two efficient cytoplasmic trilactone hydrolases required for intracellular iron release and utilization. Thus, the findings from this study strongly support a new pathway for CfrB-dependent FeEnt acquisition: after passing through outer membrane, FeEnt is hydrolyzed by the periplasmic Cee, which leads to immediate iron release/transfer in periplasm for assimilation. The Cee-mediated hydrolysis of FeEnt in periplasm may facilitate subsequent iron removal by ferric reduction and/or by the transfer of ferric iron from low-affinity DHBS products, such as monomers, to other cellular iron-binding components. If hydrolyzed Ent products are subjected ferric reduction, the FeoB, an inner membrane ferrous (Fe2+) transporter, should play a critical role in CfrB/Cee-dependent FeEnt acquisition. However, inactivation of FeoB did not affect Cee-mediated FeEnt utilization (Fig. 3B), suggesting that reductive iron release from Cee-hydrolyzed Ent products in periplasm is not a preferred pathway. Therefore, efficient hydrolysis of FeEnt by Cee may lead to dramatic loss of complex stability and direct release of Fe3+ in the periplasm. However, we cannot rule out one possibility that the linearized DHBS trimers, the initially degraded products that still have high affinity to Fe3+, may transport through inner membrane via an unknown transporter for cytoplasmic iron release. This possibility needs to be examined in the future.
High-affinity siderophore salmochelin, the glucosylated analog of Ent, has been isolated from Salmonella enterica and E. coli strains (Hantke et al., 2003). Unlike Ent that can be sequestered by host innate immunity protein lipocalins, salmochelin has low affinity for lipocalins and thus the use of salmochelin could confer an in vivo growth advantage to bacteria by resisting lipocalins, particularly in the inflamed intestine (Flo et al., 2004, Raffatellu et al., 2009). Both IroD and IroE could hydrolyze salmochelin and play a role in uptake or export of salmochelin products (Lin et al., 2005a, Zhu et al., 2005). IroD is likely the only trilactone esterase that hydrolyzes ferric forms of salmochelins to release iron in cytoplasm (Lin et al., 2005a). As an important strategy to evade host innate immunity, utilization of salmochelin by Campylobacter is still unknown. However, given the functional similarity between Cee and IroD for hydrolysis of trilactone Ent (Fig. 2), it is likely that Campylobacter could utilize salmochelin via Cee. In addition, because some FeEnt receptors, such as IroN, Cir, or Fiu from E. coli, also could recognize salmochelins or their hydrolyzed linear trimers for iron acquisition (Hantke et al., 2003, Zhu et al., 2005), we speculate that Campylobacter may employ Cee together with CfrA and/or CfrB to utilize salmochelin for iron acquisition. Examining these hypotheses will greatly improve our understanding on the role of siderophore-mediated iron scavenging in Campylobacter pathophysiology.
This study showed that CfrA- and CfrB-dependent pathways are intersected via Cee. Although the Cee is dispensable for CfrA-dependent pathway, Cee could utilize CfrA-transported FeEnt when CeuBCDE transporter system is inactivated. One intriguing finding from this study is that the CfrB-transported FeEnt in periplasm could not interact with CeuBCDE for further utilization. As shown in Fig. 3A, the NCTC 11168 derivative cfrAcee/pCfrB failed to utilize FeEnt although this strain produces functional CfrB, TonB-ExbB-ExbD energy transduction system, and CeuBCDE transporter system for taking up FeEnt. Therefore, it is likely that CfrA, TonB/ExbBD, and CeuBCDE form confined machinery for transporting FeEnt into cytoplasm. To test this model, the structure and stoichiometry of the components that are involved in active transport of FeEnt should be revealed in the future.
The findings from this study also suggest the presence of intracellular reductive iron release mechanism for FeEnt utilization in C. jejuni. For most siderophores that have higher redox potential than Ent, reduction of ferric-siderophore complex by reductase is a common route for iron acquisition (Miethke & Marahiel, 2007). However, little compelling evidence exists showing the intracellular reduction of FeEnt followed by release of Fe2+. Recently, Miethke et al. (2011) demonstrated that the siderophore-interacting protein YqiH functions as a ferric reductase to catalyze the release of iron from Ent in E. coli. In our study, isogenic cee mutant of NCTC 11168 and its parent strain displayed similar ability to utilize FeEnt utilization (Fig. 3A) although Cee appears to be the sole Ent trilactone esterase in Campylobacter (Fig. 1 and Fig. S3), strongly suggesting that the CfrA-transported FeEnt is subjected to intracellular reductive iron release. The FeEnt reduction mechanism may be important for Campylobacter who only utilizes exogenous Ent because ferric siderophore reduction provides an advantage of reutilizing intact siderophores (Miethke et al., 2011). A recent study has supported the close relationship among riboflavin biosynthesis, assimilatory ferric reduction, and iron acquisition in C. jejuni (Crossley et al., 2007); but the identity of the putative ferric reductase is still unknown. Thus, identification and characterization of ferric reductase(s) in C. jejuni is highly warranted in the future to improve our understanding of FeEnt assimilation in C.jejuni and other bacteria. We have performed extensive genome analysis but failed to find YqiH homolog in C. jejuni and C. coli (data not shown). However, Cj1377c, the gene adjacent to cee (Cj1376), encodes an iron-regulated Fe-S protein. Cj1377c contains a highly conserved domain (TIGR03224, E-value of 2e-07) represented by BoxA, a ferredoxin-NADPH reductase (Zaar et al., 2004). Because ferredoxin-NADPH reductase can function as a ferric reductase (Takeda et al., 2010, Yeom et al., 2009), we hypothesize that Cj1377c is likely a ferric reductase involved in FeEnt acquisition in C. jejuni. This speculation needs to be examined in the future.
This study showed that prevalence of cee varies between C. jejuni and C. coli. The cee gene is present in all examined C. coli strains, which is consistent with the previous finding that CfrB-dependent pathway plays a dominant role in FeEnt acquisition in C. coli (Xu et al., 2010), and the essential role of Cee in CfrB-dependent pathway observed in this work. In C. jejuni, cee is present in about half of the strains examined in this study, which is also consistent with our previous finding that C. jejuni usually has only one functional system (CfrA- or CfrB-dependent) for FeEnt acquisition (Xu et al., 2010). Since Cee could utilize the CfrA-transported FeEnt, expression of a functional Cee may provide plasticity of C. jejuni for utilizing FeEnt during evolution. Due to the difference in the prevalence of cee in C. jejuni and C. coli, the definitive role of Cee in Campylobacter pathogenesis is still not clear although Cee is required for optimal in vivo colonization of C. jejuni as observed in this study (Fig. 4). We should be cautious with regard to the competition experiments in chickens because Coward et al. (2008) showed that competition experiments sometimes could give erroneous outcomes. In addition, the importance of FeEnt acquisition for general Campylobacter colonization is challenged by the intriguing finding that 81–176, an efficient colonizer of the chicken intestine, is deficient in FeEnt acquisition due to the lack of Cee. It is likely that 81–176 could effectively use different iron acquisition systems for in vivo colonization, such as utilization of haem compounds (Pickett et al., 1992) and host iron-binding glycoproteins (Miller et al., 2008). However, given the presence of various hydrolyzed Ent products in vivo (Caza et al., 2008), we cannot rule out the possibility that 81–176 may take up hydrolyzed DHBS products of Ent in the intestine via CfrB and utilize such siderophore source without reliance on Cee.
In conclusion, identification and characterization of Cee in this study revealed novel features of FeEnt acquisition in C. jejuni. A new FeEnt acquisition model in Campylobacter is proposed (Fig. 5). This study together with our previous work suggest that Campylobacter may serve as an ideal model organism to examine FeEnt acquisition due to its small genome (1.6–1.8 Mbp), inability to synthesize any siderophores, low redundancy of iron acquisition systems, and enormous strain diversity. Future validation studies for the proposed model will provide new insights into the role of FeEnt acquisition in the evolution and pathogenesis of Campylobacter and other bacteria.
Figure 5.
Proposed new model of FeEnt acquisition system in C. jejuni. The model indicates that Cee is a periplasmic Ent esterase that is involved in both CfrA- and CfrB-dependent Ent utilization systems. Cee is an essential player in CfrB-dependent pathway but also could hydrolyze CfrA-transported FeEnt. The thinner arrow line indicates an alternative pathway for utilizing CfrA-transported FeEnt via Cee when CeuBCDE system is not functional. In periplasm, the degraded products of FeEnt by Cee may follow two journeys: immediate release/transfer of Fe3+ from the low-affinity DHBS monomers and/or dimers, and transport of linear Fe3+-(DHBS)3 into cytoplasm for utilization. In CfrA-dependent pathway, FeEnt is likely subjected to reductive iron release by an uncharacterized ferric reductase system in cytoplasm. E, extracellular environment; OM, outer membrane; PS, periplasmic space; IM, inner membrane; C, cytoplasm.
Experimental Procedures
Bacterial strains, plasmids and culture conditions
The major bacterial strains and plasmids used in this study are listed in Table S4. The C. jejuni strains were routinely grown in Müller-Hinton (MH) broth or on agar at 42°C under microaerophilic conditions (85% N2, 10% CO2, 5% O2). When iron-limited conditions were needed, media was supplemented with 20 µM of deferoxamine mesylate (DFO). E. coli strains were grown routinely in Luria-Bertani (LB) broth with shaking (250 rpm) or on agar at 37 °C overnight. When needed, culture media were supplemented with ampicillin (100 µg/ml), kanamycin (30 µg/ml), chloramphenicol (Cm) (6 µg/ml), or tetracycline (5 µg/ml).
Whole genome sequencing of C. jejuni ATCC 33560
The 454 GS FLX pyrosequencing technology (Voelkerding et al., 2009) was used to sequence the genome of C. jejuni ATCC 33560 (JL11, Table S4). The genomic DNA of ATCC 33560 was extracted using Wizard® Genomic DNA Purification Kit (Promega, Madison) and size fractioned into 400–500 bp fragments, which subsequently were blunt end repaired, ligated to specific adaptors, immobilized and amplified on the DNA Capture Beads, followed by sequencing using the PicoTiter Plate in the 454-FLX instrument, using the FLX Titanium protocol. The generated reads were de novo assembled into contigs using the 454 Newbler Assembler software. The contigs were aligned against three reference genomes, C. jejuni NCTC 11168, RM1221 and 81–176 (Parkhill et al., 2000, Hofreuter et al., 2006, Fouts et al., 2005) by using Mauve (v 2.3.0) (Darling et al., 2004, Rissman et al., 2009). C. jejuni RM1221 was chosen as a template for the contig extension. Primers targeted to the ends of each contig were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Since the contig ends used for primer design often contain unreliable sequences, the priming sites of primers were designated at least 50 bp far from each end of the contigs. The gaps between the ordered contigs were amplified by PCR and subsequently sequenced by Sanger sequencing at the Molecular Biology Resource Facility at The University of Tennessee.
Annotation and comparative genomic analysis
The assembled ATCC 33560 genome sequence draft was analyzed by the RAST server (Rapid Annotation using Subsystem Technology, http://rast.nmpdr.org/) for automatic annotation (Aziz et al., 2008). Genes present in C. jejuni NCTC 11168 and ATCC 33560 but absent in C. jejuni 81–176 were identified by both Integrated Microbial Genomes-Human Microbiome Project (IMG/HMP) system (http://www.hmpdacc-resources.org/cgi-bin/img_hmp/main.cgi) and literature search followed by manual inspection. The transcriptome profiling in response to iron (Palyada et al., 2004, Holmes et al., 2005) was used to improve the power of identification of novel gene(s) involved in iron acquisition.
Sequence analysis and homolog modeling of Cee
The alignment of protein sequences of Ent esterase from Escherichia coli CFT073, Salmonella enterica serovar Typhi Ty2, Pseudomonas aeruginosa and Bordetella pertussis Tohama I (retrieved from database xBASE (Chaudhuri et al., 2008)), and the Cj1376 homologs from different Campylobacter strains (http://watson.bham.ac.uk:8003/campydb/) was performed using ClusterW algorithm in MEGA4. The phylogenetic tree was constructed by using neighbor-joining method in MEGA4. The homolog modeling of Cj1376 (Cee) was performed by using Moe 2008 against IroE (2GZS.pdb) template (Larsen et al., 2006).
Construction of insertional mutants and complementation in trans
The cee gene (Cj1376) was inactivated in C. jejuni NCTC11168, ATCC 33560, and C. coli JL170 by insertional mutagenesis according to a protocol described in our previous publications (Akiba et al., 2006, Lin et al., 2005b, Lin et al., 2003). To construct cee mutant in C. jejuni, primer Cj1376_F and Cj1376_R (Table S5) were used to amplify 1,917-bp fragment from NCTC11168. The PCR product was cloned into pGEMT-Easy (Promega), resulting in the construct pCee. The chloramphenicol (Cm) resistance gene (Turcatti et al.) was amplified from plasmid pUOA18 (Wang & Taylor, 1990) with primers CHLF1 and CHLR1 (Table S5) using PfuUltra® High-Fidelity DNA polymerase (Stratagene). The 0.8-kb PCR product containing a Cm resistance gene was ligated to SwaI-digested pCee to obtain suicide vector pCee(Cm). Sequence analysis of the construct indicated that the Cmr cassette was inserted in the cee fragment with the same transcriptional direction. The plasmid pCee(Cm) was electroporated into C. jejuni NCTC11168 and ATCC 33560. Cmr mutant was selected on MH agar containing 6 µg/ml of Cm and the insertional mutation was confirmed by PCR using primer pairs of J1376F and J1376R (Table S5).
To construct isogenic cee mutant of C. coli JL170, primer Cj1376_1F and Cj1376_1R (Table S5) were used to amplify 2,212-bp fragment from JL170. The PCR product was cloned into pGEMT-Easy (Promega), resulting in the construct pC1376. The blunt-ended PCR product containing a Cm resistance gene described above was ligated to the HindIII-digested and T4 DNA polymerase-repaired pC1376, resulting suicide vector pC1376(Cm). Sequence analysis of pC1376(Cm) indicated that the Cmr cassette was inserted in the cee fragment with the same transcriptional direction. The pC1376(Cm) was electroporated into C. coli JL170. The isogenic cee mutant of JL170 was selected and confirmed as described above for construction of cee mutant of NCTC 11168.
The isogenic cee mutants with tet resistance, the ceuE mutants with Cm resistance, and the feoB mutant with Km resistance were constructed by standard natural transformation (Wang & Taylor, 1990) using corresponding mutant genomic DNA, which are kindly provided by Dr. Qijing Zhang (Iowa State University, US; cee::tetO DNA of NCTC11168), and Dr. Alain Stintzi (University of Ottawa, Canada; ceuE::cat DNA and feoB::km DNA of NCTC11168), respectively.
To complement C. jejuni 81–176 (JL242) that is defective to utilize FeEnt, a 2.5-kb fragment spanning from the promoter region of Cj1375-76 operon to 32 bp downstream of Cj1376 stop codon was PCR amplified from NCTC11168 using primers Cj1376F and Cj1376R (Figure S2A and Table S5). The PCR was performed using PfuUltra DNA polymerase (Stratgene) and the blunt-ended PCR product was purified and ligated to shuttle vector pRY107 (Yao et al., 1993) which was digested with SmaI prior to ligation. The ligation mix was introduced into DH5α by transformation. A transformant with a plasmid construct bearing the intact Cj1375-1376 operon (named ‘pCj1375-76’) was identified. The in-frame deletion of Cj1375 from pCj1375-76 was performed by inverse PCR with the primer Cj1376F1 and Cj1376R1 (Table S5). The PCR product was purified and transformed into E. coli competent cell Top10. A transformant containing desired plasmid construct (named pCj1376) was identified. To complement 81–176 with the cee gene from C. coli, a 2.6-kb fragment covering cee as well as its two upstream genes within the same operon was PCR amplified from C. coli JL170 using primers C1376F and C1376R (Figure S2A and Table S5). The PCR product was cloned into pRY107 as described above, generating desired plasmid pCc1376 for complementation. All resulting plasmids (pCj1375-76, pCj1376, and pCc1376) were then transferred to C. jejuni 81–176 (JL242) by tri-parental conjugation using DH5α/pRK2013 (JL48) as a helper strain (Akiba et al., 2006).
The pABC-cj1375-76 (Kindly provided by Dr. Qijing Zhang, Iowa State University), a plasmid overexpressing Cj1375-76 operon due to the presence of a strong promoter of CmeABC efflux pump (Jeon et al., 2011), was conjugatively transferred into C. jejuni 81–176 to generate JL870 for studying localization of Cee.
To complement C. jejuni RM1221 with a wild-type ceuBCDE operon, a plasmid construct containing ceuBCDE operon was created. Briefly, the 3.9-kb ceuBCDE operon was amplified from NCTC11168 using PfuUltra DNA polymerase and primers Ceu_F and Ceu_R (Table S5). Subsequently, this fragment was ligated into SmaI-digested pRY107 to obtain the plasmid pCeuBCDE. This plasmid pCeuBCDE was conjugatively transferred into C. jejuni RM1221 to create construct JL547 (Table S3).
Site-directed aa substitution mutagenesis
The two aa residues (S157 and H251) in Cee that may be critical for the enzymatic activity were replaced by alanine in vector pCj1376 (Table S4) using partial overlapping PCR as detailed in our recent publication (Zeng et al., 2009). A plasmid with a specific aa substitution in Cee was confirmed by sequencing using primer J1376F or J1376R (Table S5). Two plasmids with different aa substitutions were conjugatively transferred into C. jejuni 81-176. These constructs and the control strain 81-176/pCj1376 were used for Ent growth promotion assay to determine if S157 or H251 is required for the activity of Cee.
Enterobactin growth promotion assay
The ability of Campylobacter to use FeEnt as a sole iron source for growth was assessed using standard growth promotion assay as detailed in our previous publications (Zeng et al., 2009, Xu et al., 2010). Ent was purified from E. coli AN102 (an Ent transport mutant) as described previously (Anderson & Armstrong, 2004). The purified enterobactin was dissolved in a methanolic solution and stored at −20°C until it was used. A sterile disk containing 10 µl of Ent (5 mM) was placed upon the surface of the agar.
Production of recombinant Cee and generation of polyclonal antiserum
N-terminal histidine-tagged recombinant Cee (rCee) was produced in E. coli by using pQE-30 vector of QIAexpress Expression System (Qiagen). Briefly, a 741 bp fragment covering a 240 aa region of Cee (aa 30 to 269) was PCR amplified from C. jejuni NCTC 11168 using primer pairs rCj1376F2 and rCj1376R (Table S5). The amplified product and the vector pQE30 were both digested with BamHI and KpnI and ligated with each other. Cloning, expression, and purification of rCee peptide were performed using the procedures described in our previous publication (Lin et al., 2005b). The plasmid pCj1376N30 in the E. coli JM109 clone (JL751) producing rCee was sequenced and no mutations in the coding sequence of cee were detected. The fractions containing pure rCee were pooled and dialyzed against buffer of 50 mM Tris (pH8.0), 50 mM NaCl, 10% (v/v) glycerol. The concentration of the purified rCee was determined using BCA assay (Pierce). The purified rCee was aliquoted and stored at −80°C. Approximately 4 mg of highly purified rCee was used for the production of rabbit polyclonal antisera against Cee by Pacific Immunology Corp (Ramona, CA). Pre- and post-immune serum samples were analyzed by immunoblotting using pure rCee.
Localization of the Cee
To determine the cellular localization of Cee, the periplasmic and spheroplastic fractions were prepared using the PeriPreps™ Periplasting Kit (Epicentre) that has been successfully used in Campylobacter (Naikare et al., 2006). Approximately 10 ml of early log phase culture (OD600 = 0.1) was pelleted by centrifugation and washed one time with MEMα medium (Invitrogen). The cells were thoroughly resuspended in 50 µl of the PeriPreps Periplasting Buffer and incubated at room temperature for 10 min. Then the osmotic shock was triggered by adding 50 µl of ice cold water into the bacterial suspension, followed by 5 min incubation on ice. The cell suspension was centrifuged for 5 min at room temperature and the supernatant containing the periplasmic fraction was transferred to a clean tube while the pellets (the spheroplasts) were washed one time with MEMα medium. The whole cell lysate, cellular fractions as well as the purified rCee were used for SDS-PAGE and immunoblotting analysis using Cee specific antiserum as described in our previous publications (Zeng et al., 2009, Lin et al., 2002). To ensure the quality of periplasmic and spheroplastic fractions, antibodies directed against CmeR (cytoplasmic control, 1:200 dilution) and CmeB (inner membrane, 1:1,000 dilution) were also used as primary antibodies for immunoblotting.
Ent esterase activity assay using whole cell extracts
To prepare whole-cell extracts, Campylobacter culture (OD600nm = 0.1) were 1:50 diluted into 20 ml of MH broth containing 20 µM of DFO. Following 2-day incubation under microaerophilic conditions at 42 °C, the cells were pelleted, washed once with ice cold lysis buffer (10 mM Tris-HCl, pH 8.0; 150 mM NaCl), and re-suspended in 2 ml of the same lysis buffer. The cell suspension was sonicated on ice with 3 cycles of action (15 sec per cycle). The sonicated cell suspension was centrifuged at 13,000 rpm for 10 min at 4 °C and the supernatant was used as the raw cell extract for esterase activity analysis using following two complementary approaches.
i) PBS counter-extraction method
(O'Brien et al., 1971, Langman et al., 1972). The reaction mix containing 0.15 ml cell extract (approximately 300 µg/ml proteins) and 0.15 ml Ent solution (1.27 mM Ent, 1mM EDTA, 50 mM DTT) was incubated at 37 °C for 90 min. The reaction was stopped by adding 0.1 ml HCl (1M) and the solutions were extracted with 1ml ethyl acetate for 30 min, followed by centrifugation at 13,000 rpm for 5 min. Three hundred microliter of the organic phase was used to determine total catecholates (intact Ent plus its hydrolyzed products) based upon a millimolar extinction coefficient of Ent of 9.39 at 316 nm (O'Brien et al., 1971, Brickman & McIntosh, 1992). Another part of organic phase was counter-extracted with equal volumes of PBS (pH7.4) to quantify the hydrolyzed products that are soluble in PBS. The esterase activity was expressed as nanomoles of Ent hydrolyzed per hour per mg of cell extract. These experiments were performed in triplicate.
ii) Reverse phase HPLC
(Lin et al., 2005a). Briefly, the reaction was carried out at room temperature in 50 mM Tris buffer (pH 7.5) with 27 µM Ent, 37 µg/ml of cell extract, 2.0 mM MgCl2. Two hundred microliter of reaction mix was sampled at different time points (15 sec, 31.5 min, and 60 min), and quenched with 100 µl 2.5 N HCl in methanol. The samples were stored in −80°C. To analyze the reaction with HPLC, the samples were thawed and centrifuged at 20,000×g for 10 minutes to remove precipitate. The supernatant was analyzed by HPLC with Vydac 201TP5405 column at a gradient of 0-43.75% CH3CN (with 0.1% trifluoroacetic acid) in water (with 0.1% trifluoroacetic acid) over 10 min.
Ent esterase activity assay using purified trilactone esterases
The purified rCee was first subjected to thin layer chromatography (TLC) (Furrer et al., 2002) to evaluate its hyfrolysis activity with apo Ent. Briefly, reaction mixture at room temperature contained 9.6 µl of cyclic Ent (2.9 mg/ml), 2 µl of 100mM MgCl2, 20 µl of 50mM Tris buffer (pH 7.5), and 5 µl of 0.2mg/ml Cee protein. To prepare TLC sample for different reaction time (0, 1, 2, 5, 20, or 60 minutes), 6 µl reaction mixture was mixed with 3ul of 2.5N HCl in methanol that was used to quench the reaction. Then, 9 µl of solution for each sample was spotted at the TLC plate (Whatman PE silica gel with UV), and the TLC plate was developed with benzene:acetic acid:water=110:85:5 (v/v). The hydrolysis products were visualized by spraying plates with 1% FeCl3.
Comparative enzyme activity assays of rCee, IroE, IroD, and Fes with apo Ent and FeEnt were performed using reverse phase HPLC as described above. The His-tagged recombinant IroE, IroD, and Fes were purified from JL633, JL634, and JL637 (Table S4), respectively, as detailed in a previous publication (Lin et al., 2005a). For enzyme activity assay with apo Ent, all reactions were carried out at room temperature in 50 mM Tris buffer (pH 7.5) with 30 µM Ent, 25 nM specific esterase, and 2.0 mM MgCl2. For enzyme activity with FeEnt, all reactions were carried out at room temperature in 50 mM Tris buffer (pH 7.5) with 20 µM FeEnt, 80 nM specific esterase, and 2.0 mM MgCl2. The FeEnt complex was prepared by mixing Ent with 1.2 equivalent of FeCl3 as described by Lin et al. (2005a).
Chicken colonization experiment
Competitive assay of wild-type strain together with its isogenic cee mutant was performed in a chicken model system. One-day-old broiler chickens were obtained from a commercial hatchery (Hubbard Hatchery, Pikesville, TN). The chickens were negative for Campylobacter as determined by culturing cloacal swabs prior to use in this study. C. jejuni JL11 and NCTC11168 and their cee mutant were used in this study. Two groups of 4-day-old chickens (10 birds/group) were inoculated with a 1:1 mixture of C. jejuni JL11 and its isogenic cee mutant (group 1), or with a 1:1 mixture of C. jejuni NCTC11168 and its isogenic cee mutant (group 2). Inoculation was performed via oral gavage using a dose approximately 5 × 105 CFU of bacteria per bird. Notably, the motility of parent strain and its cee mutant was confirmed to be at a comparable level prior to challenge. For each group, 5 birds were euthanized and cecal samples were collected at 4 and 7 days postinoculation (DPI). The cecal contents from each bird were weighed and diluted in MH broth. The diluted samples were duplicate plated onto MH agar plates with Campylobacter specific selective supplements (Oxoid, United Kingdom) for total Campylobacter enumeration and onto selective plates supplemented with Cm (6 µg/ml) for enumeration of the Cm-resistant cee mutants in each sample. The number of CFU per gram of cecal contents was calculated for each chicken and was used as an indicator of the colonization level. The detection limit of the plating methods was 100 CFU/g of cecal contents. A bird from which no Campylobacter colonies were detected was assigned a conservative value of 99 CFU/g of cecal contents for the purpose of calculating means and for statistical analysis. The competitive index was calculated as the ratio between the CFU number of cee mutant and that of corresponding wild-type strain.
In parallel to the chicken experiment, the 1:1 mixtures that were used to challenge chicken were also passaged every two days in MH broth under microaerophilic condition. The culture was serially diluted at 4 and 7 DPI; the diluted samples were subjected to differential plating as described above and in vitro competitive index was determined.
Nucleotide sequence accession number
The cee sequence of C. coli JL170 have been deposited into GenBank under the accession number JQ684870. The whole genome shotgun project of C. jejuni JL11 (ATCC 3356)0) has been deposited at DDBJ/EMBL/GenBank under the accession AIJN00000000. The version described in this paper is the first version, AIJN01000000.
Supplementary Material
Acknowledgements
We thank Alain Stintzi (University of Ottawa, Canada) for providing ceuE and feoB mutant DNA, Qijing Zhang (Iowa State University) for providing CmeR antiserum and pABC-Cj1375-76 plasmid, Christopher T. Walsh (Harvard Medical School) for providing strains for production of recombinant IroE, IroD, and Fes, Hening Lin (Cornell University) for helpful discussion with HPLC analysis of Cee, Barbara Gillespie for technical support and proofreading of this manuscript, and Alice Layton (The University of Tennessee) for technical support to perform 454 sequencing. This work was supported by NIH grant 1R56AI090095-01A1 (to J.L.) and University of Tennessee AgResearch Innovation Grant (to J.L.).
References
- Akiba M, Lin J, Barton YW, Zhang Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother. 2006;57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- Anderson MT, Armstrong SK. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J Bacteriol. 2004;186:7302–7311. doi: 10.1128/JB.186.21.7302-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler AJ, Tsolis RM, van der Velden AW, Stojiljkovic I, Anic S, Heffron F. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, McIntosh MA. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J Biol Chem. 1992;267:12350–12355. [PubMed] [Google Scholar]
- Caza M, Lepine F, Milot S, Dozois CM. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect Immun. 2008;76:3539–3549. doi: 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 2008;36:D543–D546. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward C, van Diemen PM, Conlan AJ, Gog JR, Stevens MP, Jones MA, Maskell DJ. Competing isogenic Campylobacter strains exhibit variable population structures in vivo. Appl Environ Microbiol. 2008;74:3857–3867. doi: 10.1128/AEM.02835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley RA, Gaskin DJ, Holmes K, Mulholland F, Wells JM, Kelly DJ, van Vliet AH, Walton NJ. Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl Environ Microbiol. 2007;73:7819–7825. doi: 10.1128/AEM.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field LH, Headley VL, Payne SM, Berry LJ. Influence of iron on growth, morphology, outer membrane protein composition, and synthesis of siderophores in Campylobacter jejuni. Infect Immun. 1986;54:126–132. doi: 10.1128/iai.54.1.126-132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrer JL, Sanders DN, Hook-Barnard IG, McIntosh MA. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol Microbiol. 2002;44:1225–1234. doi: 10.1046/j.1365-2958.2002.02885.x. [DOI] [PubMed] [Google Scholar]
- Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, Galan JE. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, Wells JM. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology. 2005;151:243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother. 2011;66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langman L, Young IG, Frost GE, Rosenberg H, Gibson F. Enterochelin system of iron transport in Escherichia coli: mutations affecting ferric-enterochelin esterase. J Bacteriol. 1972;112:1142–1149. doi: 10.1128/jb.112.3.1142-1149.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NA, Lin H, Wei R, Fischbach MA, Walsh CT. Structural characterization of enterobactin hydrolase IroE. Biochemistry. 2006;45:10184–10190. doi: 10.1021/bi060950i. [DOI] [PubMed] [Google Scholar]
- Lefebure T, Bitar PD, Suzuki H, Stanhope MJ. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol. 2010;2:646–655. doi: 10.1093/gbe/evq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Fischbach MA, Liu DR, Walsh CT. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J Am Chem Soc. 2005a;127:11075–11084. doi: 10.1021/ja0522027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Akiba M, Sahin O, Zhang Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother. 2005b;49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Sahin O, Michel LO, Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of. Campylobacter jejuni. Infect Immun. 2003;71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Hou J, Marahiel MA. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry. 2011;50:10951–10964. doi: 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CE, Rock JD, Ridley KA, Williams PH, Ketley JM. Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J Bacteriol. 2008;190:1900–1911. doi: 10.1128/JB.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CE, Williams PH, Ketley JM. Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology. 2009;155:3157–3165. doi: 10.1099/mic.0.032425-0. [DOI] [PubMed] [Google Scholar]
- Naikare H, Palyada K, Panciera R, Marlow D, Stintzi A. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect Immun. 2006;74:5433–5444. doi: 10.1128/IAI.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien IG, Cox GB, Gibson F. Enterochelin hydrolysis and iron metabolism in Escherichia coli. Biochim Biophys Acta. 1971;237:537–549. doi: 10.1016/0304-4165(71)90274-1. [DOI] [PubMed] [Google Scholar]
- Olson CK, Ethelberg S, Pelt WV, Tauxe RV. Epidemiology of Campylobacter jejuni infections in industrialized nations. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington, DC: ASM Press; 2008. pp. 163–189. [Google Scholar]
- Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pickett CL, Auffenberg T, Pesci EC, Sheen VL, Jusuf SS. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect Immun. 1992;60:3872–3877. doi: 10.1128/iai.60.9.3872-3877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JH, Soudain SE, Gregson NA, Hughes RA. Campylobacter jejuni infection and Guillain-Barre syndrome. N Engl J Med. 1995;333:1374–1379. doi: 10.1056/NEJM199511233332102. [DOI] [PubMed] [Google Scholar]
- Richardson PT, Park SF. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology. 1995;141(Pt 12):3181–3191. doi: 10.1099/13500872-141-12-3181. [DOI] [PubMed] [Google Scholar]
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Vliet AHMV, Ketley JM. Iron Metabolism, Transport, and Regulation. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington, DC: ASM Press; 2008. pp. 591–610. [Google Scholar]
- Takeda K, Sato J, Goto K, Fujita T, Watanabe T, Abo M, Yoshimura E, Nakagawa J, Abe A, Kawasaki S, Niimura Y. Escherichia coli ferredoxin-NADP+ reductase and oxygen-insensitive nitroreductase are capable of functioning as ferric reductase and of driving the Fenton reaction. Biometals. 2010;23:727–737. doi: 10.1007/s10534-010-9339-8. [DOI] [PubMed] [Google Scholar]
- Turcatti G, Romieu A, Fedurco M, Tairi AP. A new class of cleavable fluorescent nucleotides: synthesis and optimization as reversible terminators for DNA sequencing by synthesis. Nucleic Acids Res. 2008;36:e25. doi: 10.1093/nar/gkn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Wang Y, Taylor DE. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- Xu F, Zeng X, Haigh RD, Ketley JM, Lin J. Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J Bacteriol. 2010;192:4425–4435. doi: 10.1128/JB.00478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Yeom J, Jeon CO, Madsen EL, Park W. Ferredoxin-NADP+ reductase from Pseudomonas putida functions as a ferric reductase. J Bacteriol. 2009;191:1472–1479. doi: 10.1128/JB.01473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaar A, Gescher J, Eisenreich W, Bacher A, Fuchs G. New enzymes involved in aerobic benzoate metabolism in Azoarcus evansii. Mol Microbiol. 2004;54:223–238. doi: 10.1111/j.1365-2958.2004.04263.x. [DOI] [PubMed] [Google Scholar]
- Zeng X, Xu F, Lin J. Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect Immun. 2009;77:5437–5448. doi: 10.1128/IAI.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Valdebenito M, Winkelmann G, Hantke K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology. 2005;151:2363–2372. doi: 10.1099/mic.0.27888-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.