Abstract
The stratum corneum is composed of protein-enriched corneocytes embedded in an intercellular matrix of nonpolar lipids organized as lamellar layers and give rise to epidermal permeability barrier (EPB). EPB defects play an important role in the pathophysiology of skin diseases such as eczema. The transcriptional control of skin lipid metabolism is poorly understood. We have discovered that mouse lacking a transcription factor COUP-TF interacting protein 2 (Ctip2) exhibit EPB defects including altered keratinocyte terminal differentiation, delayed skin barrier development and interrupted neutral lipid distribution in the epidermis. We adapted herein a targeted lipidomic approach using mass spectrometry, and have determined that Ctip2−/− mice (germline deletion of Ctip2 gene) display altered composition of major epidermal lipids such as ceramides and sphingomyelins compared to wildtype at different stages of skin development. Interestingly, expressions of several genes involved in skin sphingolipid biosynthesis and metabolism were altered in mutant skin. Ctip2 was found to be recruited to the promoter region of a subset of those genes, suggesting their possible direct regulation by Ctip2. Our results confirm an important role of Ctip2 in regulating skin lipid metabolism and indicate that profiling of epidermal sphingolipid could be useful for designing effective strategies to improve barrier dysfunctions.
Keywords: Bcl11b, epidermis, epidermal permeability barrier (EPB), ceramide, sphingomyelin, sphingosine, sphinganine, ceramide synthases, gene expression, de novo pathway, salvage pathway, sphingomyelin pathway, LC-tandem mass spectrometry, ultra high pressure liquid chromatography
INTRODUCTION
The exterior layer of skin, stratum corneum (SC), plays a critical role in skin permeability barrier formation and maintenance (Fuchs and Raghavan, 2002; Elias, 2004). It forms a protective barrier against external stress and environmental insults. Skin barrier defects are caused not only by altered levels of protein(s) involved in keratinocytes terminal differentiation, cross-linking of cornified envelops (CEs), but also by impaired formation and maintenance of skin lipid barrier (Elias and Menon, 1991; Nemes and Steinert, 1999; Feingold, 2007, 2009). Mammalian SC contains a large number of lipids, such as ceramides, sphingomyelins, cholesterols and fatty acids (Elias and Menon, 1991; Ishikawa et al., 2010; Feingold, 2007, 2011). Ceramides (CERs) are the most abundant lipid types in SC (50% by weight) and can be divided into at least 11 species (Oda et al., 2008; Ishikawa et al., 2010). It has been shown that reduced content of CERs leads to epidermal water loss as well as skin epidermal dysfunctions, and causes diseases such as atopic dermatitis (AD) or eczema (Imokawa, 2009; Ishikawa et al., 2010). Spingomyelins are another large group of epidermal sphingolipids, which have been reported as precursors of ceramides (Schmuth et al., 2000; Uchida et al., 2000; Holleran et al., 2006). Cholesterols and fatty acids are also major skin lipids, and cholesterol 3-sulfate (CSO4) is a ubiquitous metabolite of cholesterol, which is involved in cornified envelope formation by interacting with transglutaminase1 (Nemes et al., 2000).
Ceramide biosynthesis is regulated at multiple levels. Four pathways have been reported so far, de novo pathway, sphingomyelinase pathway, salvage pathway and exogenous ceramide recycling pathway (Kitatani et al., 2008). In de novo pathway, serine and fatty acyl-CoA are the initiators of a series of reactions. Ceramide synthases 1-6 (Lass1-Lass6) play an important role since each of them has specificities on synthesizing certain ceramides (Mizutani et al., 2009). Other enzymes such as palmitoyltransferases (Sptlc1-3), dihydroceramide desaturase (Degs1) and 3-ketodihydrosphingosine reductase (Kdsr) are also key enzymes in de novo pathway (Merrill et al., 2002). The salvage pathway has been reported recently as a complex mechanism, commencing as the catabolism of complex sphingolipids into glucosylceramide by glucosylceramide synthase (Ugcg), and followed by the formation of ceramide by acid beta-glucosidase (Gba2) (Ogretmen, 2002; Becker, 2004). These ceramides can be broken down into sphingosines by ceramidases (Asah1 and Asah2), which are reused to produce ceramides (Bernardo et al., 1995; Koch et al., 1996; Li et al., 1998; Tani et al., 2000; Mao and Obeid, 2008). In sphingomyelinase pathway, ceramide is formed by hydrolysis of sphingomyelins, and the reverse reaction is controlled by sphingomyelin synthase (Marchesini and Hannun, 2004). Exogenous short chain ceramide can also be utilized to generate sphingosine, thus leads to the synthesis of endogenous long chain ceramides (Ogretmen, 2002; Sultan et al., 2006).
COUP-TF-interacting protein 2 (Ctip2), also known as Bcl11b, is a C2H2 zinc finger protein expressed in many organs and tissues including developing mouse epidermis (Avram et al., 2000, 2002; Golonzhka et al., 2008). It has been reported that Ctip2 null mice exhibit altered epidermal proliferation and late terminal differentiation, as well as barrier defects during embryogenesis (Golonzhka et al., 2008). The barrier defects were correlated with altered surface lipid distribution in the absence of Ctip2 (Golonzhka et al., 2008).
Liquid chromatography-tandem mass spectrometry (LC/MS/MS) has been used to characterize ceramide species in mouse and human stratum corneum (Masukawa et al., 2008; Park et al., 2009; Ishikawa et al., 2010; Janssens et al., 2011; van Smeden et al., 2011). Here we adopted the LC/MS/MS methodology to elucidate the ceramide containing nonhydroxy fatty acids (N) and sphingosines (S) [CER (NS)], sphingmyelin and sphingosine profile in wildtype and Ctip2 null (Ctip2−/−) embryonic skin epidermis. The expression levels of genes encoding various enzymes involved in sphingolipid metabolism were analyzed by qRT-PCR and immuno-blotting in wildtype and Ctip2-null skin. Furthermore, Ctip2 recruitment to the promoter regions of several genes that were altered in Ctip2−/− mice was evaluated by chromatin immunoprecipitation (ChIP) analyses. Results indicated that Ctip2 control sphingolipid metabolism in the developing skin by directly or indirectly regulating expression of a subset of genes involved in the sphingolipid biosynthesis pathways.
RESULTS
Altered profile of sphingolipids in the developing epidermis of Ctip2-null mice
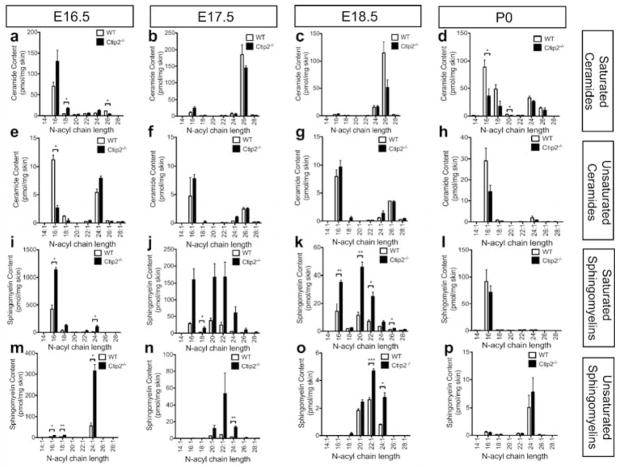
We hypothesized that impaired epidermal permeability barrier in the Ctip2-null (Ctip2−/−) mice could be at least in part due to altered skin lipid composition. In order to determine the lipid composition of embryonic skin, wildtype and Ctip2-null epidermis were isolated at different developmental stages, E16.5, E17.5 E18.5 and neonatal (P0). Lipidomic studies were then performed using LC/MS/MS technique and absolute quantities of the different skin sphingolipids were analyzed (for details see materials and methods). Both ceramides and sphingomyelins were abundant during mouse skin development, especially during the earlier stages of skin development (E16.5, E17.5) (Figure 1).
Figure 1. Liquid chromatography tandem mass-spectrometry (LC/MS/MS) analyses of mouse epidermal sphingolipids during skin development.
Epidermal sphingolipids were extracted from skin of E16.5, E17.5, E18.5 embryos and P0 wildtype and Ctip2−/− mice. Amount of saturated and unsaturated ceramides and sphingomyelins was determined according to their N-acyl chain length using LC/MS/MS analyses. The absolute content of saturated (a–d) and unsaturated (e–h) ceramides at (a, e) E16.5, (b, f) E17.5, (c, g) E18.5 and (d, h) P0 were quantified utilizing saturated and unsaturated ceramide standards, and expressed as pmol/mg of skin (for details see methods). Amount of saturated (i–l) and unsaturated (m–p) sphingomyelin was also determined at different developmental stages (i, m) E16.5, (j, n) E17.5, (k, o) E18.5 and (i, p) P0 using saturated and unsaturated sphingomyelin standards, and expressed as pmol/mg of skin (see methods). Statistical analyses were performed by student’s unpaired t-test using Graphpad Prism software. *p<0.05; **p<0.005, ***p<0.001. Data were reported as mean ± SEM (n=9). The results represent three separate experiments performed in triplicate.
Ceramide C14:0 level remained very low from E16.5-P0. Long chain ceramide C16:0 was the most abundant saturated ceramide at E16.5 and was 2-fold higher in the mutant epidermis compared with wildtype (Figure 1a). Significant induction of ceramide C18:0 was observed in the E16.5 mutant skin (Figure 1a). The amount of long chain ceramides (C16:0, C18:0, C20:0 and C22:0) was in a very low abundance at E17.5–E18.5, with no discernable difference observed between wildtype and mutant skin; however, their quantities are larger at P0, each having a ~2-fold reduction in the Ctip2−/− skin (Figure 1b–d). At E17.5 and E18.5, very long chain ceramide C26:0 became the predominant subtype in mouse skin, modestly reduced at E17.5 and 2-fold decreased at E18.5 in Ctip2−/− skin (Figure 1b–c). Modest decrease of ceramide C24:0 was also observed in Ctip2−/− epidermis at P0 (Figure 1d). Unsaturated ceramides were less abundant compared with saturated ceramides in the wildtype epidermis. Long chain unsaturated ceramide C16:1 was significantly reduced at E16.5 in the mutant epidermis, unaffected at E17.5 and E18.5, with a 2-fold decrease in P0 mutant epidermis (Figure 1e–h).
Among saturated sphingomyelins, long chain sphingomyelins C16:0, C20:0 and C22:0 were the most abundant subtypes in embryonic epidermis at most of the stages, which were highly induced in the mutant skin, especially at E17.5 and E18.5 (Figure 1j and k). Very long chain sphingomyelins (C24:1, C26:1, C28:1) were also increased in the mutant skin at E16.5–E18.5. Interestingly, unsaturated sphingomyelins were less abundant at all developmental stages; however, they showed the similar trend as saturated ones, with higher amount in embryonic mutant epidermis. As early as E16.5, long chain unsaturated sphingomyelin C24:1 was significantly elevated in Ctip2−/− skin (Figure 1m). Higher quantities of long chain sphingomyelins C20:1, C22:1, and C24:1 were noted in the mutant epidermis at E17.5 and E18.5 (Figure 1n–o).
The profiles of sphingosine and sphinganine were also determined by LC/MS/MS. Compared with ceramides and sphingomyelins, the quantity of free sphingoid bases was smaller in embryonic skin, with no significant changes observed between wildtype and mutant epidermis at all developmental stages (Figure S1).
We also performed UPLC/MS/MS analyses to determine the relative amount of cholesterols and fatty acids. Levels of cholesterol and cholesterol 3-sulfate were significantly reduced in the epidermis of Ctip2−/− mice at E16.5, but were unaltered at all later timepoints (Figure S2a, b). Although the amount of palmitic acid and stearic acid was similar between wildtype and null skin at E16.5–E18.5, their levels doubled in the P0 Ctip2−/− epidermis compared to the wildtype epidermis (Figure S2c, d). Altogether, our results indicate that epidermal lipid composition is altered in absence of transcriptional regulator Ctip2.
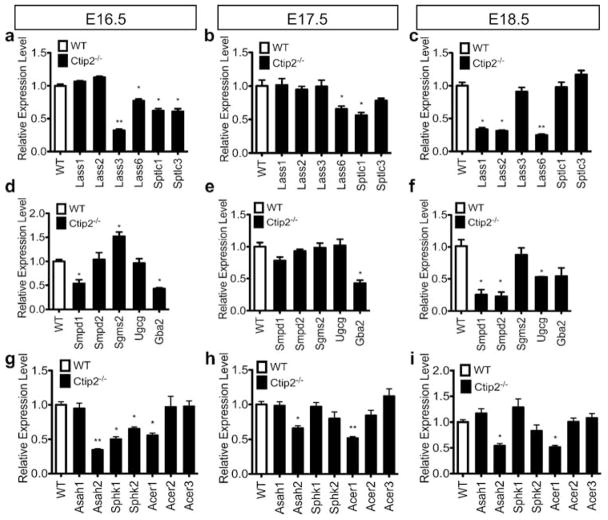
Ctip2 directly regulates the expression of a subset of genes involved in lipid metabolism during skin development
Previous studies have identified four major pathways involved in ceramide synthesis: de novo pathway, salvage pathway, sphingomyelinase pathway and exogenous ceramide-recycling pathway (Kitatani et al., 2008; Mizutani et al., 2009). We hypothesized that altered sphingolipid profiles in mutant mice could be due to altered expression of genes encoding proteins involved in the sphingolipid metabolism. To test that, we evaluated the expression of genes involved in the different ceramide biosynthesis pathways by qRT-PCR analyses in the wild type control and mutant skin (for details see materials and methods). Expressions of ceramide synthase 3 (Lass3), palmitoyltransferases (Sptlc1 and Sptlc3) were downregulated at E16.5 and E17.5, while ceramide synthases Lass1 and Lass2 were significantly reduced at E18.5 in Ctip2−/− skin (Figure 2a–c). Lass6 was consistently downregulated from E16.5 to E18.5 in the mutant skin (Figure 2a–c). Thus, genes encoding enzymes in de novo pathway were altered after ablation of Ctip2. Similarly, transcript levels of enzymes involved in sphingomyelinase pathway such as sphingomyelinases (Smpd1 and Smpd2) were significantly downregulated at E18.5, whereas sphingomyelin synthase (Sgms2) level was upregulated at E16.5 in null embryos compared to the wildtypes (Figure 2d–f). Expression of glucosylceramide synthase (Ugcg), which turns complex sphingolipids into glucosylcerarimde, was also reduced at E18.5 while that of acid beta-glucosidase (Gba2), which is involved in salvage pathway, was downregulated in skin of null mice from E16.5 to E18.5 (Figure 2d–f). Ceramidases are involved in the breaking down of short-chain ceramide, facilitating its conversion to sphingosine. In particular, reduced expression of neutral ceramidase 2 (Asah2) and alkaline ceramidase 1 (Acer1) was observed in all developmental stages in the mutant epidermis (Figure 2g–i). Levels of all other members of acid ceramidases (Asah1 and Asah3) and alkaline ceramidases (Acer2 and Acer3) were unaltered in the mutants at all stages of development (Figure 2g–i). Similarly, sphingosine kinases (Sphk1, Sphk2) are responsible for the disposal of sphingosine to sphingosine-1-phosphate, whose expression level was reduced at E16.5 in the mutant epidermis (Figure 2g). No significant changes in the relative mRNA levels of several other enzymes involved in ceramide synthesis pathways was observed between wildtype and Ctip2−/− embryonic skin (Figure S3a–c). Furthermore, protein levels of Lass2, Gba2 and epidermis-type lipoxygenase 3 (eLox3) were significantly reduced in the Ctip2 null skin compared with the wildtype skin at E18.5 (Figure 3a–b) (Golonzhka et al., 2008). Taken together, our results indicate that expression of a subset of genes encoding enzymes involved in the sphingolipid biosynthesis pathways was altered during skin organogenesis in absence of Ctip2.
Figure 2. Ctip2 regulates the expression of genes encoding lipid-metabolizing enzymes during skin development.
Expression of (a, b, c) ceramide synthases (Lass1-Lass3, Lass6), serine palmitoyltransferases (Sptlc1, Sptlc3); (d, e, f) sphingomyeinases (Smpd1, Smpd2), sphingomyelin synthase 2 (Sgms2), glucosylceramide synthase (Ugcg) and acid beta-glucosidase (Gba2); (g, h, i) N-acylsphingosine amidohydrolases (acid and neutral ceramidases: Asah1, Asah2), sphingosine kinases (Sphk1, Sphk2) and alkaline ceramidases (Acer1, Acer2) was analyzed by qRT-PCR using specific primers indicated in table S1. All values represent relative transcript level after normalization with Gapdh transcripts. Statistical analyses were performed by student’s unpaired t-test using Graphpad Prism software. *p<0.05; **p<0.005. Data were reported as mean ± SEM (n=9). The results represent three separate experiments performed in triplicate.
Figure 3. Immunoblot analyses of lipid metabolizing enzymes in the developing murine epidermis.
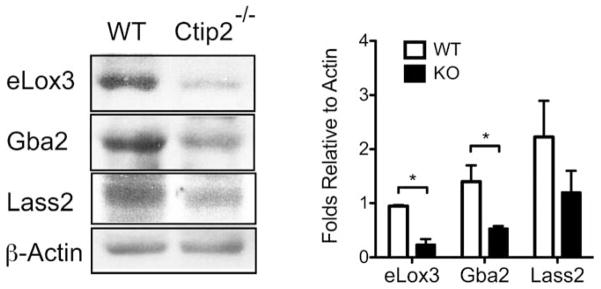
(a) Western blotting analyses was performed on protein extracted from E18.5 wildtype (WT) and Ctip2−/− mice epidermis to study the expression level of eLox3, Gba2 and Lass2 using specific antibodies. β-actin was used as an internal control. (b) Protein levels of eLox3, Gba2 and Lass2 were quantified in wildtype and mutant epidermis by densitometry analyses of western blots. All data was normalized by the corresponding β-actin levels. Statistical analyses were performed by student’s unpaired t-test using Graphpad Prism software. *p<0.05. Data were reported as mean ± SEM (n=9). The results represent three separate experiments performed in triplicate.
To investigate whether some of the lipid-metabolizing genes, which are dysregulated in the mutant skin, are direct targets of transcriptional regulator Ctip2, we conducted in vivo chromatin immunoprecipitation (ChIP) assay on neonatal (P0) mouse epidermal keratinocyte extracts. The promoter regions of several genes encoding lipid-metabolizing enzymes such as Lass1-3, 6, Sptlc1-3, Smpd1-2, Ugcg, Gba2, and eLox3 (Golonzhka et al., 2008), whose expressions were altered in the Ctip2−/− skin, were selected and primers were designed to determine possible Ctip2 binding within −6 kb upstream of transcription starting sites (TSS). Our results confirmed that Ctip2 was recruited within −1 kb promoter region of Lass2, Gba2 and eLox3, as well as between 5–6 kb promoter region of eLox3 relative to TSS (Figure 4a–c). However, we did not observe the direct recruitment of Ctip2 to the promoter regions of several other lipid-metabolizing genes (Figure S4a–k). Our results indicate that Lass2, Gba2 and eLox3 are possible direct targets of Ctip2 to regulate skin lipid metabolism. Altogether, results suggest that Ctip2 directly or indirectly regulates expression of a subset of lipid metabolizing genes and control epidermal sphingolipid biosynthesis during skin organogenesis.
Figure 4. ChIP analyses on murine keratinocytes for lipid metabolizing genes.
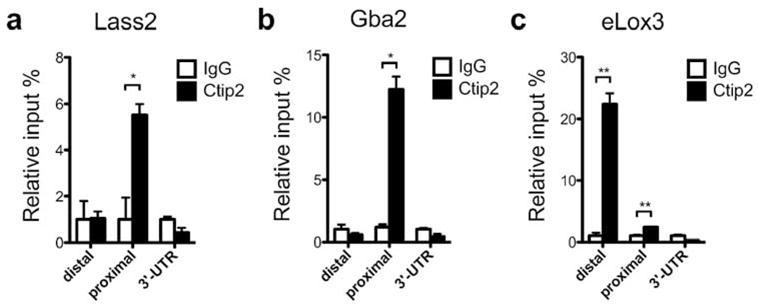
Chromatin immunoprecipitation (ChIP) assay was performed on freshly isolated neonatal mouse epidermal keratinocytes using anti-Ctip2 antibody and results were analyzed by qPCR using primers indicated in table S2. Rat IgG was used as the control. Ctip2 was recruited to the promoter (relative to transcription start site) regions of (a) Lass2, (b) Gba2 and (c) eLox3. Statistical analyses were performed by student’s unpaired t-test using Graphpad Prism software. *p<0.05; **p<0.005. The results represent three separate experiments performed in triplicate.
DISCUSSION
The establishment of an effective physical barrier protecting the living organism from its surrounding environment is the key function of mammalian skin (Hoffjan and Stemmler, 2007). The transport of most substances across the stratum corneum (SC) takes place through the lipid bilayer, suggesting its essential role for the epidermal barrier function (Jungersted et al., 2008). The major SC lipids, ceramides, cholesterol and fatty acids have distinct roles in maintaining normal barrier functions (Elias and Menon, 1991; Ishikawa et al., 2010). Ceramides as well as cholesterol were shown to have an influence on (and are influenced by) the integrity of the SC, while free fatty acids play a major role in the lipid bilayer formation and pH maintenance (Baroni et al., 2012). We have identified Ctip2 as a key regulator of skin lipid metabolism, which plays an important role in establishment of epidermal permeability barrier (EPB) during development. Ablation of Ctip2 leads to altered lipid composition in the developing mouse epidermis, such as reduced ceramide at E17.5-P0, as well as increased sphingomyelins at E16.5–E18.5. Ctip2 regulate sphingolipid homeostasis in mouse embryonic skin by modulating the expression of several key enzymes involved in lipid metabolism. Here we demonstrate that transcription factor Ctip2 is recruited to the promoter regions of a subset of genes (elox3, Lass2 and Gba2) involved in the sphingolipid biosynthesis pathways and could directly regulate their expression (Figure S5).
Skin barrier defects are caused not only by altered levels of protein(s) involved in keratinocytes terminal differentiation, cross-linking of cornified envelops (CEs), formation of intercellular junctions, but also by impaired formation and maintenance of skin lipid barrier (Furuse et al., 2002; Kirschner and Brandner, 2012). Previous studies have shown that Ctip2 regulate keratinocyte proliferation and late terminal differentiation in a cell-autonomous manner; the ultrastructural analyses of the epidermis have revealed defects in lipid disc formation, loosely packed corneocytes, as well as disorganized intercellular lamellar body membranes in Ctip2 null mice (Golonzhka et al., 2008 and unpublished data). In this study, we have focused on the role of Ctip2 in controlling epidermal lipid barrier by modulating lipid metabolism. Herein, we have applied a mass spectrometry method to systemically detect and profile ceramides, sphingomyelins, sphingosines and sphinganines simultaneously in murine epidermis. As a crucial member of stratum corneum lipids, there are few reports on profiling of skin sphingomyelins. Our present data confirm deregulated levels of both long chain and very long chain saturated and unsaturated ceramides in the mutant epidermis at different stages of skin development. Along the same line, overall levels of long chain and very long chain saturated and unsaturated sphingomyelins were significantly enhanced in the mutants at all stages analyzed. In the present study, we have specifically looked at ceramide [NS] containing nonhydroxy fatty acids (N) and sphingosines (S), but were unable to perform mass-spectrometry studies on other subclasses containing (N) and phytosphingosines (P)-[NP]; (N) and 6-hydroxy sphingosines (H)-[NH]; hydroxy fatty acids (A) and sphingosines (S)-[AS]; [AP]; [AH]; ester-linked fatty acids (E), hydroxy fatty acids (O) and (S)-[EOS]; [EOP] and [EOH], due to the lack of available deuterated ceramide internal standards for each subclasses. Additional analyses for the different subclasses will be performed in future using specific internal standard for each subclass.
Our results indicate that altered expression(s) of ceramide synthases 1-6 (Lass 1-6), sphingomyelinases (Smpd1 and Smpd2), glucosylceramide synthase (Ugcg) and that of acid beta-glucosidase (Gba2) at different developmental stages of Ctip2 null skin contribute to an overall decrease in the epidermal ceramide levels in the mutant embryonic skin. It was also shown that several ceramidases involved in ceramide metabolism (Asah2 and Acer1) are reduced in Ctip2−/− skin. As ceramide biosynthesis is a complex process regulated by multiple pathways, it is still unclear that which pathway contributes most to the reduction of epidermal ceramide content in ablation of Ctip2. Lass 1-6 have been reported to be involved in UVB-induced apoptosis and their expression was increased after UVB irradiation in normal human keratinocytes, but reduced in aged skin (Uchida et al., 2003; Jensen et al., 2005). A recent study has reported that Lass3 mutant mice die after birth due to increased skin barrier defects, characterized by increased transepidermal water loss (TEWL), loss of ultra-long chain ceramides as well as non-functional cornified envelopes (Jennemann et al., 2011). Upregulation of Sgms2 and reduced expression of Smpd1 and Smpd2 in Ctip2−/− mice may partially account for increased amount of sphingomyelins observed at later stages in the mutant skin (Figure 1j–k). Furthermore, loss of Gba2, whose expression is altered in the mutant epidermis, has been reported to lead to barrier defects in Gaucher’s disease patients (Holleran et al., 1994). Similarly, eLox3 as well as its substrate 12R-lipoxygenase (12R-LOX) are involved in the formation of oxidized ceramides in mouse epidermis; 12R-LOX deficient mice die shortly after birth due to severe barrier abnormalities with absence of protein-bound EOS ceramide (Zheng et al., 2011). Interestingly, we have reported earlier that eLox3 expression is downregulated in the Ctip2−/− embryonic skin (Golonzhka et al., 2008). Here we have furthered our studies and have shown that Ctip2 could directly regulate the expression of eLox3, Gba2 and Lass2 in mouse embryonic skin by recruitment to their promoter regions. Also, the catabolism and metabolism of sphingosines were both downregulated in Ctip2−/− epidermis, which may explain the consistency of its amount between wildtype and null mice epidermis. Altogether, we have identified Ctip2 as a key regulator of several lipid metabolizing genes and hence epidermal sphingolipid biosynthesis during skin development.
Sphingolipids are key components in skin barrier homeostasis, as well as in other cellular processes such as cell cycle arrest, apoptosis and stress responses (Hannun, 1996; Ogretmen and Hannun, 2004). Ceramide is a central metabolite, whose biosynthesis is regulated at multiple levels with spatial separation of enzymes involved in ceramide formation (Futerman and Riezman, 2005; Kitatani et al., 2008). It remains unclear that if alteration of the enzymes in one of the pathways could have an effect on other pathways, and finally modulate the biological functions. It would be interesting to systemically compare the lipid composition in healthy subjects vs patients with skin disorders such as atopic dermatitis and psoriasis. It is possible that besides Ctip2, other regulatory pathways are also involved in controlling ceramide biosynthesis. B-cell lymphoma/leukemia 11A (Bcl11a/Ctip1) protein is a homolog of Ctip2, which was found to be upregulated in Ctip2 null mice at E18.5 (Golonzhka et al., 2008). The role of Ctip1 in epidermal barrier development and maintenance is still unclear. Further investigation about the expression pattern and distribution of Ctip1, as well as its role in lipid barrier homeostasis in mouse epidermis will be performed utilizing Ctip1−/− mice.
MATERIALS AND METHODS
Mice
Floxed Citp2 allele was generated by flanking exon 4 of Ctip2 locus, which encodes for three-fourth of the open reading frame (ORF) (Golonzhka et al., 2008). Ctip2 null (Ctip2−/−) mice were generated by breeding mice containing floxed Ctip2 allele with cytomegalovirus (CMV) promoter driven Cre-recombinase transgenic mice (Golonzhka et al., 2008). OSU IACUC approval was obtained for animal experiments.
Lipid extraction
Fetus from E16.5 to E18.5 and neonatal mice were collected and genotyped (Golonzhka et al., 2008). A minimum of three wildtype and Ctip2−/− fetuses were taken from the same litter and compared between littermates. A minimum of three litters were analyzed from each developmental stages. Skin was isolated, weighed and incubated overnight in CnT-07 medium (CELLnTEC, Bern, Switzerland) containing 1 mg ml−1 dispase (Gibco, Grand Island, NY) at 4 °C to dissociate epidermis.
Raw skin lipid was extracted using Bligh and Dyer method (Folch et al., 1957; Bligh and Dyer, 1959). Briefly, skin epidermis was incubated in extraction solvent [chloroform: methanol: water (1:2:0.8), 10–15mg tissue/1ml extraction solvent] overnight at room temperature. Fresh extraction solvent was added to epidermis samples the second day and old extraction solvent was saved. Samples were sonicated for 5 cycles, 30 seconds each, and the suspension was shaked for 30min in room temperature. After centrifuged at 2000rpm for 10min, the pellets were discarded and the extraction solvent was combined. Chloroform and water were added to extraction solvent in a ratio of extraction solvent: chloroform: water (7.6: 2: 2). After mixing and centrifuging, upper phase was discarded and the lower phase (chloroform) was washed twice with chloroform: methanol: water (1: 1: 0.9). Lower phase was dried in nitrogen, weighed and saved in −20°C.
LC-MS/MS
Raw skin lipid was further purified as in previous reports (Merrilljr et al., 2005; Monette et al., 2011). 10μl sphingolipid internal standard mixture (Avanti, Alabaster, Alabama) and 1mg raw lipid were added to 1.5ml of chloroform: methanol (1:2) and sonicated in the bath sonicator for 30 seconds. After 2 hours of incubation at 48 °C, the samples were cooled on ice. Samples were then incubated for 1 hour at 37°C with 75μl 1M KOH in methanol to remove the free fatty acids and triacylglycerol, and cooled on ice. 12μl of glacial acetic acid was added to neutralize the pH. Phase separation was achieved by adding 2ml chloroform and 4ml water and samples were gently vortexed. After centrifugation for 15 min at 2000rcf, the chloroform layer was aspirated and dried under nitrogen. The samples were reconstituted in 200μl chloroform: methanol (3:1) and diluted 1:4 with acetonitrile: methanol: acetic acid (97:2:1) with 5mM ammonium acetate.
LC-tandem mass spectrometry was performed and relative quantification was based on skin weight and sphingolipid standards (Monette et al., 2011). In brief, lipids were separated by HPLC using a Supelco Discovery column (2mm × 50mm; Sigma-Aldrich) with 300μl/min flow rate. Mobile phase A: methanol: water (60:40) and mobile phase B: methanol: chloroform (60:40) both contained 0.2% (v/v) formic acid and 10 mM ammonium acetate. The column was pre-equilibrated at 100% phase A and 5μl sample was injected; 100% mobile phase A was maintained for 1min, followed by a linear increase to 40% mobile phase B in a 7 min period; followed by a linear increase to 70% mobile phase B in next 6 min, which was maintained for the remainder (6min) of the 20 min run.
Sample detection was performed using a triple-quadrupole mass spectrometer operated in positive mode (Applied Biosystems/MDS Sciex, API300) with multiple reaction monitoring. The results represent three separate experiments performed in triplicate.
UPLC-MS/MS
Ultra high pressure liquid chromatography was performed on a Shimadzu Nexera system (Shimadzu, Columbia, MD) coupled to a hybrid quadrupole-time of flight mass spectrometer (TripleTOF™ 5600, AB SCIEX) with raw skin lipids. Chromatographic separations were carried out on a Brownlee Analytical C18 column (50 × 2.1 mm, 1.9 μm; PerkinElmer, Boston, MA). The flow rate was 0.35 mL/min and mobile phases consisted of water (A) and acetonitrile (B), both with 0.1% formic acid. Elution gradient was as follows: 0 min, 25% B; 2 min, 55% B; 7 min, 100 % B; 9 min, 100% B. The column was equilibrated 3 minutes prior to each injection. Column temperature was held at 40 °C and the injection volume was 3 μL.
Mass spectrometry was performed on an AB SCIEX TripleTOF™ 5600 equipped with an electrospray ionization source. The instrument was operated in information dependent MS/MS acquisition mode with the collision energy set at 30 V and a collision energy spread of 10 V. TOF MS acquisition time was 0.15 seconds and MS/MS acquisition time was 0.10 seconds. Scan range was 70–1000 m/z for TOF MS and 40–1000 m/z for MS/MS. Ion source gas 1 and 2 and curtain gas (all nitrogen) were set at 50 and 40 and 25, respectively. Source temperature was set at 500 °C and IonSpray voltage at 5.5/−5.5 kV. A set of standards was run periodically throughout the batch to assess system variance and stability. The instrument was automatically calibrated every five runs. Stearic acid, palmitic acid, cholesterol, and cholesterol-3-sulfate were identified using authentic standards with accurate mass measurements, isotope distribution, MS/MS fragmentation, and retention time. The results represent three separate experiments performed in triplicate.
Absolute quantification
Absolute quantification of epidermal ceramides, sphingomyelin, sphingosine and sphinganine was based on comparison to a synthetic sphingolipid standard mixture as previously described (Monette et al., 2010). In brief, 10 synthetic compounds were obtained (Avanti): sphinganine C18 (860498P), sphingosine C18 (860490P), ceramide C16:0 (860516P), ceramide C22:0 (860510P), ceramide C18:1 (860519C), ceramide C24:1 (860525C), sphingomyelin C18:0 (860586P), sphingomyelin C18:1 (860587C), sphingomyelin C24:0 (860592P) and sphingomyelin C24:1 (860593P). Standard solutions were prepared, and the amount of analyte was determined based on comparison of the peak area of the analyte with that of the sphingolipid. The results represent three separate experiments performed in triplicate.
qRT-PCR
RNA was extracted and cDNA was synthesized from skin of mice fetuses and neonates (Indra et al., 2005b; Wang et al., 2011). Real-time PCR was performed on an ABI 7500 Real-Time PCR system using SYBR green methodology (Indra et al., 2005a, 2005b). Relative gene expression analysis of the qRT-PCR data was performed using Gapdh as an internal control, and delta-delta CT method was used for quantification as previously described (Wang et al., 2011). All qRT-PCR primers are indicated in Table S1. The results represent three separate experiments performed in triplicate.
Western blotting
Protein was extracted from mouse epidermis and western blotting was performed due to previous reports using specific antibodies against Lass2 (1:500), Gba2 (1:1000) and eLox3 (1:2000) (Wang et al., 2011; Santa Cruz Biotechnology, Santa Cruz, CA). Western blotting represents the results from three separate experiments performed in triplicate.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed according to Hyter et al., 2010. ChIP-DNA was amplified using ABI-7500 Real-time PCR machine with specific primers indicated in table S2. The results represent three separate experiments performed in triplicate.
Statistics
Statistical significance of differences between groups was analyzed using GraphPad Prism software using student’s unpaired t-test. All statistical analyses were performed in a double-blinded manner.
Supplementary Material
Acknowledgments
We would like to thank Dr. Taifo Mahmud, Dr. Phil Proteau and Dong Li for their help in sample preparation, and Xiaobo Liang for mouse colony maintenance. We also would like to thank Dr. Walter Vogel from College of Pharmacy, the OSU Mass Spectrometry Facility, and the Oxidative/Nitrative Stress Core Laboratory of Linus Pauling Institute for technical support in mass spectrometry analyses. We acknowledge Dr. Tory Hagen and Liam Finlay for providing two of the ceramide standards and their advice in standard solution preparation. These studies were supported by grant AR056008-03 (AI) from National Institute of Health, OHSU Medical Research Foundation grant to AI and NIEHS center grant (ES00210) to Environmental Health Sciences Center, Oregon State University. We also thank Drs. Mark Zabriskie and Gary DeLander of College of Pharmacy, Oregon State University for continuous support and encouragement.
Abbreviations
- Ctip2
chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2
- TEWL
transepidermal water loss
- Lass1-Lass6
ceramide synthases 1-6
- Sptlc1-3
palmitoyltransferase 1-3
- Degs1
dihydroceramide desaturase 1
- Kdsr
3-ketodihydrosphingosine reductase
- Smpd1 and 2
sphingomyelinase 1 and 2
- Sgms1 and 2
sphingomyelin synthase 1 and 2
- Ugcg
glucosylceramide synthase
- Gba2
acid beta-glucosidase
- Asah1 and 2
acylsphingosine amidohydrolases 1 and 2
- Acer1-Acer3
alkaline ceramidase 1-3
- Sphk1 and 2
sphingosine kinases 1 and 2
- eLox3
-
epidermis-type lipoxygenase 3
The following ceramide code is used according to previous reports (Motta et al, 1993; Robson et al, 1994)
- Ceramide EOS (or ceramide 1)
ceramide containing ester-linked fatty acids (E), -hydroxy fatty acids (O), and sphingosines (S)
- Ceramide NS (or ceramide 2)
ceramide containing nonhydroxy fatty acids (N) and sphingosines (S)
- Ceramide NP (or ceramide 3)
ceramide containing nonhydroxy fatty acids (N) and phytosphingosines (P)
- Ceramide EOH (or ceramide 4)
ceramide containing ester-linked fatty acids (E), -hydroxy fatty acids (O), and 6-hydroxy sphingosines (H)
- Ceramide AS (or ceramide 5)
ceramide containing -hydroxy fatty acids (A) and sphingosines (S)
- Ceramide AP (or ceramide 6)
ceramide containing -hydroxy fatty acids (A) and phytosphingosines (P)
- Ceramide AH (or ceramide 7)
ceramide containing -hydroxy fatty acids (A) and 6-hydroxy sphingosines (H)
- Ceramide NH
ceramide containing nonhydroxy fatty acids (N) and 6-hydroxy sphingosines (H)
- Ceramide EOP (or ceramide 9)
ceramide containing ester-linked fatty acids (E), -hydroxy fatty acids (O), and phytosphingosines (P)
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
References
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem Apr. 2000;275:10315–22. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–63. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KP. Selective Inhibition of Juxtanuclear Translocation of Protein Kinase C II by a Negative Feedback Mechanism Involving Ceramide Formed from the Salvage Pathway. Journal of Biological Chemistry. 2004;280:2606–12. doi: 10.1074/jbc.M409066200. [DOI] [PubMed] [Google Scholar]
- Bernardo K, Hurwitz R, Zenk T, Desnick RJ, Ferlinz K, Schuchman EH, et al. Purification, characterization, and biosynthesis of human acid ceramidase. J Biol Chem. 1995;270:11098–102. doi: 10.1074/jbc.270.19.11098. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Elias PM. The epidermal permeability barrier: from the early days at Harvard to emerging concepts. J Invest Dermatol. 2004;122:xxxvi–xxxix. doi: 10.1046/j.0022-202X.2004.22233.x. [DOI] [PubMed] [Google Scholar]
- Elias PM, Menon GK. Structural and lipid biochemical correlates of the epidermal permeability barrier. Adv Lipid Res. 1991;24:1–26. doi: 10.1016/b978-0-12-024924-4.50005-5. [DOI] [PubMed] [Google Scholar]
- Feingold KR. Thematic review series: skin lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. J Lipid Res. 2007;48:2531–46. doi: 10.1194/jlr.R700013-JLR200. [DOI] [PubMed] [Google Scholar]
- Feingold KR. The outer frontier: the importance of lipid metabolism in the skin. J Lipid Res. 2009;50(Suppl):S417–422. doi: 10.1194/jlr.R800039-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold KR. Lipid metabolism in the epidermis. Dermatoendocrinol. 2011;2:52. doi: 10.4161/derm.3.2.15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- Golonzhka O, Liang X, Messaddeq N, Bornert J-M, Campbell AL, Metzger D, et al. Dual Role of COUP-TF-Interacting Protein 2 in Epidermal Homeostasis and Permeability Barrier Formation. J Investig Dermatol. 2008;129:1459–70. doi: 10.1038/jid.2008.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran WM, Takagi Y, Uchida Y. Epidermal sphingolipids: metabolism, function, and roles in skin disorders. FEBS Lett. 2006;580:5456–66. doi: 10.1016/j.febslet.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Hyter S, Bajaj G, Liang X, Barbacid M, Ganguli-Indra G, Indra AK. Loss of nuclear receptor RXRα in epidermal keratinocytes promotes the formation of Cdk4-activated invasive melanomas. Pigment Cell Melanoma Res. 2010;23:635–48. doi: 10.1111/j.1755-148X.2010.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imokawa G. A possible mechanism underlying the ceramide deficiency in atopic dermatitis: Expression of a deacylase enzyme that cleaves the N-acyl linkage of sphingomyelin and glucosylceramide. Journal of Dermatological Science. 2009;55:1–9. doi: 10.1016/j.jdermsci.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, et al. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005a;132:4533–44. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- Indra AK, Mohan WS, Frontini M, Scheer E, Messaddeq N, Metzger D, et al. TAF10 is required for the establishment of skin barrier function in foetal, but not in adult mouse epidermis. Dev Biol. 2005b;285:28–37. doi: 10.1016/j.ydbio.2005.05.043. [DOI] [PubMed] [Google Scholar]
- Ishikawa J, Narita H, Kondo N, Hotta M, Takagi Y, Masukawa Y, et al. Changes in the Ceramide Profile of Atopic Dermatitis Patients. Journal of Investigative Dermatology. 2010;130:2511–4. doi: 10.1038/jid.2010.161. [DOI] [PubMed] [Google Scholar]
- Janssens M, van Smeden J, Gooris GS, Bras W, Portale G, Caspers PJ, et al. Lamellar Lipid Organization and Ceramide Composition in the Stratum Corneum of Patients with Atopic Eczema. Journal of Investigative Dermatology. 2011;131:2136–8. doi: 10.1038/jid.2011.175. [DOI] [PubMed] [Google Scholar]
- Kitatani K, Idkowiak-Baldys J, Hannun YA. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal. 2008;20:1010–8. doi: 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J, Gärtner S, Li CM, Quintern LE, Bernardo K, Levran O, et al. Molecular cloning and characterization of a full-length complementary DNA encoding human acid ceramidase. Identification Of the first molecular lesion causing Farber disease. J Biol Chem. 1996;271:33110–5. doi: 10.1074/jbc.271.51.33110. [DOI] [PubMed] [Google Scholar]
- Li CM, Hong SB, Kopal G, He X, Linke T, Hou WS, et al. Cloning and characterization of the full-length cDNA and genomic sequences encoding murine acid ceramidase. Genomics. 1998;50:267–74. doi: 10.1006/geno.1998.5334. [DOI] [PubMed] [Google Scholar]
- Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2008;1781:424–34. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesini N, Hannun YA. Acid and neutral sphingomyelinases: roles and mechanisms of regulation. Biochem Cell Biol. 2004;82:27–44. doi: 10.1139/o03-091. [DOI] [PubMed] [Google Scholar]
- Masukawa Y, Narita H, Shimizu E, Kondo N, Sugai Y, Oba T, et al. Characterization of overall ceramide species in human stratum corneum. The Journal of Lipid Research. 2008;49:1466–76. doi: 10.1194/jlr.M800014-JLR200. [DOI] [PubMed] [Google Scholar]
- Merrill AH. De Novo Sphingolipid Biosynthesis: A Necessary, but Dangerous, Pathway. Journal of Biological Chemistry. 2002;277:25843–6. doi: 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- Merrilljr A, Sullards M, Allegood J, Kelly S, Wang E. Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–24. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Mizutani Y, Mitsutake S, Tsuji K, Kihara A, Igarashi Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie. 2009;91:784–90. doi: 10.1016/j.biochi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Monette JS, Gómez LA, Moreau RF, Bemer BA, Taylor AW, Hagen TM. Characteristics of the rat cardiac sphingolipid pool in two mitochondrial subpopulations. Biochemical and Biophysical Research Communications. 2010;398:272–7. doi: 10.1016/j.bbrc.2010.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monette JS, Gómez LA, Moreau RF, Dunn KC, Butler JA, Finlay LA, et al. (R)-α-Lipoic acid treatment restores ceramide balance in aging rat cardiac mitochondria. Pharmacological Research. 2011;63:23–9. doi: 10.1016/j.phrs.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta S, Monti M, Sesana S, Caputo R, Carelli S, Ghidoni R. Ceramide composition of the psoriatic scale. Biochim Biophys Acta. 1993;S1182:147–51. doi: 10.1016/0925-4439(93)90135-n. [DOI] [PubMed] [Google Scholar]
- Nemes Z, Demény M, Marekov LN, Fésüs L, Steinert PM. Cholesterol 3-sulfate interferes with cornified envelope assembly by diverting transglutaminase 1 activity from the formation of cross-links and esters to the hydrolysis of glutamine. J Biol Chem. 2000;275(4):2636–46. doi: 10.1074/jbc.275.4.2636. [DOI] [PubMed] [Google Scholar]
- Nemes Z, Steinert PM. Bricks and mortar of the epidermal barrier. Exp Mol Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- Oda Y, Uchida Y, Moradian S, Crumrine D, Elias PM, Bikle DD. Vitamin D Receptor and Coactivators SRC2 and 3 Regulate Epidermis-Specific Sphingolipid Production and Permeability Barrier Formation. J Investig Dermatol. 2008;129:1367–78. doi: 10.1038/jid.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B. Biochemical Mechanisms of the Generation of Endogenous Long Chain Ceramide in Response to Exogenous Short Chain Ceramide in the A549 Human Lung Adenocarcinoma Cell Line. ROLE FOR ENDOGENOUS CERAMIDE IN MEDIATING THE ACTION OF EXOGENOUS CERAMIDE. Journal of Biological Chemistry. 2002;277:12960–9. doi: 10.1074/jbc.M110699200. [DOI] [PubMed] [Google Scholar]
- Park H, Haynes CA, Nairn AV, Kulik M, Dalton S, Moremen K, et al. Transcript profiling and lipidomic analysis of ceramide subspecies in mouse embryonic stem cells and embryoid bodies. The Journal of Lipid Research. 2009;51:480–9. doi: 10.1194/jlr.M000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson KJ, Stewart ME, Michelsen S, Lazo ND, Downing DT. 6-Hydroxy-4-sphingenine in human epidermal ceramides. J Lipid Res. 1994;35:2060–8. [PubMed] [Google Scholar]
- Schmuth M, Man MQ, Weber F, Gao W, Feingold KR, Fritsch P, et al. Permeability barrier disorder in Niemann-Pick disease: sphingomyelin-ceramide processing required for normal barrier homeostasis. J Invest Dermato. 2000;l115:459–66. doi: 10.1046/j.1523-1747.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- van Smeden J, Hoppel L, van der Heijden R, Hankemeier T, Vreeken RJ, Bouwstra JA. LC/MS analysis of stratum corneum lipids: ceramide profiling and discovery. J Lipid Res. 2011;52:1211–21. doi: 10.1194/jlr.M014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan I, Senkal CE, Ponnusamy S, Bielawski J, Szulc Z, Bielawska A, et al. Regulation of the sphingosine-recycling pathway for ceramide generation by oxidative stress, and its role in controlling c-Myc/Max function. Biochemical Journal. 2006;393:513. doi: 10.1042/BJ20051083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani M, Okino N, Mori K, Tanigawa T, Izu H, Ito M. Molecular cloning of the full-length cDNA encoding mouse neutral ceramidase. A novel but highly conserved gene family of neutral/alkaline ceramidases. J Biol Chem. 2000;275:11229–34. doi: 10.1074/jbc.275.15.11229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.