Abstract
Glutamate (Glu) is a major excitatory neurotransmitter in brain and has been shown to decrease in the early stages of Alzheimer’s disease (AD). Using glutamate amine exchange saturation transfer (GluCEST) method, we imaged the change in [Glu] in APP-PS1 transgenic mouse model of AD at high spatial resolution. Compared to wild-type controls, AD mice exhibited notable reduction of GluCEST contrast (~30%) in all areas of the brain. The change in [Glu] is further validated through proton magnetic resonance spectroscopy (1HMRS). A positive correlation was observed between GluCEST contrast and 1HMRS measured Glu/total creatine (Glu/tCr) ratio. This method potentially provides a novel noninvasive biomarker for diagnosing the disease in preclinical stages and enables the development of disease modifying therapies for AD.
Keywords: Alzheimer’s disease, Brain, Glutamate, Magnetic Resonance Spectroscopy, Magnetic Resonance Imaging, Chemical Exchange Saturation Transfer Imaging
Introduction
Glutamate (Glu) is the major free amino acid present in the brain and functions as an excitatory neurotransmitter. The central role of Glu in learning, memory and cognition is well reported (1,2) and has been shown to decrease in Alzheimer’s disease (AD) pathology (3-8). The biochemical changes in AD are thought to occur earlier than structural changes. Detection of the disease in its incipient stages enhances the accuracy of diagnosis, and accelerates research in development of drugs that could modify disease progression leading to the delay or complete prevention of clinical symptoms (9-11). According to a recent review (12), a delay in onset of AD by 5 years could translate into a 50% reduction in the disease prevalence and delay by 10 years may virtually eliminate the disease. Another study estimated that delaying the clinical onset of the disease by 1 year would reduce the prevalence in 2050 by 9 million cases (13). These numbers clearly underscore the significance of the early detection of the disease.
Magnetic resonance imaging (MRI) and proton MR spectroscopy (1HMRS) are widely used in studying the structural and biochemical changes in AD brain during disease progression (3-8,14,15). Previous 1HMRS studies on human brain have depicted progressive decrease in hippocampal Glu concentration from cognitive control to mild cognitive impairment (MCI) to full blown AD (3,4,8). Although, 1HMRS has been widely used to monitor the Glu changes in AD it suffers from poor spatial resolution and long acquisition time. Positron emission tomography (PET) studies have measured the distribution of the Glu receptor; a noncompetitive and highly selective antagonist for the metabotropic Glu receptor subtype 5, 3-(6-Methyl-pyridin-2-ylethynyl)-cyclohex-2-enone-O-11C-methyloxime (11C-ABP688) was evaluated for its potential as a PET agent (16). Despite their high specificity to Glu receptors, the major shortcomings of PET is radiation exposure, low resolution, imaging logistics due to short half-lives of radio ligands, and limited applicability to functional studies. These findings underscore the importance of noninvasive and high resolution imaging methods for mapping Glu in AD.
Recently, we have demonstrated the feasibility of mapping Glu in healthy brain using the technique known as chemical exchange saturation transfer (CEST) by exploiting its amine proton exchange with the bulk water (GluCEST) (17). Briefly, in GluCEST, the exchangeable amine protons of a Glu can be selectively irradiated by application of the radio-frequency (RF) pulse and their saturated magnetization exchange with the bulk water leads to reduction in the bulk water signal in a concentration dependent manner. Due to the inherent nature of the GluCEST technique, it offers about two orders of magnitude higher sensitivity compared to 1HMRS (17). Previously, the CEST technique has been used to map pH (18), protein and peptide in brain tumors (19), glycogen in liver (20), proteoglycan in knee cartilage (21) and myo-inositol (mIns) in brain (22).
In previous study, it has been shown that the GluCEST is a large fraction of the MTRasymmetry signal at 3ppm with the relative amount depending on the saturation power used (17). Since GluCEST is a large part of the MTRasymmetry at 3ppm it offers a possibility to see changes in [Glu]. For the consistency, hereafter, the MTRassymetry at 3ppm is referred as GluCEST.
In the current study, for the first time, we report the mapping of the Glu distribution in the brain of APP-PS1 transgenic mouse model of AD as well as age matched wild type (WT) mice at high resolution by utilizing GluCEST technique. The results also demonstrate a strong correlation between Glu changes measured via GluCEST and 1HMRS. Finally, the potential implication of the GluCEST in detecting early AD pathology is discussed in detail.
Materials and Methods
Animal Preparation
Six APP-PS1 mice and six WT mice age spanning from 18 to 20 months were used in this study (from Wyeth Research) (table 1). Mice were transferred to a 9.4T horizontal bore small animal MR scanner (Varian, Palo Alto, CA) and placed in a 20-mm diameter commercial quadrature proton coil (m2m Imaging Corp., Cleveland, OH). Animals were kept under anesthesia (1.5% isoflurane in 1 liters/min oxygen) and their body temperature maintained with the air generated and blowing through a heater (SA Instruments, Inc., Stony Brook, NY). Respiration and body temperature were continuously monitored using a MRI compatible small animal monitor system (SA Instruments, Inc., Stony Brook, NY). The Institutional Animal Care and Use Committees of the University of Pennsylvania approved experimental protocols.
Table 1.
Describes the age, GluCEST contrast, Glu/tCr ratio for both wild type and APP-PS1 mice. The Cramer-Rao Lower Bound (CRLB) values for the Glu fitting were less than 20%.
| Wild Type (n=6) | APP-PS1 (n=6) | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Age (months) |
GluCEST | Glu/tCr | CRLB | Age (months) |
GluCEST | Glu/tCr | CRLB |
| 18 | 27.326 | 1.700 | 9 | 18 | 20.793 | 1.174 | 7 |
| 20 | 23.945 | 1.527 | 5 | 20 | 17.567 | 1.024 | 9 |
| 18 | 28.766 | 1.757 | 9 | 20 | 17.023 | 1.020 | 12 |
| 19 | 25.698 | 1.366 | 8 | 19 | 18.059 | 1.128 | 8 |
| 19 | 26.612 | 1.573 | 17 | 19 | 19.203 | 1.192 | 7 |
| 19.5 | 25.998 | 1.546 | 13 | 18 | 21.871 | 1.199 | 16 |
Imaging Protocols
GluCEST imaging of the mouse brain was performed using a custom-programmed segmented RF spoiled gradient echo (GRE) readout pulse sequence with a frequency selective continuous wave saturation preparation pulse. The sequence parameters were: field of view =20×20 mm2, slice thickness = 2 mm, flip angle=15 degree, GRE readout TR=6.2 ms (128 segments), TE =2.9 ms, matrix size=128×128. For every 8 s one saturation pulse and 128 acquisition segments were applied. CEST images were collected using a 1 second saturation pulse at peak B1 of 250 Hz and frequencies ranging ±5 ppm from bulk water in step size of 0.2 ppm. B1 and B0 field maps were also acquired and used to correct the GluCEST contrast as described previously (17). CEST imaging was performed on two different brain slices. For magnetization transfer ratio (MTR) mapping, same brain slices were imaged at 20 ppm with 250 Hz saturation power and 1s saturation duration. Image at 100 ppm was also collected and considered as magnetization off image. The total imaging time was around 30 min.
1H Magnetic Resonance Spectroscopy
Single voxel spectra (SVS) were performed with stimulated echo acquisition mode (STEAM) using a vendor (Varian) provided pulse sequence with the following parameters: voxel size = 3.5 mm × 3.5 mm × 2 mm (Voxel volume 24.5 μL), spectral width = 4 kHz, number of points = 4006, averages = 264, TE = 8 ms, Tm = 7 ms, and TR = 5 s. Water suppression was achieved using the variable pulse power and optimized relaxation delays method (VAPOR). Localized shimming was performed to obtain localized water line width values of 0.075 ppm or less. Unsuppressed water spectrum was also acquired using the same parameter for the purpose of normalization.
Glu concentration relative to total creatine (tCr) was measured using LC model (23). LC model quantifies data through the recursive fitting of prior knowledge basis sets. A least squares algorithm is used to optimize the fit with each iteration and quality of the final fit is determined in terms of Cramer-Rao Lower Bound (CRLB) a measure of the variance in the error.
Image Processing
All image processing and data analysis were performed using software routines written in MATLAB (version 7.5, R2007b). Acquired images were corrected for B0 and used to generate GluCEST contrast map using Equation [1].
| Eq [1] |
where S−ve and S+ve are the B0 corrected MR signals at -3ppm and +3 ppm respectively. To account for and minimize the contribution from direct saturation and magnetization transfer effects, we used S-ve instead of S0 for normalization (24). GluCEST contrast was further corrected for B1 and mapped as false-colors onto anatomical proton image. z-spectra were obtained from the CEST images by plotting normal image intensity as a function of resonance offset of saturation pulse. The CEST asymmetry curves were also generated by plotting the relative water signal difference at frequency offsets from 0 to 4.8ppm.
Similarly, MTR maps were computed by
Where M0 is the magnetization off and Msat is the magnetization with a saturation pulse applied at 20 ppm.
Results
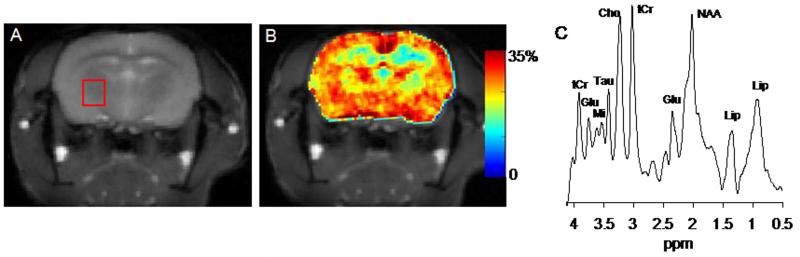
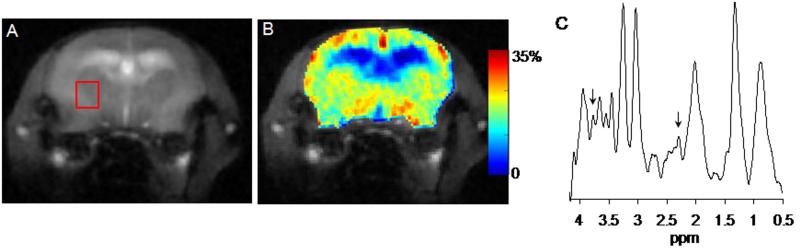
The brain anatomical images and corresponding GluCEST maps and MRS spectra from WT and APP-PS1 mice are shown in figures 1 (A-C) and 2 (A-C). The anatomical proton images display the typical structural changes such as large ventricular dilation in AD mouse (Fig. 2A). The GluCEST maps illustrate the regional distribution of Glu in different regions of brain both in WT and AD mouse (Figs. 1B & 2B). The higher GluCEST contrast was observed in the gray matter compared to the white matter, which is due to the difference in the Glu concentration and is consistent with the previous studies on rat brain and human brain (17). The mean GluCEST contrast from the WT and APP-PS1 mice brain over the chosen voxel was 26.4 ± 1.6 % and 19.1 ± 1.9 %. To further validate the change in GluCEST contrast is due to alter Glu concentration we performed 1HMRS spectra for the voxel shown on the anatomical images (Figs. 1A & 2A). This voxel was placed in an area devoid of cerebral spinal fluid. 1HMRS spectra show decreased Glu peak amplitude in APP-PS1 mice compared to WT mice (arrows, Figs. 1C & 2C). Decreased N-acetyl aspartate (NAA) peak signal amplitude and increased mIns peak signal amplitude can also be seen from 1HMRS spectra, which is characteristic of AD pathology. The mean Glu/tCr in WT and APP-PS1 mice was 1.58±0.13 and 1.12±0.08, respectively. Comparative analysis showed ~28% decreased GluCEST (p<0.01) contrast and ~29% decrease (p<0.01) in Glu/tCr in APP-PS1 mice brain than in that of WT mice over the chosen voxel (table 1).
Figure 1.
GluCEST mapping and 1H MRS of wild type mouse. A. The anatomical brain image from a wild type (WT) mouse. B. The corresponding GluCEST map. The GluCEST map shows the regional distribution of Glu in brain, higher GluCEST contrast is observed in gray matter compared to white matter. C. The 1HMRS spectra from the voxel placed in (A) shows the major brain metabolites. The glutamate peak is labeled at 2.3 and 3.75 ppm respectively. (NAA = N-acetyl aspartate, Glu = glutamate, tCr = total creatine, Cho = choline, Tau = taurine, Mi = Myo-inositol, Lip = lipid)
Figure 2.
GluCEST mapping and 1H MRS of age matched transgenic mouse model of Alzheimer’s disease (APP-PS1). The anatomical brain image and corresponding GluCEST map are shown in figures A and B. Ventricular dilation is characteristic of AD can be clearly seen in anatomical Image (A). Figure C shows the 1HMRS spectra for the voxel placed in (A). The GluCEST map shows the decreased GluCEST contrast along with concomitant decrease in glutamate as shown in spectra (C, arrows) compared to WT.
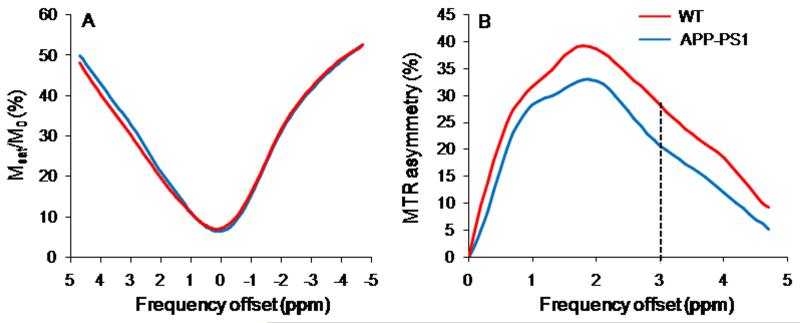
The z-spectra and MTRasymmetry curve obtained from WT and APP-PS1 mouse is shown in figure 3A and 3B. The 3 ppm line on the MTRasymmetry curves corresponds to GluCEST. These curves are rather broad and showed a maximum CEST contrast at ~2ppm which may be due to the broad asymmetry of Glu due to its faster exchange rate as well as chemical shift averaging effect that shifts the line to higher field towards water resonance.
Figure 3.
z-spectra (A) and MTRasymmetry curves (B) from WT and APP-PS1 mice for the voxel shown in figures 1A and 2A. The dotted line at 3ppm in MTRasymmetry curves reflects the GluCEST contrast.
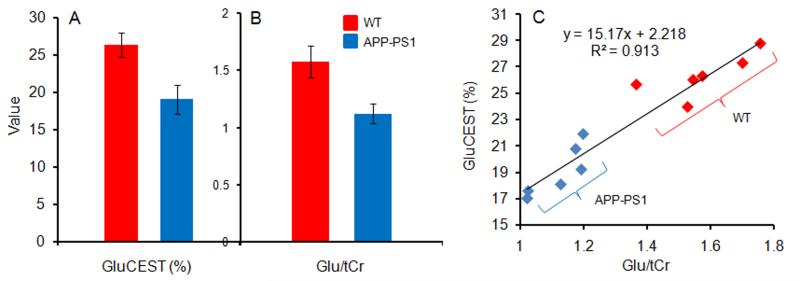
The bar graphs show the average GluCEST contrast (%) and Glu/tCr both in WT and APP-PS1 mice (Figs. 4A & 4B). The plot in figure 4C shows the correlation between Glu/tCr measured from 1HMRS and GluCEST contrast. An excellent positive correlation (R2=0.91) was observed with a slope of ~15% GluCEST per Glu/tCr ratio.
Figure 4.
The bar graphs show the mean GluCEST contrast (%) (A) and Glu/tCr ratio from WT and AD mice for the voxel shown in figures 1A and 2A (B). The graph (C) shows a strong positive correlation between GluCEST and Glu/tCr.
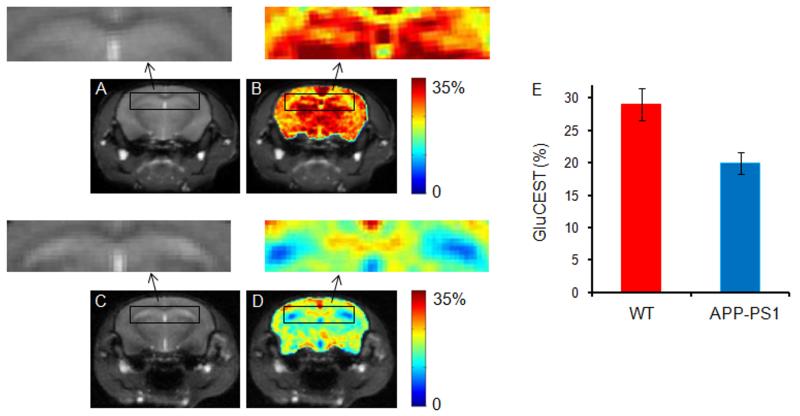
Hippocampus is the primary structure associated with the early loss of the pyramidal neurons and their synapses in AD pathology, which control the learning and cognitive function (25). The GluCEST maps of the brain slice showing hippocampus regions both in WT and AD mice were also obtained (Figs. 5A-E). In AD mice brain, an average of ~31 % decreased GluCEST contrast was found in hippocampus compared to WT.
Figure 5.
Brain GluCEST map across the hippocampus region. A. Anatomical brain image from a wild type (WT) mouse. B. The corresponding GluCEST map. C & D. Anatomical image and corresponding GluCEST map from the age matched AD mouse. Significantly decreased GluCEST contrast in hippocampus was observed in APP-PS1 mice compared to WT mice. The hippocampus regions are shown in rectangular box both in WT and AD mouse (arrows). E. The bar graphs show the mean hippocampus GluCEST contrast in WT and AD mice respectively.
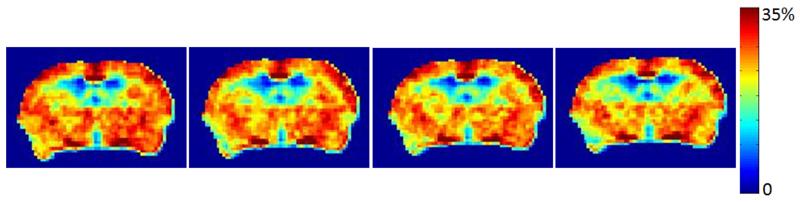
We also performed the reproducibility measurement on two mice which were imaged for GluCEST at different time points (Fig. 6). The GluCEST images were highly reproducible (Fig. 6) with the intra-covariance less than 3%.
Figure 6.
shows the reproducibility of GluCEST mapping in a WT mouse. Less than 3% intra-covariance was found.
The other factor that may contribute to GluCEST is magnetization transfer effect from bound water pool. No significant change in MTR contrast was observed in AD mice (48.3 ± 1.5) compared to WT mice (48.0 ± 1.1).
Discussion
In the current study, we have shown that high resolution imaging of Glu alteration in transgenic mouse model of AD is possible using GluCEST technique. Significantly decreased GluCEST contrast in APP-PS1 mice was observed compared to WT mice and results were further validated through comparable changes of Glu/tCr on 1HMRS.
In this work, we used APP-PS1 mouse model as this model appears to match most closely with the neurochemical and neurological profile in human AD (7). Compared to WT mice, reduced signals from Glu, NAA and increased mIns signals were found in APP-PS1 mice, which is consistent with the previously reported studies (7,26). The above discussion shows that it is indeed possible to track the [Glu] changes in transgenic animal models of AD using 1HMRS. However, using 1HMRS, Glu cannot be imaged at high resolution and thus heterogeneous distribution of Glu cannot be easily monitored. On the other hand, the GluCEST technique maps the distribution of Glu in the brain at high resolution.
Hippocampus is the primary region affected in the early AD pathology and is associated with the learning and memory. In the current study, more profound decrease in GluCEST contrast in the hippocampus was observed compared to other regions which clearly suggests its involvement in the early disease process. In an earlier MRS study, progressive decrease in hippocampus Glu/tCr in APP-PS1 mice was observed with advancing age (7). In addition, they have observed significant (p<0.01) reduction in hippocampus Glu/tCr in APP-PS1 compared to age matched WT mice. In the current study, we found ~31% decreased GluCEST contrast in hippocampus of APP-PS1 mice compared to WT mice, which is consistent with the previously reported study in human AD where ~35% decreased hippocampus [Glu] is observed (4). Same study has also shown 15-20% change in the Glu concentration in MCI compared to control (4). Due to high reproducibility of GluCEST mapping it may feasible to image the change in Glu concentration at high spatial resolution from control to MCI to full onset of AD and may provide a definite diagnostic marker at early stage.
In the current study, strong positive correlation between GluCEST and 1HMRS measured Glu/tCr concentration suggests that the observed change in GluCEST contrast is due to change in [Glu]. It is also worth monitoring the change in Glu level at different age of APP-PS1 mouse. The other factor which may contribute to the GluCEST is the change in MT effect from bound pool of water. However, in the current study, we did not observed any apparent change in the MTR contrast. Moreover, any decrease in MT effect will overestimate the GluCEST contrast from AD pathology.
Since most patients with MCI ultimately develop AD, it is reasonable to regard neuroimaging findings that are sensitive to the changes in the brains of individuals with MCI as markers of early or “prodromal” AD pathologic changes. In human, gradual decrease in Glu concentration has been shown from control to MCI to full onset of AD (3,4). It has been suggested that 1HMRS quantified Glu can be used as a biomarker to detect the early changes in AD pathology at the stage of MCI and may serve as a tool to monitor the therapeutic response. However, due to large voxel size (8 cm3) it is difficult to monitor the regional changes in [Glu]. Mapping of Glu through GluCEST technique potentially facilitate monitoring the regional changes in Glu concentration at high resolution (~200 micron) during disease course and in response to treatment. GluCEST technique also does not require any exogenous contrast administration and is relatively easy to translate into clinical setting.
It is worth of discussing some technical details of the GluCEST method. Precise saturation of exchangeable protons requires perfect B0 homogeneity. B0 inhomogeneity could interfere with the selective saturation frequency and makes the two selective frequencies asymmetric to bulk water. In that case, the CEST effect would reflect differences between direct saturation and magnetization transfer effects. The CEST effect is also highly dependent on saturation B1 amplitude (27). With fixed saturation duration, a higher B1 can induce a higher CEST effect (28), and B1 homogeneity is a critical requirement for quantitative CEST imaging. Artifacts induced by B0 and B1 field inhomogeneity and the corresponding correction algorithms have been previously discussed in detail (29). In the current study, we used methods described previously to correct the B1 and B0 induced artifacts due to small B0 and B1 variations. For proof of principle purposes, in the current study only a single slice with a two dimensional imaging sequence was imaged. However, it would be relatively straightforward to implement a three dimensional acquisition of GluCEST map.
Conclusions
The findings of this preliminary study suggest that using GluCEST it is feasible to obtain in vivo high-resolution maps of altered regional Glu concentration in AD pathology. The large changes of GluCEST observed in this study from fully developed AD model coupled with the previously published MRS results from AD and MCI patients suggest that it has sufficient dynamic range to detect changes from MCI stage of the disease. Given that these molecular changes are associated with incipient stages of the disease, this method has the potential to detect changes before structural alterations in the brain. Further, GluCEST mapping could serve as in vivo surrogate marker for evaluating therapeutic efficacy in trials of therapeutic agents designed to reduce neurotoxicity during early stages of AD. The noninvasive and quantitative aspects of GluCEST mapping potentially could be exploited in enabling the drug discovery efforts in preclinical as well as in clinical models of AD.
Acknowledgements
We thank Dr. John Q. Trojanowski and Dr. Virginia M.Y. Lee for stimulating discussion and support. Our thanks are due to Drs. Weixia Liu and Steve Pickup for their technical assistance in using the 9.4 T research scanners.
Funding
This work was supported by the National Center for Research Resources and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grant Number P41-EB015893.
Abbreviations
- Glu
Glutamate
- AD
Alzheimer’s disease
- MRI
Magnetic resonance imaging
- 1HMRS
Proton MR Spectroscopy
- MCI
Mild cognitive impairment
- CEST
Chemica exchange saturation transfer
- GluCEST
Glutamate CEST
- PET
Positron emission tomography
- RF
radio-frequency
- mIns
Myo-Inositol
- WT
Wild type
- GRE
Gradient echo
- SVS
Single voxel spectra
- STEAM
stimulated echo acquisition mode
- VAPOR
variable pulse power and optimized relaxation
- MTR
magnetization transfer ratio
- NAA
N-acetyl aspartate
References
- 1.Francis PT. Glutamatergic systems in Alzheimer’s disease. Int J Geriatr Psychiatry. 2003;18(Suppl 1):S15–21. doi: 10.1002/gps.934. [DOI] [PubMed] [Google Scholar]
- 2.Francis PT, Sims NR, Procter AW, Bowen DM. Cortical pyramidal neurone loss may cause glutamatergic hypoactivity and cognitive impairment in Alzheimer’s disease: investigative and therapeutic perspectives. J Neurochem. 1993;60(5):1589–1604. doi: 10.1111/j.1471-4159.1993.tb13381.x. [DOI] [PubMed] [Google Scholar]
- 3.Kantarci K, Reynolds G, Petersen RC, Boeve BF, Knopman DS, Edland SD, Smith GE, Ivnik RJ, Tangalos EG, Jack CR., Jr Proton MR spectroscopy in mild cognitive impairment and Alzheimer disease: comparison of 1.5 and 3 T. AJNR Am J Neuroradiol. 2003;24(5):843–849. [PMC free article] [PubMed] [Google Scholar]
- 4.Rupsingh R, Borrie M, Smith M, Wells JL, Bartha R. Reduced hippocampal glutamate in Alzheimer disease. Neurobiology of aging. 2011;32(5):802–810. doi: 10.1016/j.neurobiolaging.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Hattori N, Abe K, Sakoda S, Sawada T. Proton MR spectroscopic study at 3 Tesla on glutamate/glutamine in Alzheimer’s disease. Neuroreport. 2002;13(1):183–186. doi: 10.1097/00001756-200201210-00041. [DOI] [PubMed] [Google Scholar]
- 6.Antuono PG, Jones JL, Wang Y, Li SJ. Decreased glutamate + glutamine in Alzheimer’s disease detected in vivo with (1)H-MRS at 0.5 T. Neurology. 2001;56(6):737–742. doi: 10.1212/wnl.56.6.737. [DOI] [PubMed] [Google Scholar]
- 7.Marjanska M, Curran GL, Wengenack TM, Henry PG, Bliss RL, Poduslo JF, Jack CR, Jr., Ugurbil K, Garwood M. Monitoring disease progression in transgenic mouse models of Alzheimer’s disease with proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 2005;102(33):11906–11910. doi: 10.1073/pnas.0505513102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohanakrishnan P, Fowler AH, Vonsattel JP, Jolles PR, Husain MM, Liem P, Myers L, Komoroski RA. Regional metabolic alterations in Alzheimer’s disease: an in vitro 1H NMR study of the hippocampus and cerebellum. J Gerontol A Biol Sci Med Sci. 1997;52(2):B111–117. doi: 10.1093/gerona/52a.2.b111. [DOI] [PubMed] [Google Scholar]
- 9.Ewers M, Walsh C, Trojanowski JQ, Shaw LM, Petersen RC, Jack CR, Jr., Feldman HH, Bokde AL, Alexander GE, Scheltens P, Vellas B, Dubois B, Weiner M, Hampel H. Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampel H, Wilcock G, Andrieu S, Aisen P, Blennow K, Broich K, Carrillo M, Fox NC, Frisoni GB, Isaac M, Lovestone S, Nordberg A, Prvulovic D, Sampaio C, Scheltens P, Weiner M, Winblad B, Coley N, Vellas B. Biomarkers for Alzheimer’s disease therapeutic trials. Prog Neurobiol. 2010;95(4):579–593. doi: 10.1016/j.pneurobio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, Herholz K, Bokde AL, Jessen F, Hoessler YC, Sanhai WR, Zetterberg H, Woodcock J, Blennow K. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9(7):560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 12.DeKosky ST, Marek K. Looking backward to move forward: early detection of neurodegenerative disorders. Science. 2003;302(5646):830–834. doi: 10.1126/science.1090349. [DOI] [PubMed] [Google Scholar]
- 13.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 14.Vemuri P, Jack CR., Jr Role of structural MRI in Alzheimer’s disease. Alzheimers Res Ther. 2010;2(4):23. doi: 10.1186/alzrt47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P. Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain. 2009;132(Pt 8):2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ametamey SM, Treyer V, Streffer J, Wyss MT, Schmidt M, Blagoev M, Hintermann S, Auberson Y, Gasparini F, Fischer UC, Buck A. Human PET studies of metabotropic glutamate receptor subtype 5 with 11C-ABP688. J Nucl Med. 2007;48(2):247–252. [PubMed] [Google Scholar]
- 17.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nat Med. 2012;18(2):302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature medicine. 2003;9(8):1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 19.Jones CK, Schlosser MJ, van Zijl PC, Pomper MG, Golay X, Zhou J. Amide proton transfer imaging of human brain tumors at 3T. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2006;56(3):585–592. doi: 10.1002/mrm.20989. [DOI] [PubMed] [Google Scholar]
- 20.van Zijl PC, Jones CK, Ren J, Malloy CR, Sherry AD. MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST) Proc Natl Acad Sci U S A. 2007;104(11):4359–4364. doi: 10.1073/pnas.0700281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, Haris M, Cai K, Kassey VB, Kogan F, Reddy D, Hariharan H, Reddy R. Chemical exchange saturation transfer magnetic resonance imaging of human knee cartilage at 3 T and 7 T. Magn Reson Med. 2011 doi: 10.1002/mrm.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haris M, Cai K, Singh A, Hariharan H, Reddy R. In vivo mapping of brain myo-inositol. Neuroimage. 2011;54(3):2079–2085. doi: 10.1016/j.neuroimage.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14(4):260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Gilad AA, Bulte JW, van Zijl PC, McMahon MT. High-throughput screening of chemical exchange saturation transfer MR contrast agents. Contrast Media Mol Imaging. 2010;5(3):162–170. doi: 10.1002/cmmi.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu Y, Gage FH. Adult Hippocampal Neurogenesis and its Role in Alzheimer’s Disease. Mol Neurodegener. 2011;6(1):85. doi: 10.1186/1750-1326-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen SQ, Wang PJ, Ten GJ, Zhan W, Li MH, Zang FC. Role of myo-inositol by magnetic resonance spectroscopy in early diagnosis of Alzheimer’s disease in APP/PS1 transgenic mice. Dement Geriatr Cogn Disord. 2009;28(6):558–566. doi: 10.1159/000261646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward KM, Balaban RS. Determination of pH using water protons and chemical exchange dependent saturation transfer (CEST) Magn Reson Med. 2000;44(5):799–802. doi: 10.1002/1522-2594(200011)44:5<799::aid-mrm18>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 28.Winter PM, Cai K, Chen J, Adair CR, Kiefer GE, Athey PS, Gaffney PJ, Buff CE, Robertson JD, Caruthers SD, Wickline SA, Lanza GM. Targeted PARACEST nanoparticle contrast agent for the detection of fibrin. Magn Reson Med. 2006;56(6):1384–1388. doi: 10.1002/mrm.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun PZ, Farrar CT, Sorensen AG. Correction for artifacts induced by B(0) and B(1) field inhomogeneities in pH-sensitive chemical exchange saturation transfer (CEST) imaging. Magn Reson Med. 2007;58(6):1207–1215. doi: 10.1002/mrm.21398. [DOI] [PubMed] [Google Scholar]