Abstract
Fas binding to Fas-associated death domain (FADD) activates FADD-caspase-8 binding to form death-inducing signaling complex (DISC) that triggers apoptosis. The Fas-Fas association exists primary as dimer in the Fas-FADD complex and the Fas-FADD tetramer complexes have the tendency to form higher order oligomer. The importance of the oligomerized Fas-FADD complex in DISC formation has been confirmed. This study sought to provide structural insight for the roles of Fas death domain (Fas DD) binding to FADD and the oligomerization of Fas DD-FADD complex in activating FADD-procaspase-8 binding. Results show Fas DD binding to FADD stabilized the FADD conformation, including the increased stability of the critical residues in FADD death effector domain (FADD DED) for FADD-procaspase-8 binding. Fas DD binding to FADD resulted in the decreased degree of both correlated and anti-correlated motion of the residues in FADD and caused the reversed correlated motion between FADD DED and FADD death domain (FADD DD). The exposure of procaspase-8 binding residues in FADD that allows FADD to interact with procaspase-8 was observed with Fas DD binding to FADD. We also observed different degrees of conformational and motion changes of FADD in the Fas DD-FADD complex with different degrees of oligomerization. The increased conformational stability and the decreased degree of correlated motion of the residues in FADD in Fas DD-FADD tetramer complex were observed compared to those in Fas DD-FADD dimer complex. This study provides structural evidence for the roles of Fas DD binding to FADD and the oligomerization degree of Fas DD-FADD complex in DISC formation to signal apoptosis.
Keywords: Fas-FADD binding, DISC, oligomeric Fas-FADD complex, molecular dynamics, conformational and dynamical motion analysis
INTRODUCTION
Fas death receptor-activated signaling pathways regulate apoptosis in a variety of cells including cholangiocarcinoma, Jurkat cells and osteoclasts (1, 2). Dysfunction of Fas and its signaling pathways is associated with diseases in both mice and humans (3–6). Fas-activated signaling pathway has been well characterized. The Fas death receptor has the ability to self-associate or bind with other target proteins including calmodulin (CaM) and the Fas-associated death domain (FADD) (7–11). Upon triggering by Fas ligand (FasL), the clustering of Fas, FADD, and procaspase-8 that is FADD downstream protein forms death-inducing signaling complex (DISC) (Figure S1) that triggers apoptosis (12–14). Fas death domain (Fas DD) binds to FADD through homotypic interactions, which leads to activation of caspase-8 (15–18). The activation of caspase-8 initiates a set of reactions that leads to cell death. In the DISC formation, Fas binding to the C-terminal death domain of FADD (FADD DD) activates the binding of procaspase-8 to the N-terminal death effector domain of FADD (FADD DED) (15–19).
The structure of Fas DD consists of six amphipathic α-helices antiparallel to one another and the hydrophobic residues of the side chains form the core of the protein (10). FADD is composed of the N-terminal death effector domain and the C-terminal death domain, which are structurally similar to one another (Figure S2) (20–23). Both FADD DD and FADD DED adopt a six α-helical bundle structure that is characteristic of a structural family of “death motifs” (Figure S2) (23). The FADD DD is engaged with receptor, while the FADD DED contains the binding site for procaspase-8 and/or -10 (Figure S1) (15, 23, 24). The crystal structure of the Fas DD-FADD DD complex that has a tetrameric arrangement of four FADD DDs bound to four Fas DDs has been solved (19). Compared to the solution structure of the monomeric Fas DD (10), oligomeric Fas DD experiences an opening in which helix α6 shifts and merges with helix α5 to form a relatively long helix called stem helix, and a new helix named C-helix at the C-terminal of Fas DD is formed (Figure S3. A, B and C) (19). The opening of Fas to have the hydrophobic residues exposed is crucial for Fas binding to FADD (Figure S3. D)(19). The interaction region between death domains of Fas and FADD for Fas-FADD death domain complex has been identified (19). The interfaces for the Fas-FADD interactions are formed by the hydrophobic patch surrounded by polar residues in the helices α1 and α6 of FADD DD (α7 and α12 of full length FADD) and the hydrophobic residues in Fas DDs exposed only upon Fas opening (Figure S3. D) (19). The open forms of Fas are in an unstable equilibrium state and the opening of Fas allows the formation of a Fas-Fas bridge with another open Fas molecule through the stem helices and C-helices of the two Fas molecules to stabilize the open form and to form a stable Fas-Fas dimer (Figure S3. E) (19). The Fas-Fas association in a dimer behavior predominantly exists in Fas-FADD complex (Figure S3. E) (19). The crystal structure of Fas-FADD complex is a tetramer and the tetramer has the tendency to form higher oligomers (19). The formation of Fas-Fas bridge and Fas binding to FADD foster the DISC formation and the clustering to form higher oligomers (19). A recent study further demonstrates the importance of the oligomerized Fas-FADD complex in DISC formation (25). Fas binding to FADD activates FADD-caspase-8 binding to form the DISC (15–18). The caspase-8 binding site in FADD has been identified, showing that the α1 and α4 helices of the FADD DED interact with the death effector domain 2 of procaspase-8, and residues S12, R38, D44, and E51 in FADD DED domain are important for FADD binding to procaspase-8 (Figure S1) (26). The experimental studies have demonstrated the importance of Fas binding to FADD in facilitating FADD recruitment of procaspase-8 to form the DISC and the importance of the oligomerized Fas-FADD complex in DISC formation. However, structural insight for the roles of Fas binding to FADD and the oligomerization degree of Fas-FADD complex in activating FADD-procaspase-8 binding remain unclear.
This study to provides structural insight for the role of Fas binding to FADD and the oligomerization of Fas-FADD complex in activating FADD-procaspase-8 binding. We determined the conformational and motion changes of FADD resulted from binding to Fas for the Fas-FADD complex in either dimer or tetramer behavior and compared to those of single FADD without binding to Fas with molecular dynamics (MD) simulations. We evaluated the structural characteristics of the caspase-8 binding site in FADD by interactions with Fas at the different oligomerizations. Results from this study provide structural insight into the roles of Fas binding to FADD and the oligomerization degree of Fas-FADD complex in DISC formation to signal apoptosis.
MATERIALS AND METHODS
Fas DD-FADD dimer and tetramer complexes construction
The crystal structure of the complex of Fas death domain-FADD death domain with a tetrameric arrangement has been solved (PDBID: 3EZQ) (19). The NMR structure of the full length FADD that includes both death domain (DD) and death effector domain (DED) has also been solved (PDBID: 2GF5) (26). The interactions of Fas DD with FADD DD activate FADD DED binding to procaspase-8 (15–19, 26). To determine the roles of the interactions of Fas DD with FADD and the oligomerization degree of Fas DD-FADD complex in FADD binding to procaspase-8, we constructed the Fas DD-FADD complex by first geometrically fitting the α7 helix of the full length FADD (α1 helix for FADD DD only) to the α1 helix of FADD DD in the crystal structure of Fas DD-FADD DD complex using VMD software (27). Then, we removed the FADD DD in the initial crystal structure of Fas DD-FADD DD complex. This fitting method to obtain a protein complex has been used in previous studies (19, 28, 29). Fas opening that forms Fas-Fas bridge and allows Fas to bind to FADD is the key to obtain a stabilized Fas-FADD complex (19). Therefore, the Fas-FADD dimer complex is a minimal requirement for a stable Fas-FADD complex. Studies have showed that Fas-FADD complexes tend to form higher order oligomers for the DISC formation (19, 25). To evaluate the effect of oligomerization degree of Fas DD-FADD complex on FADD conformational changes and on FADD binding to procaspase-8, we constructed the Fas DD-FADD complexes in dimer and in tetramer based on the crystal structure of Fas DD-FADD DD in tetramer and the NMR structure of full length FADD. The constructed Fas DD-FADD complexes in dimer and tetramer were energy minimized using AMBER 9 MD package (30).
Molecular dynamics simulations
To examine the effect of Fas DD binding to FADD and the oligomerization degree of Fas DD-FADD complex in FADD binding to procaspase-8, we performed 150 ns molecular dynamics (MD) simulations for the three systems of single full length FADD, Fas DD-FADD complex in dimer and Fas DD-FADD complex in tetramer using AMBER 9 MD package (30). The AMBER force field was used for the simulated systems. The MD simulations were performed in a periodic box. One nm of solvent between the protein and the box boundaries was ensured to reduce potential artifacts arising from periodicity. The periodic box was filled with TIP3P water molecules (31) and 150 mM NaCl (physiological salt concentration). Additional ions of Na+ or Cl− were added to the system to neutralize the charge of the protein or protein complex. The simulated system size for single full length FADD is 9.6 nm × 6.4 nm × 6.1 nm, for Fas DD-FADD complex in dimer is 9.1 nm × 10.1 nm × 9.6 nm, for Fas DD-FADD complex in tetramer is 13.0 nm × 10.7 nm × 9.7 nm. The MD simulation protocol was identical for the three systems, which was similar to that of our previous studies (29, 32–36). The MD simulation protocol included: 1) steepest descent minimization for the solvent with the protein and ions restrained but with water mobile; 2) equilibration of water with mobile water molecules but with the protein and ions restrained at constant number-pressure-temperature (NpT) at 50K and 1 atm for 20 ps; 3) the warm up of the system via a series of 10 ps constant number-volume-temperature (NVT) MD simulations at 50, 100, 150, 200, 250, and 300K with SHAKE constraints and 2 fs time steps; 4) production simulation at NpT of 300K and 1 atm for the assigned time length of 150 ns in this study. In the production simulations, SHAKE constraints with relative tolerance of 1×10−5 were used on all hydrogen-heavy atom bonds to permit a dynamics time step of 2 fs. Electrostatic interactions were calculated by the particle-mesh Ewald method (PME) (37). The Lennard-Jones cutoffs were set at 1.0 nm. Root mean square deviation (RMSD) of protein or protein complex was calculated over time to ensure that the system reached equilibration during the MD simulations. The simulation trajectories after the initial equilibration were used for the conformational, motion and structural analyses.
Conformational and dynamical motion analyses
We performed a series of analyses to better understand the effect of Fas DD binding to FADD and the oligomerization degree of Fas DD-FADD complex on FADD binding to procaspase-8. The root mean-square deviation (RMSD) of protein backbone atoms was analyzed to determine the systems’ equilibration tendencies and its convergence. Root mean square fluctuations (RMSF) of the proteins were calculated on a residue-by-residue basis, and averaged over the production simulation trajectories after the systems reached equilibrium to observe the conformational fluctuation of protein domains. Dynamical cross-correlation maps (normalized covariance matrices) between residues of FADD were analyzed to gain insight into the degree of correlated motions of the residues in FADD for single FADD or FADD in Fas DD-FADD complexes. Principal component analysis (PCA) (38) was used to decompose a protein motion over a trajectory into a few principal motions which are described by eigenvectors and eigenvalues. The first PCA mode is usually interpreted as the direction of the largest conformational fluctuation of the system during MD simulations (39, 40). These analyses were performed using the ptraj program of AMBER, Matlab (41) was used to generate the cross-correlation plots, and porcupine plots (42) were used to visualize the collective dynamic modes calculated from PCA analysis. Previous studies shows that the α1 and α4 helices of the FADD DED are critical for FADD-procaspase-8 binding, and the C-terminal helix of FADD (helix α12) in the single full length FADD is located at the interface of the FADD DD and FADD DED, which could block the key residues in FADD for binding to procaspase-8 to signal apoptosis (26, 43). The results of overlaying the structure of full length FADD onto the FADD DD in the crystal structure of Fas-FADD complex show that helix α12 in FADD has to shift to avoid the steric clash with the newly formed C-helix of Fas DD in Fas-FADD complex, which could expose procaspase-8 binding residues in FADD for FADD-procaspase-8 interactions (19). From the MD simulations trajectories, we calculated the distance between the end of α12 helix (Ca atom of residue GLY191) and the midpoint between the ends of α1 helix (Ca atom of residue SER1) and α4 helix (Ca atom of residue LEU50) of FADD in FADD DED for single FADD, FADD in Fas-FADD dimer complex and FADD in Fas-FADD tetramer complex. The results help to determine whether Fas binding to FADD results in the exposure of procaspase-8 binding residues in FADD and whether the position of α12 helix relative to the helices of α1 and α4 in FADD is affected by the oligomerization degree of Fas-FADD complex.
Statistical methods
To compare the effect of Fas DD binding to FADD and the oligomerization degree of Fas DD-FADD complex on FADD conformation changes, both the average values and the standard deviations of the analyzed variables were calculated. Due to the tendency of adjacent snapshots from the MD trajectories to be correlated with each other, the autocorrelation time (44, 45) for the studied quantities were obtained to re-sample the trajectories into statistically independent periods in order to calculate the standard deviations for each quantity. With the obtained decorrelation times, bootstrap analysis (46) was performed following an analysis protocol similar to that of Chen and Pappu and our published study (29, 32–35). With the mean and standard deviation between the observables for the simulated systems, we used the Student’s t-test (47) with 95% confidence to compare whether the effects of Fas DD binding to FADD and the oligomerization degree of Fas DD-FADD complex on FADD conformational changes were significant.
RESULTS & DISCUSSION
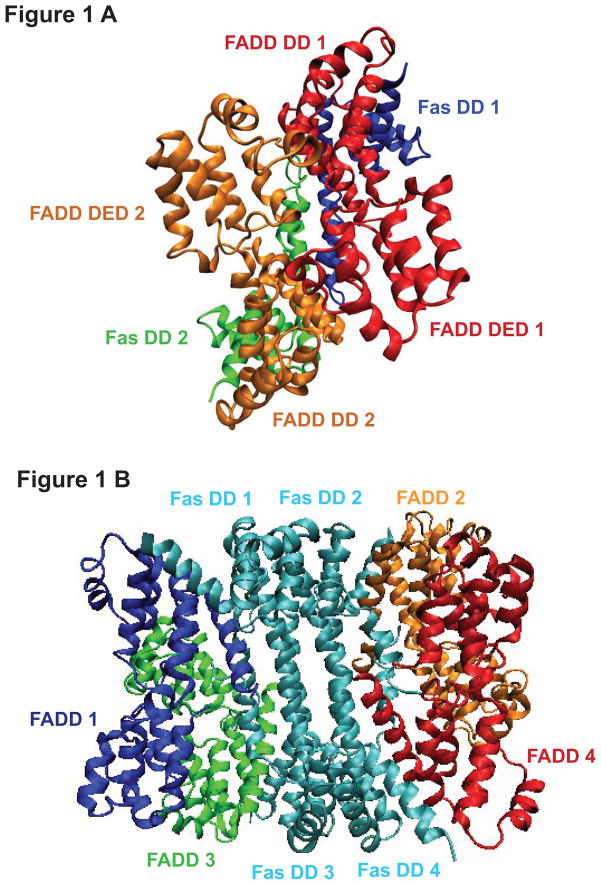
Because the Fas -FADD dimer complex is a minimal requirement for a stable Fas-FADD complex (19) and we are interested in understanding the role of the oligomerization degree of Fas-FADD complex in activating FADD-procaspase-8 binding, we constructed the Fas DD-FADD complexes in dimer and in tetramer based on the crystal structure of Fas DD-FADD DD in tetramer (PDBID: 3EZQ) (19) and the NMR structure of full length FADD (PDBID: 2GF5) (26). The constructed Fas DD-FADD complexes in dimer and tetramer were energy minimized using AMBER 9 MD package (30). The minimized Fas DD-FADD complexes in dimer and tetramer were shown as Figure 1A and Figure 1B. The RMSD of Fas DD protein core (res 225–318) and FADD from the Fas DD-FADD complexes either in dimer or tetramer over 150 ns MD simulations showed that the systems reached initial equilibration after the first 90 ns (Figure S4). The last 60 ns simulations trajectories of the 150 ns MD simulations were used for the conformational and dynamical motion analyses of FADD by binding to Fas DD either in dimer or tetramer complexes
FIGURE 1.
The constructed complex of Fas death domain with full length FADD. (A) Fas DD-FADD complex in dimer (red: FADD 1; orange: FADD 2; blue: Fas DD 1; green: Fas DD 2); (B) Fas DD-FADD complex in tetramer (blue: FADD 1; orange: FADD 2; green: FADD 3; red: FADD 4; cyan: Fas DD 1, Fas DD 2, Fas DD 3 and Fas DD 4).
Conformational changes in FADD
RMSF comparison for single FADD and FADD in the Fas DD-FADD complexes
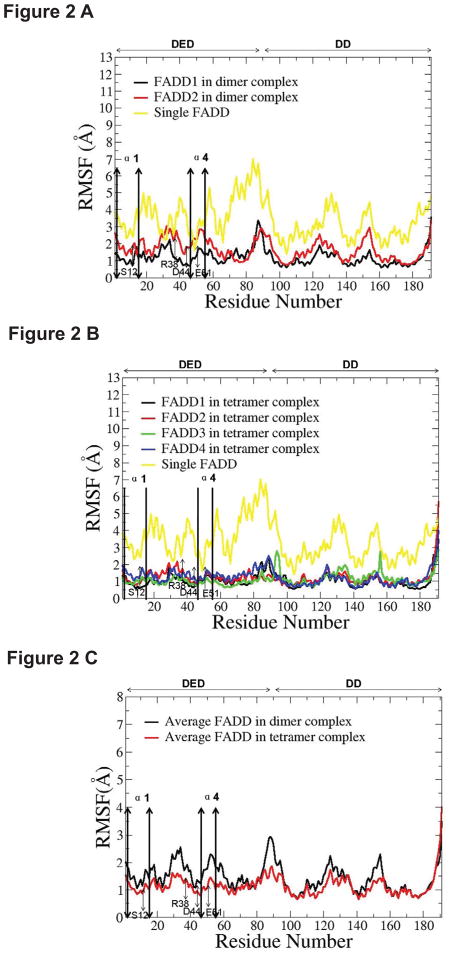
For FADD in Fas DD-FADD dimer complex, the RMSF result showed that the fluctuations of the two individual FADD in dimer complex were consistently lower than those of a single FADD (Figure 2A, Figure S5 A). The α1 and α4 helices of the FADD DED have been identified to interact with the death effect domain 2 of procaspase-8, and the residues of S12, R38, D44, and E51 in FADD DED domain are important for FADD binding to procaspase-8 (26). Based on the RMSF results, the flexibility of the α1 and α4 helices and key residues (S12, R38, D44, and E51) in each FADD DED of dimer complexes was consistently lower than those in a single FADD DED (Figure 2A, Figure S5 A), indicating that, in dimer complexes, FADD DEDs were more stable than a single FADD DED. The stabilized FADD DED conformation could facilitate the FADD-procaspase-8 binding through α1 and α4 helices and key residues on FADD DEDs. The FADD DD was also stabilized by the binding to Fas DD. The similar observations were observed for the comparison of FADDs in Fas DD-FADD tetramer complex with a single FADD (Figure 2B, Figure S5 B).
FIGURE 2.
The root mean squared fluctuation (RMSF) comparison. (A) RMSF of two FADDs in dimer complex compared to single FADD; (B) RMSF of four FADDs in tetramer complex and single FADD; (C) the average RMSF of the two FADDs in Fas DD-FADD dimer complex compared to the average RMSF of the four FADDs in Fas DD-FADD tetramer complex.
The comparison of the average RMSF of the two FADDs in Fas DD-FADD dimer complex with the average RMSF of the four FADDs in Fas DD-FADD tetramer complex showed the increased conformational stability for FADDs in Fas DD-FADD tetramer complex compared to that of the FADDs in Fas DD-FADD dimer complex (Figure 2C). Particularly, the flexibility of the α1 and α4 helices and key residues in FADD DEDs of tetramer complexes was consistently lower than those in FADD DEDs of dimer complexes (Figure 2C), indicating that FADDs in Fas DD-FADD tetramer complex could be more preferable than that in dimer complex for binding to procaspase-8 to form the DISC. The changed degree of conformational flexibility of FADD could result in changed electrostatic distribution, van der Waals energy, polar solvation energy and entropy of FADD, which could directly affect the electrostatic interactions and van der Waals interactions between FADD and procaspase-8, further affecting FADD recruitment of procaspase-8 to signal apoptosis.
Position of α12 helix in FADD DD relative to the helices of α1 and α4 in FADD DED (Relative position changes between FADD DD and FADD DED)
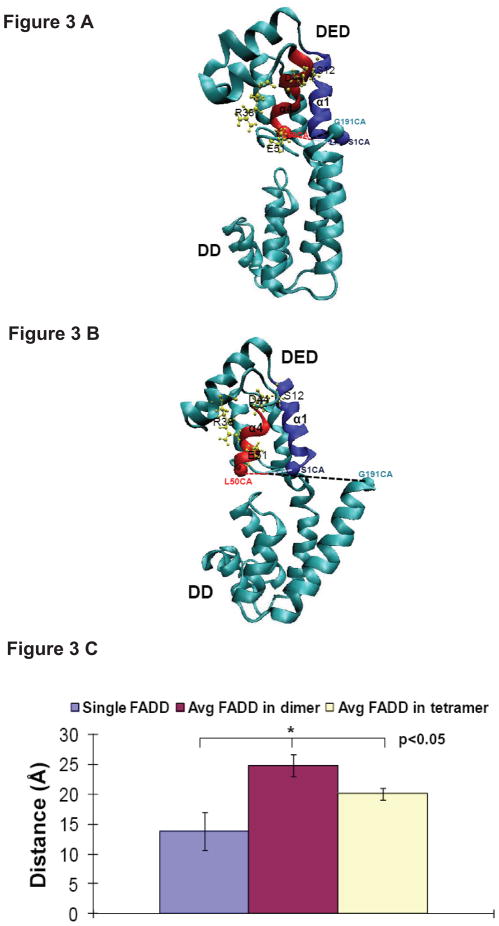
The α1 and α4 helices of the DED of FADD are critical for FADD-procaspase-8 binding (26). NMR structure of the single FADD (PDBID: 2GF5) (26) shows that the end of α12 helix of FADD DD (FADD C terminus) is located at the interface of the FADD DED and FADD DD and is close to the α1 and α4 helices of the FADD DED, which could block α1 and α4 helices from binding to procaspase-8 (Figure 3A). Based on the crystal structure of the Fas DD-FADD DD tetrameric complex (PDBID: 3EZQ) and NMR structure of the full-length FADD (PDBID: 2GF5) (26), the study shows that the C-terminal helix (α12 helix) of FADD has to shift to avoid a steric clash with the newly formed C-helix of Fas DD in Fas DD-FADD complex when the structure of the full-length FADD was overlaid onto the FADD DD in Fas DD-FADD DD complex. This could cause relative position changes between FADD DED and FADD DD (19). A representative structure of the full length FADD from Fas DD-FADD tetrameric complex from our equilibrated MD simulations is shown as Figure 3B, illustrating that Fas-FADD binding could expose the α1 and α4 helices of FADD DED that will facilitate FADD binding to procaspase-8 (Figure 3A and B). We calculated the distance between the end of α12 helix in FADD DD and the midpoint of the ends of the α1 and α4 helices of FADD DED in single FADD and FADDs in Fas DD-FADD complexes either in dimer or in tetramer complex based on the last 60 ns simulation trajectories of the 150 ns MD simulations (Figure 3C). The results showed that Fas DD binding to FADD resulted in significantly increased distance between the end of α12 helix in FADD DD and the midpoint of the ends of α1 and α4 helices for Fas DD-FADD complexes in either dimer or tetramer compared to a single FADD, and the distance of α12 helix away from α1 and α4 helices of FADD was significantly larger in Fas DD-FADD dimer complex compared to that in Fas DD-FADD tetramer complex (Figure 3C). The results clearly demonstrated that Fas binding to FADD could expose the α1 and α4 helices of FADD to allow FADD to interact with the death effector domain 2 of procaspase-8, and Fas-FADD dimer complex formation was sufficient for the exposure of procaspase-8 binding site in FADD that allows FADD to interact with procaspase-8 for DISC formation compared to Fas-FADD tetramer complex formation (Figure 3C).
FIGURE 3.
Definition and visualization of the distance between the end of α12 helix (Ca atom of residue GLY191) and the midpoint between the ends of α1 helix (Ca atom of residue SER1) and α4 helix (Ca atom of residue LEU50) of FADD in single FADD (A) and in Fas DD-FADD complex (B); (C) The comparison among the distance between the end of α12 helix (Ca atom of residue GLY191) and the midpoint between the ends of α1 helix (Ca atom of residue SER1) and α4 helix (Ca atom of residue LEU50) of FADD for single FADD, the average distance of two FADDs in dimer complexes and the average distance of four FADDs in tetramer complexes (unit: Å).
Dynamical motion changes in FADD
Dynamical cross-correlation analysis
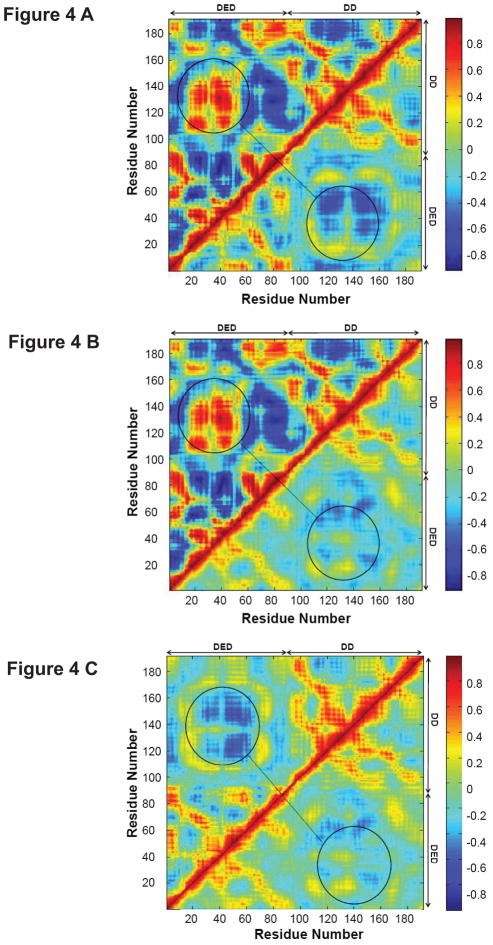
We calculated the dynamical cross-correlation maps to compare the degrees of correlated motion of the residues of a single FADD, FADDs in Fas DD-FADD dimer complex and FADDs in Fas DD-FADD tetramer complex (Figure 4, Figure S6 and Figure S7). The comparison of the dynamical cross-correlation maps of a single FADD (top left in Figure 4A and B), FADDs in Fas DD-FADD dimer complex (bottom right in Figure 4A, Figure S6 A and B) and FADDs in Fas DD-FADD tetramer complex (bottom right in Figure 4B, Figure S7 A, B, C and D) showed that Fas DD binding to FADD resulted in an overall lesser degree of both correlated (red) and anticorrelated (blue) movement for the residues of FADD and caused the reversed correlated motion between FADD DED and FADD DD (circled regions) compared to that of a single FADD. The dynamical motion changes of FADD by binding to Fas DD could directly contribute to the conformational changes of FADD, which could affect the electrostatic interactions and van der Waals interactions of FADD with procaspase-8 and the recruitment of procaspase-8 to signal apoptosis. We also compared the dynamical cross-correlation map of FADD in Fas DD-FADD dimer complex (top left of Figure 4C) to that of FADD in Fas DD-FADD tetramer complex (bottom right in Figure 4C). The results showed the lesser degree of correlated motion of the residues of FADD in Fas DD-FADD tetramer complex compared to that of FADD in Fas-FADD dimer complex, indicating a more stabilized structure for FADD in Fas-FADD tetramer complex compared to that in Fas-FADD dimer complex.
FIGURE 4.
Dynamical cross-correlation maps to compare the degree of correlated motion of the residues in FADD (red: correlation between residues; blue: anticorrelation between residues). (A) Single FADD (top left) and FADD in Fas DD-FADD dimer complex (bottom right, data averaged from the two FADDs in the dimer complex); (B) single FADD (top left) and FADD in Fas DD-FADD tetramer complex (bottom right, data averaged from the four FADDs in the tetramer complex); (C) FADD in Fas DD-FADD dimer complex (top left, data averaged from the two FADDs in the dimer complex) and FADD in Fas DD-FADD tetramer complex (bottom right, data averaged from the four FADDs in the tetramer complex). (Circled regions show the degree of correlated motion between res 10–70 in FADD DD and res 100–170 in FADD DED)
Principal component analysis
We performed principal component analyses (PCA) to describe the dynamics of FADD and to visualize the essential mode of motion by generating porcupine plots using the MD trajectory (Figure S8 and Figure S9). PCA results for a single FADD showed that the residues in different helixes of FADD DD were moving together but the residues in different helixes of FADD DED were moving away from each other (Figure S8 A). These observations were significantly different from those occurred for the two FADDs in the Fas DD-FADD dimer complex (Figure S8 B and C) and those observed for the four FADDs in the Fas DD-FADD tetramer complex (Figure S9). Procaspase-8 binds to the α1 and α4 helices of the FADD DED and the residues (S12, R38, D44, and E51) in FADD DED are critical for FADD-procaspase-8 binding as experimentally observed (26). The dynamical motion changes of FADD by binding to Fas DD could directly affect the relative position changes between FADD DED and FADD DD, and the overall conformational changes of FADD, further resulting in the changed electrostatic distribution, van der Waals energy, polar solvation energy, and entropy of FADD, which could directly affect FADD-procaspase-8 interactions for DISC formation to signal apoptosis.
CONCLUSIONS
Experimental studies have shown that the α1 and α4 helices of the FADD DED interact with the death effector domain 2 of procaspase-8, and residues S12, R38, D44, and E51 in FADD DED domain are important for FADD binding to procaspase-8 (26). The results from this study demonstrated that Fas DD binding to FADD stabilized the overall conformation of FADD, including the increased stability of the α1 and α4 helices and the critical residues (S12, R38, D44, and E51) in FADD DED that will affect the electrostatic interactions and van der Waals interactions of FADD with procaspase-8 and facilitate the recruitment of procaspase-8 to signal apoptosis. Fas binding to FADD resulted in a decreased degree of both correlated and anti-correlated motion of the residues in FADD and caused a reversed correlated motion between FADD DED and FADD DD. Dynamical motion changes of residues in FADD by binding to Fas DD caused the relative position changes between FADD DD and FADD DED to expose the α1 and α4 helices of the FADD DED to enhance FADD binding to procaspase-8. Experimental studies also showed that the Fas -FADD dimer complex is a minimal requirement for a stable Fas-FADD complex and Fas binding to FADD fosters the DISC formation and the clustering to form higher oligomers (19). A recent experimental study further demonstrates the importance of the oligomerized Fas-FADD complex in DISC formation (25). However, structural mechanisms for these experimental observations remain unknown. We observed the different degree of the conformational and motion changes of FADD in the Fas-FADD complexes with the different degree of oligomerization. The increased conformational stability and the decreased degree of the correlated motion of the residues in FADDs in the Fas-FADD tetramer complex were observed compared to those in the Fas-FADD dimer complex. The results also showed that Fas-FADD dimer complex formation was sufficient for the exposure of the procaspase-8 binding site in FADD that allows FADD to interact with procaspase-8. This study provides structural and dynamic mechanisms for the roles of Fas DD binding to FADD and the oligomerization of Fas DD-FADD complex in activating FADD-procaspase-8 binding for DISC formation to signal apoptosis observed in experimental studies (19, 25). The structural information should facilitate the identification of novel strategies and targets to regulate DISC formation and, thereby, modulate apoptosis.
Supplementary Material
Acknowledgments
Y.H. Song acknowledges NIH K25 (K25CA140791) award. H. Chao’s initial participation of this project is acknowledged.
This work was supported by an NIH K25 award (5K25CA140791) to Y. H. Song and a Veterans Administration Merit Award to JMM.
ABBREVIATIONS
- FADD
Fas-associated death domain
- DISC
death-inducing signaling complex
- DD
death domain
- DED
death effector domain
- CaM
calmodulin
- MD
molecular dynamics
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- PCA
principal component analysis
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figure S1 is a cartoon figure to show the protein interactions underlying death-inducing signaling complex (DISC) formation (19). Figure S2 shows the secondary structure of FADD with the highlights of the key residues important for FADD binding to procaspase-8 (26). Figure S3 was cited from “FL Scott et al. Nature 457, 1019-1022, doi:10.1038/nature07606” (19), which shows Fas–FADD and Fas–Fas interactions in Fas–FADD DD complex. Root mean squared deviation (RMSD) of Fas DD protein core (res 225–318) and FADD over the 150ns MD simulation showed that the simulated systems reached the initial equilibration after MD simulation for 90 ns (Figure S4). The comparison of RMSF of a single FADD with the average RMSF of the two FADDs in the Fas DD-FADD dimer complex (Figure S5 A) and with the average RMSF of the four FADDs in the Fas DD-FADD tetramer complex (Figure S5 B) showed that Fas DD binding to FADD stabilized FADD conformation. The comparison of the dynamical cross-correlation maps for the degree of correlated motion of the residues of FADD in single FADD with FADDs in Fas DD-FADD dimer complex and with FADD in Fas DD-FADD tetramer complex showed that Fas DD binding to FADD resulted in an overall lesser degree of both correlated (red) and anticorrelated (blue) movement for the residues of FADD and caused the reversed correlated motion between FADD DED and FADD DD (circled regions) compared to that of single FADD (Figure S6 and Figure S7). Comparison of PCA results for the principal motion of single FADD, FADDs in Fas DD-FADD dimer complex and FADDs in Fas DD-FADD tetramer complex showed that Fas DD binding to FADD caused significantly different dynamical motion for the residues of FADDs in Fas DD-FADD complexes compared to those in single FADD (Figure S8 and Figure S9).
References
- 1.Wu X, McKenna MA, Feng X, Nagy TR, McDonald JM. Osteoclast apoptosis: the role of Fas in vivo and in vitro. Endocrinology. 2003;144:5545–5555. doi: 10.1210/en.2003-0296. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Xu J, Jhala N, Pawar P, Zhu ZB, Ma L, Byon CH, McDonald JM. Fas-mediated apoptosis in cholangiocarcinoma cells is enhanced by 3,3′-diindolylmethane through inhibition of AKT signaling and FLICE-like inhibitory protein. Am J Pathol. 2006;169:1833–1842. doi: 10.2353/ajpath.2006.060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puck J, Straus S, Deist FL, Rieux-Laucat F, Fischer A. Primary Immunodeficiency Diseases: A Molecular and Genetic Approach. New York: Oxford University Press; 1999. Inherited Disorders with Autoimmune and Defective Lymphocyte Regulation. [Google Scholar]
- 4.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 5.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IA, Debatin KM, Fischer A, de Villartay JP. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 6.Bi LL, Pan G, Atkinson TP, Zheng L, Dale JK, Makris C, Reddy V, McDonald JM, Siegel RM, Puck JM, Lenardo MJ, Straus SE. Dominant inhibition of Fas ligand-mediated apoptosis due to a heterozygous mutation associated with autoimmune lymphoproliferative syndrome (ALPS) Type Ib. BMC Med Genet. 2007;8:41. doi: 10.1186/1471-2350-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Pawar P, Pan G, Ma L, Liu H, McDonald JM. Calmodulin binding to the Fas-mediated death-inducing signaling complex in cholangiocarcinoma cells. Journal of cellular biochemistry. 2008;103:788–799. doi: 10.1002/jcb.21447. [DOI] [PubMed] [Google Scholar]
- 8.Wu X, Ahn EY, McKenna MA, Yeo H, McDonald JM. Fas binding to calmodulin regulates apoptosis in osteoclasts. The Journal of biological chemistry. 2005;280:29964–29970. doi: 10.1074/jbc.M500710200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn EY, Lim ST, Cook WJ, McDonald JM. Calmodulin binding to the Fas death domain. Regulation by Fas activation. The Journal of biological chemistry. 2004;279:5661–5666. doi: 10.1074/jbc.M311040200. [DOI] [PubMed] [Google Scholar]
- 10.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 11.Imtiyaz HZ, Zhang Y, Zhang J. Structural requirements for signal-induced target binding of FADD determined by functional reconstitution of FADD deficiency. J Biol Chem. 2005;280:31360–31367. doi: 10.1074/jbc.M504138200. [DOI] [PubMed] [Google Scholar]
- 12.Ashkenazi A, V, Dixit M. Apoptosis control by death and decoy receptors. Current opinion in cell biology. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 13.Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell death and differentiation. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 14.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nature reviews. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 15.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 16.Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Chang HY, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 18.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 19.Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberstadt M, Huang B, Chen Z, Meadows RP, Ng SC, Zheng L, Lenardo MJ, Fesik SW. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 21.Jeong EJ, Bang S, Lee TH, Park YI, Sim WS, Kim KS. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. The Journal of biological chemistry. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
- 22.Berglund H, Olerenshaw D, Sankar A, Federwisch M, McDonald NQ, Driscoll PC. The three-dimensional solution structure and dynamic properties of the human FADD death domain. Journal of molecular biology. 2000;302:171–188. doi: 10.1006/jmbi.2000.4011. [DOI] [PubMed] [Google Scholar]
- 23.Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T, Gavathiotis E, Wei Y, Werner MH. The structure of FADD and its mode of interaction with procaspase-8. Molecular cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 24.Chinnaiyan AM, Tepper CG, Seldin MF, O’Rourke K, Kischkel FC, Hellbardt S, Krammer PH, Peter ME, Dixit VM. FADD/MORT1 is a common mediator of CD95 (Fas/APO-1) and tumor necrosis factor receptor-induced apoptosis. The Journal of biological chemistry. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, Robinson CV, Siegel RM, Walz T, Wu H. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nature structural & molecular biology. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T, Gavathiotis E, Werner MH. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 28.Puklin-Faucher E, Gao M, Schulten K, Vogel V. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. The Journal of cell biology. 2006;175:349–360. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan D, Song Y. Role of Altered Sialylation of the I-like Domain of β1 Integrin in the Binding of Fibronectin to β1 Integrin: Thermodynamics and Conformational Analyses. Biophys J. 2010;99:208–217. doi: 10.1016/j.bpj.2010.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of computational chemistry. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 32.Yan Q, Murphy-Ullrich JE, Song Y. Structural insight into the role of thrombospondin-1 binding to calreticulin in calreticulin-induced focal adhesion disassembly. Biochemistry. 2010;49:3685–3694. doi: 10.1021/bi902067f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suever JD, Chen Y, McDonald JM, Song Y. Conformation and free energy analyses of the complex of calcium-bound calmodulin and the Fas death domain. Biophysical journal. 2008;95:5913–5921. doi: 10.1529/biophysj.108.130542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Pan D, Bellis SL, Song Y. Effect of altered glycosylation on the structure of the I-like domain of beta1 integrin: A molecular dynamics study. Proteins. 2008;73:989–1000. doi: 10.1002/prot.22126. [DOI] [PubMed] [Google Scholar]
- 35.Lee SJ, Song Y, Baker NA. Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+-lipid interactions and finite size effects. Biophysical journal. 2008;94:3565–3576. doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Guallar V, Baker NA. Molecular dynamics simulations of salicylate effects on the micro- and mesoscopic properties of a dipalmitoylphosphatidylcholine bilayer. Biochemistry. 2005;44:13425–13438. doi: 10.1021/bi0506829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darden T, York D, Pedersen LG. Particle mesh Ewald: An Nâlog(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Hayward S, Kitao A, Go N. Harmonicity and anharmonicity in protein dynamics: a normal mode analysis and principal component analysis. Proteins. 1995;23:177–186. doi: 10.1002/prot.340230207. [DOI] [PubMed] [Google Scholar]
- 39.Hayward S, Kitao A, Hirata F, Go N. Effect of solvent on collective motions in globular protein. Journal of molecular biology. 1993;234:1207–1217. doi: 10.1006/jmbi.1993.1671. [DOI] [PubMed] [Google Scholar]
- 40.Berendsen HJ, Hayward S. Collective protein dynamics in relation to function. Current opinion in structural biology. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 41.MathWorks, T. MATLAB
- 42.Tai K, Shen T, Borjesson U, Philippopoulos M, McCammon JA. Analysis of a 10-ns molecular dynamics simulation of mouse acetylcholinesterase. Biophysical journal. 2001;81:715–724. doi: 10.1016/S0006-3495(01)75736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh N, Nagata S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. The Journal of biological chemistry. 1993;268:10932–10937. [PubMed] [Google Scholar]
- 44.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University Press; New York: 1987. [Google Scholar]
- 45.Lee S, Song Y, Baker NA. Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+-lipid interactions and finite size effects. Biophysical journal. 2008 doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman & Hall; New York: 1998. [Google Scholar]
- 47.Bailey NTJ. Statistical methods in biology. Cambridge University Press; New York, NY: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.