TO THE EDITOR
Epidermal genetic mosaicism is evident as stripes of affected skin which typically appear in S or V-shaped whorled, streaked, and linear patterns called lines of Blaschko (Blaschko, 1901). These patterns represent dorsoventral migratory pathways of neuroectoderm during embryogenesis (Moss et al., 1993). Mosaic lesions result from somatic mutation during development, with timing of such events determining the extent and distribution of skin involvement. Epidermal nevi (EN) are common cutaneous mosaic disorders seen in 0.1–0.3% of births, and fall into two classes: keratinocytic epidermal nevi (KEN) and organoid epidermal nevi, which includes nevus sebaceus (NS) and follicular nevi (Solomon and Esterly, 1975). NS comprises approximately half of EN, and typically appears on the scalp as a yellowish-orange linear plaque with hyperkeratosis, acanthosis, a markedly increased number of sebaceous lobules and abortive hair follicles with resulting alopecia (Figure 1a–d). In contrast to KEN, in which neoplasia is rare, tumors develop in nearly 14% of all NS cases, and in more than 23% of affected adults (Cribier et al., 2000), suggesting that the mutation(s) causing NS also increase risk of tumorigenesis (Figure 1e–g).
Figure 1. Clinical and microscopic features of nevus sebaceus.
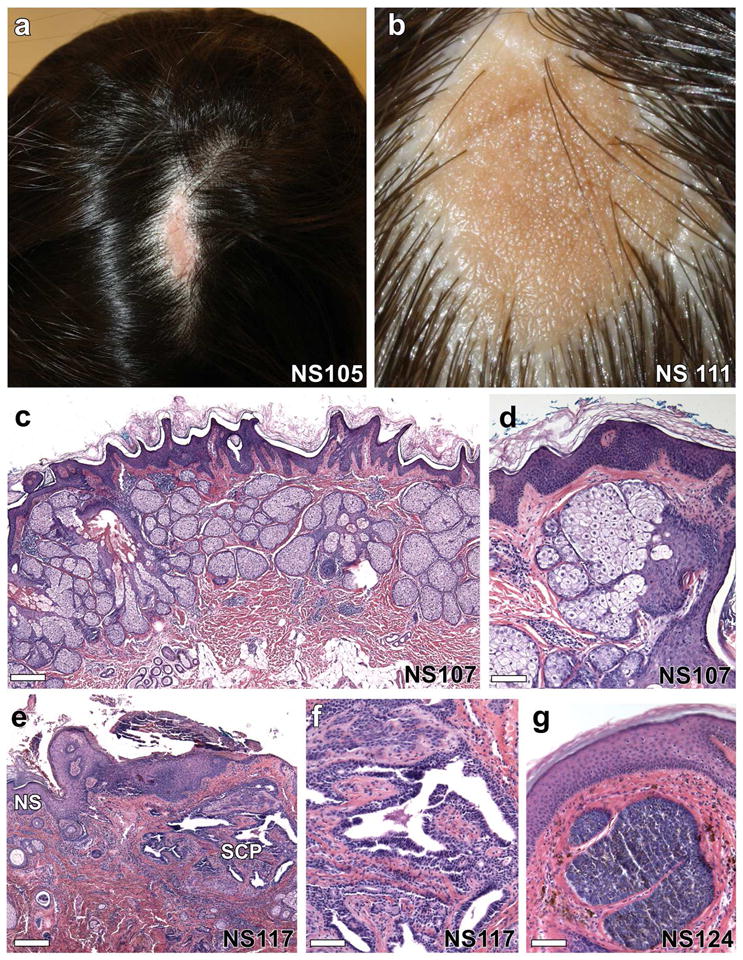
(a, b) Solitary, well-demarcated lesions on the scalp of two individuals show alopecia and a yellow-orange waxy appearance. (c) On histological examination, there is epidermal acanthosis, papillomatosis, and hyperkeratosis with dramatic increase in the number of sebaceous lobules and abortive hair follicles, scale = 288 μm, which is more evident at higher magnification (d), scale = 85 μm. (e–g) Up to 20% of nevus sebaceus lesions develop tumors including syringocystadenomas, trichoblastomas, trichilemmomas and tubular apocrine adenomas. (e) Nevus sebaceus (NS) with syringocystadenoma papilliferum (SCP) composed of villous structures lined by a columnar epithelium with stromal plasma cells, scale = 570 μm, most evident at higher magnification, (f), scale = 92 μm. (g) A trichoblastoma arising within a nevus sebaceus shows a well-circumscribed nodule of basaloid cells with a dense fibrocytic stroma, scale = 92 μm.
Recently, somatic mosaicism has been identified in KEN using SNaPshot assays to identify mutations in MAPK pathway genes including FGFR3, HRAS, KRAS, NRAS and PIK3CA. Activating Ras mutations, including HRAS p.Gly13Arg and KRAS p.Gly12Asp, were the most common and accounted for 39% of KEN, with HRAS mutations predominating (Hafner et al., 2012). Similar approaches have been employed in NS, identifying HRAS p.Gly13Arg in 91% of lesions and KRAS p.Gly12Asp in 5% of lesions (Groesser et al., 2012). We present an independent, complementary approach to genetic pathogenesis in NS in which we employed whole exome sequencing to characterize the spectrum of de novo coding mutations present within NS lesions.
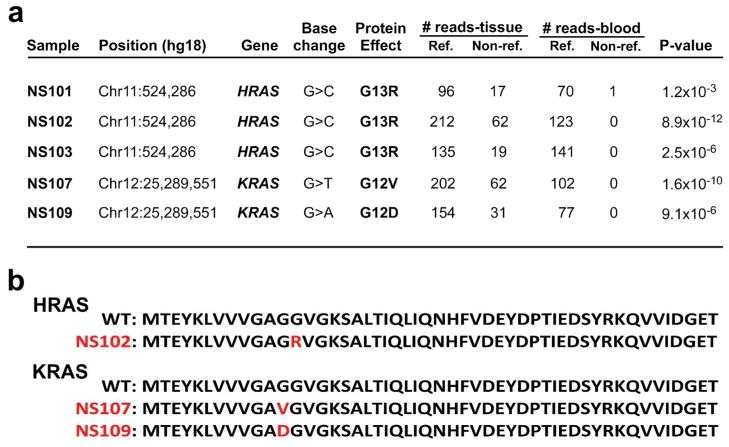
For our study cohort, we identified five individuals with nevus sebaceus and performed exome sequencing of paired DNA from blood and NS tissue in each (Supplementary Table 1). Genetic variants were annotated and compared to identify de novo somatic mutations present solely within nevus sebaceus tissue and not in germline DNA. Via this analysis, we identified two genes with recurrent somatic mutations: three NS samples had an identical somatic mutation in HRAS (p.Gly13Arg), and the remaining two had a somatic mutation at the Gly12 residue of KRAS (p.Gly12Asp and p.Gly12Val, respectively) (Figure 2, Supplementary Figure 1). Furthermore, no genes other than HRAS and KRAS were found to be mutated in more than one lesion. Notably, one sample showed a concurrent HRAS p.Gly13Arg and a BRAF p.Arg347Gln mutation which has not previously been described as a germline or somatic mutation. This BRAF mutation was the only other somatic mutation found in exome sequences of these five NS samples.
Figure 2. Exome sequencing reveals somatic HRAS and KRAS mutations in nevus sebaceus tissue.
(a) HRAS and KRAS mutation annotation, including genomic position, nucleotide change, protein consequence, and number of reference and non-reference reads obtained from paired sequencing of tissue and blood in 5 independent, unrelated nevus sebaceus cases. Significance of the mutant allele frequency difference between tissue and blood DNA was calculated with a one-tailed Fisher’s exact test. When corrected for multiple testing, 2.4×10−6 is the threshold for genome wide significance. In each case, HRAS and KRAS mutations showed the lowest P-value. (b) Alignment of the N-termini of HRAS and KRAS reveals identical residues through position 94, with an overall 95% identity and 99% similarity. The first 50 amino acids are shown for the wild-type and each mutant protein, with mutant residues indicated in red.
To examine whether these mutations were present uniquely in the epidermis, and not in the underlying dermis, we used laser capture microdissection to prepare DNA independently from epidermis and dermis of NS lesions and then performed Sanger sequencing with HRAS- or KRAS-specific primers. We found the mutant allele in an approximately equimolar ratio in epidermis and sebaceous lobules, but it was entirely absent in dermis and blood (Supplementary Figures 2 and 3). Via Sanger sequencing, we evaluated 11 additional paired samples for HRAS and KRAS mutations, and all were found to have the same somatic p.Gly13Arg HRAS mutation (Supplementary Table 2; Supplementary Figure 3).
Prior studies have reported that up to 40% of NS lesions exhibit loss of heterozygosity (LOH) on chromosome 9q, inclusive of PTCH (Xin et al., 1999). Examination of exome data from our discovery cohort found no evidence of LOH in NS lesions at this locus or elsewhere (Supplementary Figure 4).
Recognizing that HRAS and KRAS are oncogenes, that observed mutations could serve as an initiating event in multistep carcinogenesis (Knudson, 2001), and that tumors frequently develop in NS, we sought to determine if tumors arise specifically in HRAS or KRAS mutation-positive lesions. Neoplasms arising within NS are typically benign and consist primarily of trichoblastomas, syringocystadenoma papilliferum, trichilemmomas, and tubular apocrine adenomas (Cribier et al., 2000), although occasional basal cell carcinomas and rarely more aggressive malignant tumors have been reported (Moody et al., 2012). We identified 11 archival NS specimens containing tumors, isolated DNA from the NS portion of the lesion, and found that all samples had an HRAS p.Gly13Arg mutation (Supplementary Table 2, Supplementary Figure 5). The spectrum of additional genetic events necessary for tumorigenesis in NS lesions remains unknown.
In total, 27 nevus sebaceus samples were evaluated, with 25 harboring an identical HRAS mutation (p.Gly13Arg), and two exhibiting a KRAS mutation in the adjacent paralogous residue (p.Gly12Asp or p.Gly12Val). The occurrence of multiple, tightly clustered somatic mutations in adjacent residues of these highly homologous proteins is definitive proof of a role for these mutations in nevus sebaceus and suggests a gain of function mechanism. Indeed, expression of the KRAS p.Gly12Asp allele within murine hair follicles reproduces features of NS including abortive hair follicles and epidermal and sebaceous gland hyperplasia (Lapouge et al., 2011; Mukhopadhyay et al., 2011).
Missense mutations at codons 12 and 13 of HRAS and KRAS, respectively, are common in malignancies, including squamous cell carcinoma, and lead to constitutive activation of Ras by blocking the activity of GTPase activating proteins (Grewal et al., 2011). Ras isoforms are central regulators of the mitogen-activated protein kinase (MAPK) pathway which controls cell proliferation, differentiation and survival. Rare inherited disorders caused by germline mutations in Ras and other MAPK pathway members, known as “RASopathies,” include Costello, Noonan, and cardio-facio-cutaneous syndrome. These show variable cutaneous features, particularly hyperkeratosis, palmoplantar keratoderma, papillomas, and hair abnormalities in addition to craniofacial and systemic findings. KEN and NS have not been reported in affected individuals. Notably, HRAS p.Gly13Arg and KRAS p.Gly12Asp have not been reported as germline mutations in these or other disorders, and KRAS p.Gly12Asp leads to embryonic lethality in mice, suggesting that both mutations grossly disrupt embryonic development and are thus likely to be found primarily in mosaic states (Tuveson et al., 2004). Our findings confirm the predominance of HRAS mutations in NS, including those with tumors (Groesser et al., 2012), and provide evidence that HRAS and KRAS mutations are sufficient to cause NS without genome instability, LOH, or secondary mutation.
The marked sebaceous hyperplasia observed NS, which are found almost exclusively on the scalp and face, is not seen in KEN, which appear primarily on the torso, despite identical underlying somatic HRAS and KRAS mutations. This suggests that body site determines phenotype and is supported by a report of a contiguous linear nevoid lesion extending from the scalp to the neck with transition in clinical and histologic appearance from KEN on the upper back and neck to NS on the scalp (Waltz et al., 1999). The specific determinants of such site-specific phenotypes are unknown, though distinct epithelial-mesenchymal interactions are a possible cause.
Supplementary Material
Acknowledgments
We thank Gerald Goh, Emily Hast, Vincent Klump, Akdes Serin and Jade Wititsuwannakul for technical assistance, Ben Yu for careful discussion of data and potential mechanisms, and Murat Gunel for helpful conversations. JLL is the recipient of a NIH-NHLBI Medical Student Research Fellowship. This work was supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award to KAC who is also the recipient of a NIH/NIAMS K08 award (K08AR056305), and by the Yale Center for Mendelian Genomics (NIH U54 HG006504). This report was concurrently submitted with the publication of Bryan K. Sun, Andrea Saggini, Kavita Y. Sarin, Jinah Kim, Latanya Benjamin, Philip E. LeBoit, and Paul A. Khavari (2012) Mosaic activating RAS mutations in nevus sebaceus and nevus sebaceus syndrome.
Abbreviations used
- EN
epidermal nevi
- KEN
keratinocytic epidermal nevi
- NS
nevus sebaceus
- SNV
single nucleotide variation
- SNP
single nucleotide polymorphism
- LOH
loss of heterozygosity
- SCP
syringocystadenoma papilliferum
- TAA
tubular apocrine adenoma
- TB
trichoblastoma
- TL
trichilemmoma
Footnotes
Work was done in New Haven, Connecticut.
Conflict of Interest
The authors report no conflict of interest.
References
- Blaschko A. Die Nervenverteilung in der Haut in ihrer Beziehung zu den Erkrankungen der Haut. Braumüller, Wien-Leipzig 1901 [Google Scholar]
- Cribier B, Scrivener Y, Grosshans E. Tumors arising in nevus sebaceus: A study of 596 cases. J Am Acad Dermatol. 2000;42:263–8. doi: 10.1016/S0190-9622(00)90136-1. [DOI] [PubMed] [Google Scholar]
- Grewal T, Koese M, Tebar F, et al. Differential Regulation of RasGAPs in Cancer. Genes Cancer. 2011;2:288–97. doi: 10.1177/1947601911407330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groesser L, Herschberger E, Ruetten A, et al. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nature genetics. 2012 doi: 10.1038/ng.2316. [DOI] [PubMed] [Google Scholar]
- Hafner C, Toll A, Gantner S, et al. Keratinocytic epidermal nevi are associated with mosaic RAS mutations. J Med Genet. 2012;49:249–53. doi: 10.1136/jmedgenet-2011-100637. [DOI] [PubMed] [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–62. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Lapouge G, Youssef KK, Vokaer B, et al. Identifying the cellular origin of squamous skin tumors. Proc Natl Acad Sci U S A. 2011;108:7431–6. doi: 10.1073/pnas.1012720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MN, Landau JM, Goldberg LH. Nevus sebaceous revisited. Pediatric dermatology. 2012;29:15–23. doi: 10.1111/j.1525-1470.2011.01562.x. [DOI] [PubMed] [Google Scholar]
- Moss C, Larkins S, Stacey M, et al. Epidermal mosaicism and Blaschko’s lines. J Med Genet. 1993;30:752–5. doi: 10.1136/jmg.30.9.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Krishnaswami SR, Yu BD. Activated Kras alters epidermal homeostasis of mouse skin, resulting in redundant skin and defective hair cycling. The Journal of investigative dermatology. 2011;131:311–9. doi: 10.1038/jid.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon LM, Esterly NB. Epidermal and other congenital organoid nevi. Curr Probl Pediatr. 1975;6:1–56. doi: 10.1016/s0045-9380(75)80010-7. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–87. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Waltz KM, Helm KF, Billingsley EM. The Spectrum of Epidermal Nevi: A Case of Verrucous Epidermal Nevus Contiguous with Nevus Sebaceus. Pediatric dermatology. 1999;16:211–3. doi: 10.1046/j.1525-1470.1999.00056.x. [DOI] [PubMed] [Google Scholar]
- Xin H, Matt D, Qin JZ, et al. The sebaceous nevus: a nevus with deletions of the PTCH gene. Cancer Res. 1999;59:1834–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.