Abstract
Cellular membranes undergo continuous remodeling. Exocytosis and endocytosis, mitochondrial fusion and fission, entry of enveloped viruses into host cellsand release of the newly assembled virions, cell-to-cell fusion and cell division, and budding and fusion of transport carriers all proceed via topologically similar, but oppositely ordered, membrane rearrangements. The biophysical similarities and differences between membrane fusion and fission become more evident if we disregard the accompanying biological processes and consider only remodeling of the lipid bilayer. The forces that determine the bilayer propensity to undergo fusion or fission come from proteins and inmost cases from membrane-bound proteins. In this review, we consider the mechanistic principles underlying the fusion and fission reactions and discuss the current hypotheses on how specific proteins act in the two types of membrane remodeling.
Mechanics of fusion and fission
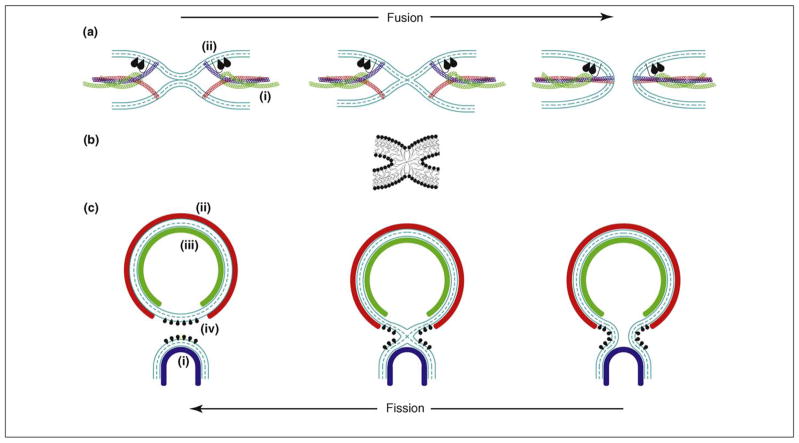
Membrane fusion occurs when two initially separate and apposed membranes merge into one by undergoing a sequence of intermediate transformations that seem to be conserved between disparate biological fusion reactions (Figure 1a) [1,2]. This membrane rearrangement begins with local merger of only the contacting monolayers of the two membranes, while the distal monolayers remain separate. The initial lipid bridge between the membranes is referred to as the fusion stalk (Figure 1b) and signifies the first stage of fusion, called hemifusion [1]. Stalk evolution ultimately leads to merger of the distal monolayers, resulting in the formation of a fusion pore that connects the volumes initially separated by the membranes and completes the membrane unification. The fusion pore must expand to a greater or smaller extent, depending on the specific biological context, for example, passage of small neurotransmitter molecules in the case of synaptic-vesicle exocytosis or a larger nucleocapsid in virus–cell fusion or the much larger nuclei in cell-to-cell fusion events.
Figure 1.
Pathways and protein-driven mechanisms of membrane remodeling. (a) Protein-mediated membrane fusion. The fusion reaction (from left to right) is driven by membrane curvature, which can be generated by (i) force transmission from the SNARE complex folding into a four-helix bundle through a sufficiently rigid helical link to the transmembrane domain [53] and/or (ii) shallow hydrophobic insertions of the C2 domains of synaptotagmin or Doc2 [21,22,32,33]. The fusion pathway consists of membrane dimpling leading to the generation of curvature stresses and the establishment of point-like intermembrane contacts (left panel), fusion stalk formation (middle panel), and fusion pore formation and expansion (right panel). (b) A membrane stalk, a common intermediate structure of fusion and fission. See [73] for details regarding the computation of the stalk configuration presented. (c) Protein-mediated membrane budding and fission. The fission reaction (from right to left) is driven by the generation of a strongly curved membrane neck whose elastic energy is relaxed as a result of membrane splitting. The neck formation and stressing can be mediated by (i) membrane adhesion on a dome-like protein scaffold formed by the ESCRT-III complex (blue) [60], and/or (ii) membrane scaffolding by the outer membrane coat (e.g. COPI, COPII and viral protein ectodomain coats, red) [3] and/or (iii) scaffolding by the inner membrane coat (e.g. viral matrix, core or capsid protein coat, green), and/or (iv) by the action of shallow hydrophobic insertions.
Membrane fission (Figure 1c) – division of an initially continuous membrane into two separate ones – proceeds via the formation of a membrane neck, which is reminiscent of a fusion pore except that it narrows rather than expands. Theoretical analysis [3] and a recent experimental study [4] substantiate a scenario in which fission begins with self-merger of the inner monolayer of the neck membrane, which generates a fission stalk analogous to the fusion stalk (Figure 1b,c). Subsequent self-merger of the outer monolayer of the membrane neck completes the fission process.
The fundamentally common feature of fusion and fission in these pathways is the formation of a membrane stalk at an intermediate stage of the reaction, which is followed by stalk decay. Obviously, stalk formation requires transient disruption of the membrane structure and hence is opposed by the powerful hydrophobic forces working to maintain continuity and integrity of any lipid assembly [5]. This leads to a currently open question about the transient structures preceding stalk formation. A candidate for such a structure in the case of fusion is a point-like protrusion characterized, according to estimations, by relatively low energy [6]. An alternative hypothesis [7], substantiated by recent numerical work [8], is that the pre-stalk fusion intermediate involves just one lipid molecule, which splays its two hydrocarbon chains such that they insert into opposing membranes, hence building a nascent lipid bridge between the membranes. In principle, a similar mechanism could work in the initial stages of the fission reaction. This chain-splaying mechanism has been demonstrated by numerical simulations under conditions of partial dehydration of the intermembrane contact, thus implying that the action of strong forces pushes the membranes together. The physical factors that facilitate these specific types of pre-stalk intermediates or other local membrane discontinuities should promote both fusion and fission reactions.
The evident distinction between fusion and fission is the reverse sequences of shapes adopted by the membranes and the opposite character of the overall topological transformation of the membrane surface. As a result of fission, the membrane splits into two smaller ones that are, on average, more strongly bent and characterized by greater curvatures. By contrast, as a result of fusion the merged membrane can partially relax the bending of the initial membranes by reducing the overall membrane curvature. This effect should be especially pronounced for fusion of small, and thus strongly bent, membrane compartments such as intracellular transport intermediates with a cross-section diameter of 50–100 nm. Hence, the forces favoring membrane bending should promote membrane fission, whereas the factors driving membrane unbending should have the opposite effect and support membrane fusion.
In addition, another geometric feature, membrane self-connectivity, changes in opposite directions as a result of fusion and fission. Fusion leads to unification of two initially separate membranes into a fully self-connected one. After fusion, the lipids and all membrane-bound molecules and molecular complexes can redistribute over the entire unified membrane area instead of being limited within one of the initial smaller membranes. By contrast, fission results in separation of one membrane into two unconnected membranes, thereby reducing the degree of membrane self-connectivity. Thus, the physical factors favoring membrane self-connectivity facilitate fusion, whereas fission is supported by forces that promote separation of the membrane surface into spatially disconnected compartments.
In this review, we discuss the mechanisms by which proteins implement these general mechanistic principles for membrane remodeling. We first formulate the energy requirements for any membrane remodeling reaction and then suggest that membrane curvature and the related elastic stresses are universal factors driving membrane fusion and fission. We overview the mechanisms of curvature generation and consider their realization by specific proteins in membrane remodeling.
Proteins as energy generators for membrane remodeling
Membrane remodeling, either by fusion or fission, can occur if two physical requirements are fulfilled. First, the process must be energetically favorable overall. The system free energy before remodeling has to be higher than that after, which means that remodeling must result in relaxation of the free energy. In other words, the remodeling process must go energetically ‘downhill’. Fulfillment of this requirement makes remodeling generally feasible. Second, the energies of the intermediate structures formed transiently in the course of remodeling and representing kinetic barriers must be low enough to be overcome by system thermal fluctuations within a biologically relevant time. Estimations based on experimental investigations of the electrical breakdown of membranes [9,10] and theoretical estimations [11] indicate that the feasible kinetic barriers constitute 40kBT or less (where kBT≈4×10−21 J is the product of the Boltzmann constant and the absolute temperature), which is equivalent to hydrolysis of a few ATP molecules.
Membrane remodeling is driven and controlled by proteins that provide the required energy. Although proteins could serve as building blocks for intermembrane connections that mediate membrane fusion or fission [12], experiments on diverse biological fusion reactions and modeling results indicate that, at least in most cases, proteins serve as indirect mediators rather than direct players in membrane remodeling [2,13]. Thus, we must consider how proteins can generate the conditions for bilayer remodeling by changing the structure and physical state of lipid bilayers. Proteins might act by modifying the lipid composition of membrane monolayers, thus resulting in membrane deformations and remodeling [14]. Specifically, it has been proposed that protein-driven segregation of phosphatidylinositol 4,5-biphosphate into domains with energetically unfavorable boundaries drives membrane fission in yeast endocytosis [15]. However, the majority of the mechanisms suggested in the literature have a different underlying idea. Proteins deform and generate elastic stresses within the membrane regions that are committed to undergo fusion or fission. Relaxation of the related elastic energy that results from remodeling provides the driving force for overall remodeling and lowers the intermediate energy barriers, thereby guaranteeing a fast rate of the reaction [16,17].
Analysis and theoretical substantiation of specific mechanisms based on this idea require the application of physics and mathematics, along with computational approaches (Box 1). When considering shape changes, the membrane and its leaflets can be regarded as continuous macroscopic surfaces and described by the theory of membrane elasticity, the central notion of which is the elastic energy of membrane monolayers. The generation of membrane discontinuity, by contrast, is a local event, which should be characterized on a microscopic and/or molecular level (Box 1).
Box 1. Membrane elastic energy.
The elastic energy that plays the major role in membrane remodeling is generated by three types of deformations: membrane bending, and stretching and tilting of the lipid hydrocarbon chains. Detailed reviews of this subject are available [13,19] and so here we highlight only a few of the major implications of membrane elasticity theory.
Bending energy depends on the curvature of the membrane surface (Figure I). The curvature describes the shape of each small membrane element which can be characterized by radii R1 and R2 of two arcs lying in the surface plane and oriented in two specific directions referred to as the principal directions [74]. The two principal curvatures are defined as the inverse radii c1=1/R1 and c2=1/R2. To describe the lipid bilayer shapes, we use the sum J=c1+c2 and the product K=c1· c2 of the principal curvatures, referred to as the total and Gaussian curvatures, respectively [75,76]. In general, both the total and Gaussian curvatures are different at different points of the membrane surface. Generation of both the total and Gaussian curvatures requires free energies (F), which, in simple cases, are given by and FK=κ̄ ∮ KdA, respectively, where integration is performed over the entire membrane area. The value k≈20kBT is the lipid bilayer bending modulus; κ̄ is the modulus of the Gaussian curvature, which, in most cases, is negative and can be substantially influenced by the lipid and protein compositions of the membrane.
The energy FB of the total curvature J depends on the membrane shape and thus its value varies at the different stages of membrane remodeling from the initial membrane deformations through the actual fusion or fission events. Fusion decreases the energy FBof the overall total curvature, whereas fission increases this energy.
The energy of the Gaussian curvature, FK, varies only with changes in membrane self-connectivity and thus remains constant during the initial deformation associated with fusion and fission. Membrane fusion results in FK variation by −4πκ̄, whereas fission results in an opposite energy change of 4πκ̄. This means that all changes in the membrane elastic properties resulting in increased (less negative) values of the Gaussian curvature modulus κ̄ would favor fusion and suppress fission, whereas decreased (more negative) values of κ̄ would promote fission and restrain fusion. The κ̄ value of a lipid bilayer depends on the lipid and protein compositions of the constituent monolayers through the tendency of each of the monolayers to bend spontaneously (see [77] and references therein).
The second relevant elastic energy of the membrane is associated with lateral tension that arises on membrane stretching and can drive expansion of the fusion pore [78,79]. The generation of lateral tension requires action on the membrane of external forces tending to expand the membrane area globally or locally.
The third elastic energy to be considered is the energy of tilting of the hydrocarbon chains of lipid molecules with respect to the membrane plane during fission and fusion stalk formation [80,81]. This deformation is required for packing of the lipid hydrocarbon chains in the middle of the stalk and contributes similarly to the energies of both fusion and fission stalks [3,73].
Figure I.
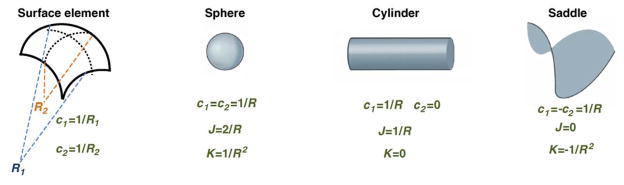
Geometric definition of membrane curvature and examples of basic shapes. The two principle curvatures of a surface element, c1 and c2, are defined as inverse radii of the two arcs that represent the surface cross-sections in two perpendicular directions called the principle directions [74]. Lipid bilayers having properties of two-dimensional fluids are described by the two combinations of the principle curvatures: their sum J=c1+c2, called the total curvature, and their product K=c1c2, called the Gaussian curvature. The basic curved shapes are a spherical shape for which c1=c2; a cylindrical shape with c1=0 such that K=0; and a saddle-like shape characterized by c1=–c2 and, hence, J=0.
Driving fusion by curvature generation
A common property of many proteins involved in endo- and exocytosis is their ability to strongly bend lipid bilayers [14,18,19]. Accordingly, an attractive idea is that specialized proteins drive membrane fusion through the generation of membrane curvature. Specifically, it has been proposed that curvature-producing proteins encircle a lipid bilayer spot and bend the membrane around it in a cylindrical or truncated conical belt (Figure 1a) [17,20–22]. As a result, the protein-free spot covering this belt bulges and adopts the shape of a spherical segment of large curvature (Figure 1a). This membrane bulge bridges the remaining gap between the membranes. The bent bilayer region at the top of the bulge is committed to fusion because its curvature and the related elastic energy relax gradually in the course of stalk and fusion pore formation [17]. There are two important requirements for this mechanism of membrane fusion. First, the membrane-bending proteins must remain at the periphery of the fusion site. If they were allowed into the bulging region, they would stabilize its curvature, cancel the tendency of this curvature to relax, and hence remove the driving force of the fusion reaction. Second, the curvature generated within this lipid bilayer spot must be sufficiently large. According to estimations [17] for curvature radii varying between 10 and 20 nm (i.e. two to three times smaller than the external radius of intracellular vesicles), the initially accumulated elastic energy, which is released in the course of fusion, is in the range between 10kBT and 20kBT. Such energies are comparable with those of fusion stalks and nascent fusion pores [13] and hence can considerably promote fusion.
Hydrophobic insertion (wedging) mechanism
Proteins can generate membrane curvature via different mechanisms (Figure 2) [18,19]. These include induction of lipid asymmetry of the membrane bilayer by flippases and lipid-modifying enzymes [14], molding of the membrane surface by rigid protein scaffolds [18,19,23–25], and insertion of hydrophobic protein domains into the lipid bilayer matrix [18,19,21,26,27]. The latter is likely to be the most common mechanism. The essence of this mechanism lies in expansion of the polar head region of one of the membrane monolayers by shallow insertions in its matrix of small hydrophobic or amphipathic protein domains [27]. The list of proteins proven to generate or with potential to generate large membrane curvatures via the hydrophobic insertion mechanism continues to increase. It includes proteins with amphipathic N-terminal α-helices such as epsins [26], small G proteins [28,29] and proteins containing bin– amphiphysin–Rvs with N-terminal amphipathic helices (N-BAR) domains [30,31]; proteins capable of inserting small hydrophobic loops into lipid monolayers, such as C2-domain containing synaptotagmin-1 [17,21,22,32] and DOC2 [33,34]; proteins with long and flexible hydrophobic domains that can lie shallowly under the monolayer surface, such as hairpin loops of reticulons and Yop1 proteins [35–38]; and dynamin family proteins whose plekstrin homology (PH) domains are anchored within lipid bilayers by small hydrophobic loops [39,40].
Figure 2.
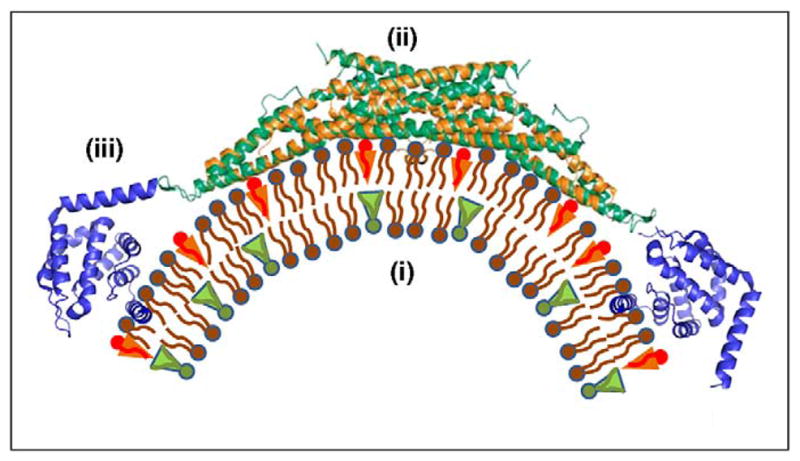
Mechanisms that generate membrane curvature. Factors producing membrane curvature include: (i) enrichment of the two membrane monolayers in lipid molecules of different effective shapes such that each monolayer has an intrinsic tendency to bend [14]; (ii) attachment to the membrane surface of intrinsically bent protein domains such as BAR domains [18,19], which mold the membranes into curved shapes; and (iii) shallow insertion into one of the membrane monolayers of hydrophobic or amphipathic protein domains, which splays the lipid molecules and cause local membrane bending [18,19,27].
It has been proposed that two of these proteins, synaptotagmin-1 [21,22,32] and DOC2 [33], which serve as Ca2+ sensors of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-dependent fusion, drive membrane fusion via the generation of a strongly curved membrane bulge [17]. Upon Ca2+ binding, small regions of the C2 domains of these proteins insert into the outer membrane leaflet to approximately the depth of the lipid glycerol backbones [41], thus facilitating the generation of membrane curvature with a radius of ~9 nm[21]. The suggested fusogenic action pathway of these proteins includes: (i) gathering of the proteins into a ring-like area around the fusion side by interaction with SNARE complexes; (ii) Ca2+-driven shallow embedding of the C2 domains into the lipid matrix and related bending of the lipid area containing the insertions into a cylindrical or truncated cone covered by a protein-free end cap of less than 10 nm in radius; and (iii) fusion of this end cap with the target membrane driven by relaxation of the bending elastic energy [17].
Other fusion proteins might also use the hydrophobic insertion mechanism to merge membranes by generating membrane curvature. Research on diverse viral fusion proteins has emphasized the functional importance of their relatively short (up to 30 residues, but often much shorter and sometimes discontinuous) amphipathic domains, referred to as fusion peptides (or fusion loops) [42–46]. Conformational changes in the fusion protein on its activation enable fusion peptides to reach and insert into the target membrane. Subsequently, the protein refolds into a rigid rod-like post-fusion conformation with two membrane- interacting regions of the protein, the fusion peptide and transmembrane domain, located at the same end of the rod. This refolding is thought to pull the viral and target membranes together [47,48]. However, the fusion peptide is clearly more than just a hold on a target membrane, because some amino acid substitutions within the fusion peptide inhibit fusion without lowering peptide–membrane binding. Moreover, synthetic peptides corresponding to the fusion peptide regions of viral fusion proteins induce fusion between lipid bilayers [43]. The fusogenic properties of the fusion peptides seem to depend on the ability of the peptide to obliquely insert into membranes [44]. Membrane deformations induced by fusion peptide insertion depend on the insertion depth and the specific conformation of the membrane-inserted peptide.
In addition, other amphipathic regions in the ectodomains of viral fusion proteins might interact with membranes under fusion conditions [49–52]. According to a recent analysis of the crystal structure of the extracellular domain of the gp41 HIV envelope protein subunit, conserved hydrophobic residues in the linker region connecting the ectodomain to the transmembrane domain are positioned for shallow insertion into the viral membrane (as opposed to fusion peptide insertion into the target membrane). Estimates suggest that these insertions could generate bulging of the viral membrane, thus facilitating its fusion [52].
Force transmission mechanism
Another possible mechanism of curvature generation at the membrane fusion site is suggested by a recent X-ray investigation of the assembly of neuronal SNARE complexes [53]. The SNARE complex consists of syntaxin and synaptobrevin, whose SNARE motifs are connected by linker regions to the C-terminal transmembrane domains spanning the two opposed membranes, and synaptosomal-associated protein (SNAP)25 anchored in the plasma membrane by means of palmitoyl chains. SNARE complex assembly begins at the N terminus and proceeds in a zipper-like fashion towards the transmembrane domains, thus forming a stable four-helix bundle, called the core SNARE complex, between the membranes. This zippering explains how generation of a SNARE complex brings the membranes together, but does not explain the mechanism of membrane fusion per se [53]. New results show that SNARE assembly tends to proceed beyond the core SNARE complex and folds into continuous helical structures that propagate through the linker region into the transmembrane domains. This finding suggests that the SNARE complex tends to adopt the conformation of a rigid rod, with the transmembrane domains of syntaxin and synaptobrevin in the same membrane. Because such a conformation is possible only as result of membrane fusion, and thus is a post-fusion complex, folding of the SNARE motifs and the membrane linker helices must develop forces that promote fusion. The generation of these forces can proceed according to the following scenario. Zippering of SNAREs into the core SNARE complex strongly bends the linker regions (Figure 1a). Propagation of the helical conformation into the linker regions renders them sufficiently rigid to strongly resist this bending. As a result, the linker regions tend to unbend by curving the two membranes towards each other, thus generating strongly bent membrane spots committed to fusion (Figure 1a).
Within this mechanism, the force that bends the membranes is generated by SNARE complex formation and is transmitted to the membrane through the rigidified linker regions of the protein. For this mechanism to be effective, the linker regions must become more rigid than the lipid bilayer. To date, however, studies examining the relationship between the effectiveness of SNARE-mediated fusion and the flexibility of the linker between the core SNARE and the transmembrane domain have yielded inconclusive results [54–58]. Hence, a thorough understanding of the feasibility of the force transmission mechanism of SNARE-mediated fusion requires additional experimentation.
It should be noted that the force transmission mechanism of membrane bending through the SNARE linker regions and the hydrophobic insertion mechanism by synaptotagmin or DOC2 C2 domains are completely compatible and could provide mutual reinforcement given that both use curvature generation as a fundamental feature of the fusion process.
Driving fission by curvature generation
During fission, bending energy accumulates owing to protein-driven narrowing of the membrane neck. It is thought that relaxation of this energy, resulting from splitting of the membrane neck into two separate membranes, drives fission [3].
Scaffolding mechanism of membrane fission
For some fission processes, the formation of a membrane neck seems to involve membrane scaffolding by protein complexes. For example, protein coats or scaffolds play an important role in the budding and release of newly assembled enveloped viruses. Interestingly, a major role in this budding–fission process can be played both by viral proteins (matrix, core or capsid) that assemble under cell membranes and by lattices of viral glycoprotein ectodomains that assemble at the outer leaflet of the plasma membrane [59]. Assembly of a rigid protein coat on the membrane surface can generate a membrane neck that emerges from the coat aperture (Figure 1c). Continuous self-assembly of the coat, accompanied by closure of its aperture, results in narrowing of the membrane neck, accumulation of the elastic stresses and ultimately in neck fission [3]. A similar scenario was recently suggested for membrane fission by endosomal sorting complex required for transport (ESCRT)-III complexes [60]. According to structural data [61], two ESCRT-III subunits, CHMP2 and CHMP3 (charged multivesicular body proteins) self-assemble into cylindrical structures with hemispherical dome-like end caps. Membrane attachment to the protein dome produces a narrowing membrane neck just above the dome (Figure 1c). The narrowness and the related elastic energy of the neck are determined by the affinity of the membrane for the protein dome. According to estimations, this affinity is sufficiently large to drive neck fission [60].
Membrane scaffolding also has been proposed for membrane fission driven by dynamin-1 (see [25,62] for reviews and [4,63] for more recent developments in this field). The discovery of dynamin self-assembly into helical structures on membrane surfaces and conformational changes of dynamin oligomers upon GTP hydrolysis have stimulated a series of mechanochemical models of dynamin action [62]. These models propose that the formation of helical dynamin oligomers scaffolds the membrane into a cylindrical shape, which loses its stability and undergoes fission as a result of narrowing and/or stretching of the dynamin helix resulting from GTP hydrolysis [64] and/or detachment of GDP–dynamin from the membrane surface [4].
Hydrophobic insertion mechanism of membrane fission
Recent studies of protein-driven membrane rearrangements support the hypothesis that insertion of their amphipathic and small hydrophobic domains into the membrane matrix constitutes the major factor used by many proteins, including the BAR-domain proteins and dynamin family proteins, for membrane fission.
The endophilin N-BAR domains form crescent-like dimers bound to membrane surfaces, mainly by inserting their amphipathic N-terminal helices into the lipid matrix [30,31]. Whereas N-BAR domain proteins normally shape membranes into long tubules, elevated concentrations of these proteins drive the formation of spherical vesicles of a few tens of nanometer in radius in a process that requires membrane scission [30]. N-BAR dimers do not undergo further oligomerization. Thus, membrane fission in this system cannot be related to membrane scaffolding by large protein assemblies, but should be driven by the mechanism by which N-BAR generates membrane bending, the hydrophobic insertion mechanism.
Recent studies suggest that the hydrophobic insertion mechanism also plays a primary role in dynamin-mediated fission. This is essentially distinct from mechanochemical models of dynamin action, which suggest that fission is driven by membrane stresses generated by scaffolds formed by oligomerized dynamin. Indeed, recent work indicates that long dynamin oligomers do not trigger fission [4]. In experiments on dynamin-mediated fission in tension-free membranes under physiological conditions of the constant presence of GTP, membrane fission occurs in the absence of long membrane tubules, thereby confirming that long dynamin spirals were not formed [63]. However, it has been proved that membrane insertion of the hydrophobic loops of the dynamin PH domains into the membrane matrix is critical for fission [40]. This factor was never taken into account in previous attempts to understand dynamin action. A role for a limited dynamin assembly [63] might be in increasing the local membrane concentration of the hydrophobic loops by enhancement of dynamin binding.
Based on these recent findings, we propose that insertion of amphipathic and hydrophobic protein domains plays a primary role in membrane fission driven by many proteins, including dynamin. This hypothesis can be supported by a straightforward physical mechanism for the role of membrane insertions in fission. Membrane fission can be stimulated if the membrane modulus of Gaussian curvature κ̄ adopts sufficiently negative values (Box 1). One of the factors producing negative κ̄ for a bilayer is a tendency of the membrane monolayers to bulge in the direction of the polar groups, which is described as a positive spontaneous curvature of the monolayers (e.g. [13]). The hydrophobic insertions are very effective in increasing the spontaneous curvature of membrane monolayers [27] and hence must drastically favor the membrane fission reaction by generating more negative κ̄ for the bilayer. Note that proteins providing the hydrophobic insertions must be bound to the outer monolayer of the membrane neck undergoing fission, because their presence on the inner monolayer would sterically interfere with monolayer self-merging.
Driving remodeling by membrane tension generation
Membrane remodeling can also be driven by tension generated in lipid bilayers by protein scaffolds. Although membrane deformations by protein coats have mostly been discussed for vesicle budding and fission, lateral organization of the proteins at the membrane surface into coat-like assemblies can also drive fusion [64]. To drive expansion of fusion pores, the fusion protein coat must have a tendency to develop a negative curvature and be much more rigid than the underlying lipid bilayer. To relieve elastic stresses, the protein coat deforms the membrane into a bulge primed for hemifusion and generates lateral tension that opens and expands the fusion pore until the pore reaches the dimension of the coat itself [64]. Many assumptions of the fusion coat hypothesis remain unsubstantiated. However, its most unexpected prediction, that fusion proteins located too far from the fusion site to be directly involved in fusion process might still control the fusion reaction by generating long-range membrane stresses, has been confirmed in some experimental systems [65,66].
Concluding remarks
The shared mechanistic principles of membrane rearrangements in fusion and fission and the functional complementarity of these processes raise the possibility that the same protein modules can be utilized to drive both types of membrane remodeling. Attempts to identify such universal membrane remodelers and to understand the principles of their organization on the membrane necessary for fusion and for fission are just beginning. The important challenge in these efforts will be to distinguish proteins that drive lipid rearrangements from proteins that operate upstream or downstream of the remodeling event or affect remodeling indirectly by regulating the action of other proteins.
Recent studies revealed an intriguing overlap between proteins controlling fusion and fission. Dynamin family members, which are key players in numerous membrane fission events, are also involved in fusion events [25]. Fusion and fission of the dynamic mitochondrial membranes are driven by dynamin family members [67], with Dnm1 and Drp1 serving as master regulators of membrane division [68] and Fzo1 and Mgm1 involved in fusion of the outer and inner mitochondrial membranes, respectively [69]. The dynamin-like GTPases Vps1 and atlastin are involved in fusion of yeast vacuoles [70] and homotypic endoplasmic reticulum (ER) fusion [71,72], respectively.
Recent data indicate that Eps15 homology (EH)-domain (EHD)-containing proteins, which comprise a class of highly conserved eukaryotic ATPases implicated in clathrin- independent endocytosis and recycling from endosomes, might generate membrane fission and fusion [24]. Structural studies have demonstrated that EHD2 deforms liposomes in a nucleotide-independent manner into 20-nm-diameter tubules by oligomerization into ring-like structures. Frequent observations of a complex network of connected tubules with an extensive surface area imply that considerable EHD2-driven fusion occurs between tubulated liposomes [24]. Experiments in which mutant EHD2 was overexpressed in HeLa cells suggested that EHD2-mediated ATP hydrolysis is involved in the breakdown of tubular structures in vivo and thus drives membrane fission [24].
Future research addressing the specific mechanisms by which structurally related proteins can drive the two oppositely directed reactions of membrane remodeling, along with advances in understanding the molecular mechanisms of action of specific fusion and fission proteins and the physics of lipid bilayer rearrangements, should provide novel approaches for controlling and directing transformations of cell membranes in normal and pathological conditions.
Acknowledgments
Financial support for MMK from the Israel Science Foundation (ISF) and Marie Curie Network “Virus Entry”, for HMM from the Medical Research Council, UK, and for LVC from the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health and NIAID, and a National Institutes of Health Intramural Biodefense Research grant is gratefully acknowledged.
References
- 1.Chernomordik LV, Kozlov MM. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 2.Sapir A, et al. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev Cell. 2008;14:11–21. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozlovsky Y, Kozlov M. Membrane fission: model for intermediate structures. Biophys J. 2003;85:85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashkirov PV, et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanford C. The hydrophobic effect: formation of micelles and biological membranes. Wiley & Sons; 1973. [Google Scholar]
- 6.Efrat A, et al. Point-like protrusion as a prestalk intermediate in membrane fusion pathway. Biophys J. 2007;92:L61–63. doi: 10.1529/biophysj.106.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holopainen JM, et al. Evidence for the extended phospholipid conformation in membrane fusion and hemifusion. Biophys J. 1999;76:2111–2120. doi: 10.1016/S0006-3495(99)77367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smirnova YG, et al. Solvent-exposed tails as pre-stalk transition states for membrane fusion at low hydration. J Am Chem Soc. 2010;132:6710–6718. doi: 10.1021/ja910050x. [DOI] [PubMed] [Google Scholar]
- 9.Abidor IG, et al. Electrical breakdown of BLM: main experimental facts and their qualitative discussion. Bioelectrochem Bioenerg. 1979;6:37–52. [Google Scholar]
- 10.Weaver JC, Mintzer RA. Decreased bilayer stability due to transmembrane potentials. Phys Lett. 1981;86A:57–59. [Google Scholar]
- 11.Kuzmin PI, et al. A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci U S A. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson MB, Chapman ER. The fusion pores of Ca2+- triggered exocytosis. Nat Struct Mol Biol. 2008;15:684–689. doi: 10.1038/nsmb.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chernomordik LV, Kozlov MM. Protein–lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 14.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010 doi: 10.1016/ j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, et al. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlov MM. Fission of biological membranes: interplay between dynamin and lipids. Traffic. 2001;2:51–65. doi: 10.1034/j.1600-0854.2001.020107.x. [DOI] [PubMed] [Google Scholar]
- 17.McMahon HT, et al. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 18.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 19.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 20.Kozlov MM, Chernomordik LV. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys J. 1998;75:1384–1396. doi: 10.1016/S0006-3495(98)74056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens S, et al. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 22.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 23.Frost A, et al. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daumke O, et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 25.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 26.Ford MG, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 27.Campelo F, et al. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95:2325– 2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MC, et al. Sar1p N-terminal helix initiates membrane curvature and completes the fission of aCOPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Lundmark R, et al. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J. 2008;414:189–194. doi: 10.1042/BJ20081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallop JL, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 32.Hui E, et al. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca2+-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groffen AJ, et al. Doc2b is a high-affinity Ca2+ sensor for spontaneous neurotransmitter release. Science. 2010;327:1614–1618. doi: 10.1126/science.1183765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedrich R, et al. DOC2B, C2 domains, and calcium: a tale of intricate interactions. Mol Neurobiol. 2010;41:42–51. doi: 10.1007/s12035-009-8094-8. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 36.Shibata Y, et al. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 37.Shibata Y, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voeltz GK, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 39.Burger KN, et al. Dynamin is membrane-active: lipid insertion is induced by phosphoinositides and phosphatidic acid. Biochemistry. 2000;39:12485–12493. doi: 10.1021/bi000971r. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran R, et al. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009;20:4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrick DZ, et al. Position of synaptotagmin I at the membrane interface: cooperative interactions of tandem C2 domains. Biochemistry. 2006;45:9668–9674. doi: 10.1021/bi060874j. [DOI] [PubMed] [Google Scholar]
- 42.Lai AL, et al. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 43.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 44.Charloteaux B, et al. The “Tilted Peptide Theory” links membrane insertion properties and fusogenicity of viral fusion peptides. Protein Pept Lett. 2009;16:718–725. doi: 10.2174/092986609788681724. [DOI] [PubMed] [Google Scholar]
- 45.Qiang W, et al. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc Natl Acad Sci U S A. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roche S, et al. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 47.Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weissenhorn W, et al. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nieva JL, Suarez T. Hydrophobic-at-interface regions in viral fusion protein ectodomains. Biosci Rep. 2000;20:519–533. doi: 10.1023/a:1010458904487. [DOI] [PubMed] [Google Scholar]
- 50.Sackett K, Shai Y. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochemistry. 2002;41:4678–4685. doi: 10.1021/bi0255322. [DOI] [PubMed] [Google Scholar]
- 51.Spadaccini R, et al. Structural characterization of the transmembrane proximal region of the hepatitis C virus E1 glycoprotein. Biochim Biophys Acta. 2010;1798:344–353. doi: 10.1016/j.bbamem.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 52.Buzon V, et al. Crystal structure of a late fusion intermediate of HIV-1 gp41. PLoS Pathog. 2010;6(5):e1000880. doi: 10.1371/journal.ppat. 1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein A, et al. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deak F, et al. Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J Neurosci. 2006;26:6668–6676. doi: 10.1523/JNEUROSCI.5272-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kesavan J, et al. v-SNARE actions during Ca2+-triggered exocytosis. Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 56.McNew JA, et al. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 57.Van Komen JS, et al. The polybasic juxtamembrane region of Sso1p is required for SNARE function in vivo. Eukaryot Cell. 2005;4:2017– 2028. doi: 10.1128/EC.4.12.2017-2028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, et al. Functional analysis of conserved structural elements in yeast syntaxin Vam3p. J Biol Chem. 2001;276:28598–28605. doi: 10.1074/jbc.M101644200. [DOI] [PubMed] [Google Scholar]
- 59.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221– 232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabrikant G, et al. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sever S, et al. Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic. 2000;1:385–392. doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- 63.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozlov MM, Chernomordik LV. The protein coat in membrane fusion: lessons from fission. Traffic. 2002;3:256–267. doi: 10.1034/j.1600-0854.2002.030403.x. [DOI] [PubMed] [Google Scholar]
- 65.Leikina E, et al. Influenza hemagglutinins outside of the contact zone are necessary for fusion pore expansion. J Biol Chem. 2004;279:26526–26532. doi: 10.1074/jbc.M401883200. [DOI] [PubMed] [Google Scholar]
- 66.Lenz O, et al. Trimeric membrane-anchored gp41 inhibits HIV membrane fusion. J Biol Chem. 2005;280:4095–4101. doi: 10.1074/jbc.M411088200. [DOI] [PubMed] [Google Scholar]
- 67.Hoppins S, et al. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 68.Lackner LL, Nunnari JM. The molecular mechanism and cellular functions of mitochondrial division. Biochim Biophys Acta. 2009;1792:1138–1144. doi: 10.1016/j.bbadis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta. 2009;1793:20–26. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 70.Peters C, et al. Mutual control of membrane fission and fusion proteins. Cell. 2004;119:667–678. doi: 10.1016/j.cell.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 71.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 73.Kozlovsky Y, Kozlov M. Stalk model of membrane fusion: solution of energy crisis. Biophys J. 2002;88:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spivak M. A Comprehensive Introduction to Differential Geometry. Brandeis University; 1970. [Google Scholar]
- 75.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch. 1973;28c:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 76.Helfrich W. Elasticity and thermal undulations of fluid films of amphiphiles. In: Charvolin J, et al., editors. Liquids at Interfaces. Proceedings of the Les Houches Summer School, Session XLVIII. Elsevier; 1990. pp. 212–237. [Google Scholar]
- 77.Siegel DP, Kozlov MM. The Gaussian curvature elastic modulus of N-monomethylated dioleoylphosphatidylethanolamine: relevance to membrane fusion and lipid phase behavior. Biophys J. 2004;87:366–374. doi: 10.1529/biophysj.104.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Safran SA, et al. Polymer-induced membrane contraction, phase separation, and fusion via Marangoni flow. Biophys J. 2001;81:659–666. doi: 10.1016/S0006-3495(01)75730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shillcock JC, Lipowsky R. Tension-induced fusion of bilayer membranes and vesicles. Nat Mater. 2005;4:225–228. doi: 10.1038/nmat1333. [DOI] [PubMed] [Google Scholar]
- 80.Hamm M, Kozlov M. Tilt model of inverted amphiphilic mesophases. Eur Phys J B. 1998;6:519–528. [Google Scholar]
- 81.Hamm M, Kozlov M. Elastic energy of tilt and bending of fluid membranes. Eur Phys J E. 2000;3:323–335. [Google Scholar]