Abstract
Reactive oxygen species induce oxidative damage in DNA precursors, i.e. dNTPs, leading to point mutations upon incorporation. Escherichia coli mutT strains, deficient in the activity hydrolysing 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate (8-oxo-dGTP), display more than a 100-fold higher spontaneous mutation frequency over the wild-type strain. 8-oxo-dGTP induces A to C transversions when misincorporated opposite template A. Here, we report that DNA pol III incorporates 8-oxo-dGTP ≍ 20 times more efficiently opposite template A compared with template C. Single, double or triple deletions of pol I, pol II, pol IV or pol V had modest effects on the mutT mutator phenotype. Only the deletion of all four polymerases led to a 70% reduction of the mutator phenotype. While pol III may account for nearly all 8-oxo-dGTP incorporation opposite template A, it only extends ≍ 30% of them, the remaining 70% being extended by the combined action of pol I, pol II, pol IV or pol V. The unique property of pol III, a C-family DNA polymerase present only in eubacteria, to preferentially incorporate 8-oxo-dGTP opposite template A during replication might explain the high spontaneous mutation frequency in E. coli mutT compared with the mammalian counterparts lacking the 8-oxo-dGTP hydrolysing activities.
Introduction
Excess oxidation is a major threat to the genomic integrity of most living organisms. Reactive oxygen species (ROS) are produced by normal cellular respiration, cellular injury or by exposure to environmental carcinogens and radiation. ROS generate a variety of altered purines and pyrimidines in DNA (Bjelland and Seeberg, 2003; Kamiya, 2003), thereby playing important roles in mutagenesis, carcinogenesis and ageing (Ames, 1983; Jackson and Loeb, 2001). It should be emphasized, however, that oxidized bases in DNA are introduced not only by direct oxidation of DNA but also by incorporation of oxidized deoxynucleoside triphosphates (dNTPs) into DNA by DNA polymerases (pols) (Michaels and Miller, 1992; Sekiguchi and Tsuzuki, 2002; Nakabeppu et al., 2006; Katafuchi and Nohmi, 2010).
7,8-Dihydro-8-oxo-dGTP (8-oxo-dGTP), a major form of oxidized dGTP in the cellular nucleotide pool, is a mutagenic substrate for DNA synthesis and the incorporation results in A:T to C:G mutations (Treffers et al., 1954; Yanofsky et al., 1966; Akiyama et al., 1989; Tajiri et al., 1995). When incorporated opposite A in the template DNA, 8-oxo-G can pair with incoming dCMP in the next round of DNA replication, then causing A:T to C:G mutations (Michaels and Miller, 1992; Kasai, 2002). To counteract the mutagenic 8-oxo-dGTP, Escherichia coli possesses a sanitizing enzyme, i.e. MutT, to hydrolyse 8-oxo-dGTP to the monophosphate form. When the mutT gene is inactivated, the mutation frequency of A:T to C:G transversions increases more than a 100-fold over the wild-type level (Yanofsky et al., 1966; Maki and Sekiguchi, 1992; Fowler et al., 2003). The high spontaneous A:T to C:G mutations in the mutT strain are almost completely suppressed when the mutT cells are cultured in anaerobic conditions, indicating the essential role of oxygen in the mutagenesis (Fowler et al., 1994; Sakai et al., 2006; Setoyama et al., 2011).
In mammals including humans, enzymes that possess similar activities to E. coli MutT are identified (Mo et al., 1992). Expression of human MTH1 (mutT homologue-1) cDNA in E. coli mutT strain significantly suppresses the frequency of spontaneous mutations (Sakumi et al., 1993; Furuichi et al., 1994). Suppressive effects are also observed when mouse or rat cDNA is expressed in the mutT cells (Cai et al., 1995; Kakuma et al., 1995). These observations imply that the mammalian proteins may sanitize the nucleotide pools in the organisms, thereby reducing the spontaneous mutagenesis and carcinogenesis. In fact, deletion of the Mth1 gene results in high frequency of tumour formation in several organs of mice (Tsuzuki et al., 2001). However, deletion of the mouse Mth1 gene enhances the spontaneous mutation frequency only twofold, which is a striking difference from the strong mutator effects of E. coli mutT (Egashira et al., 2002). Although mammalian cells possess more than one enzyme to sanitize oxidized nucleotides (Bessman et al., 1996; Cai et al., 2003; Ishibashi et al., 2003), there may be other reasons to account for the marked difference in spontaneous mutagenesis between E. coli mutT and the mammalian counterparts.
In this study, we explored the potential involvement of the different E. coli DNA polymerases in the mutT mutator phenotype. E. coli possesses five pols, i.e. pol I (A family), pol II (B family), pol III (C family), pol IV (Y family) and pol V (Y family) (Nohmi, 2006), and pol III holoenzyme (pol III HE) is mainly responsible for the chromosome replication (McHenry, 2011). Pol III HE is a large dimeric complex, which is composed of pol III core complex including ε proofreading subunit, sliding clamp (β subunit) and the clamp loader (McHenry, 2011). Although A-, B- and Y-family pols are present in mammals, C-family pols are present only in eubacteria (Ito and Braithwaite, 1991; Filee et al., 2002). We disrupted the genes encoding pols I, II, IV and V, and characterized their mutator phenotypes. We also examined the biochemical properties of pol III*, the HE without β subunit (McHenry, 1988), incorporating 8-oxo-dGTP into DNA and effects of addition of β subunit on extension reaction upon incorporation of 8-oxo-dGTP by pol III* in vitro. Our results indicate that pol III HE may be responsible for the misincorporation of 8-oxo-dGTP into DNA and suggest that the erroneous nature of pol III HE uniquely present in bacteria might account for the strong mutator effects of mutT in E. coli.
Results
Deletion of the genes encoding pols in a mutT background
To examine what pols are involved in the high spontaneous mutations in a mutT background, we deleted the genes encoding pol I (polA), pol II (polB), pol IV (dinB) or pol V (umuDC) in a mutT background. Deletion of mutT increased median frequency of rifampicin-resistant mutations more than 100 times without any damaging treatments to DNA (Fig. 1). None of single deletions of the pol genes reduced the median frequency of the mutT-deficient strain. Since pols IV and V preferentially incorporate 8-oxo-dGTP opposite template A in vitro (Yamada et al., 2006), we deleted both the dinB and umuDC genes and examined the spontaneous mutation frequency. However, the deletion of genes encoding two Y-family pols did not decrease the median frequency. We also deleted both the polA and dinB genes and examined the mutation frequency. Although pol I Klenow fragment (KF) and pol IV possess ability to extend primer DNA having 8-oxo-dGMP at the termini in vitro (see below and Fig. S3), deletion of the polA and dinB genes did not reduce the mutator phenotype substantially. Interestingly, when we deleted polA, umuDC and either polB or dinB, the spontaneous mutation frequency decreased by 40%. When we deleted all four pol genes, i.e. polA, polB, dinB and umuDC, the mutation frequency decreased by 70%. It should be emphasized, however, that the resulting penta mutant YG6376, i.e. ΔmutT, ΔpolA, ΔpolB, ΔdinB and ΔumuDC, still manifested more than 50 times higher spontaneous mutation frequency than the wild-type strain AB1157 (74 ± 20 × 10−8, YG6376 versus 1.3 ± 0.3 × 10−8, AB1157). These results suggest that pol III plays an important role in high spontaneous mutations in the mutT background, and also that other pols might have additional and redundant roles in the mutagenesis.
Fig. 1.
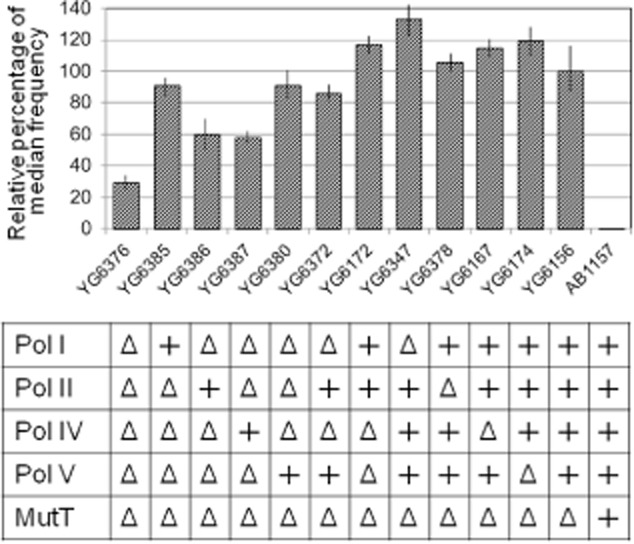
Mutation frequency of mutT strains with deficiency of DNA polymerase(s). Relative values (percentage) and the standard deviations of median frequency of rifampicin resistance mutations of mutT derivatives of E. coli are presented. Mutation assays were conducted at 30°C for strains with ΔKF and at 37°C for the other strains. The frequencies of strain YG6156 (ΔmutT) at 30°C and 37°C were set as 100%. The average mutation frequencies of YG6156 were 184 ± 58 × 10−8 at 30°C (n = 12) and 185 ± 57 × 10−8 at 37°C (n = 9). The mutation frequency of AB1157 at 37°C (the wild-type strain) was 1.3 ± 0.3 × 10−8 (n = 3). n represents number of repeated experiments. Mutation frequencies of other strains are presented in Table S1. The table under the graph indicates which polymerases are deficient (Δ) and proficient (+) in each strain.
Incorporation of 8-oxo-dGTP into DNA by pol III* in vitro
Next, we examined the specificity of pol III* incorporating 8-oxo-dGTP into DNA in vitro (Fig. 2). Pol III* preferentially incorporated 8-oxo-dGTP opposite template A in DNA. To examine the specificity quantitatively, we conducted kinetic analyses incorporating 8-oxo-dGTP into DNA (Fig. S1). Pol III* incorporated 8-oxo-dGTP opposite template A at concentration range of 8-oxo-dGTP from 0.05 to 10 μM (Fig. S1A). In contrast, pol III* incorporated 8-oxo-dGTP opposite template C at higher concentrations of 8-oxo-dGTP, i.e. 10 to 500 μM (Fig. S1B). The finc values for incorporation, i.e. the ratio between efficiency (Vmax/Km) of incorporation of 8-oxo-dGTP and that of incorporation of normal dNTP, were 5.6 × 10−2 and 2.9 × 10−3, respectively, opposite template A and template C (Table 1). It indicates that pol III* incorporates 8-oxo-dGTP opposite template A about 20 times more efficiently than opposite template C. It is remarkable that the apparent Km value for incorporation of 8-oxo-dGTP opposite template A was 2.5 μM, which is similar to the values for incorporation of normal dTTP and dGTP opposite templates A and C respectively (1.6 μM and 3.2 μM). The apparent Km value for incorporation of 8-oxo-dGTP opposite template C was 212 μM.
Fig. 2.
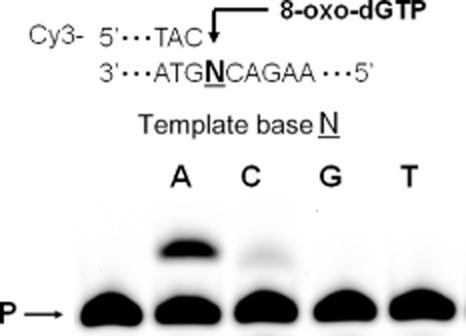
Incorporation of 8-oxo-dGTP by pol III*. The Cy3-labelled 18-mer primer/36-mer template (sequences 1, 0.1 μM) was treated with pol III* (1 nM) in the presence of 100 μM 8-oxo-dGTP. The reaction mixtures were incubated at room temperature for 1 min. The samples were analysed by denaturing polyacrylamide gel electrophoresis and visualized by the Molecular Imager as described in Experimental procedures. The alphabets shown in the figure indicate as follows: N, template base; A, adenine; C, cytosine; G, guanine; T, thymine; P, primer.
Table 1.
Kinetic parameters for 8-oxo-dGTP insertion catalysed by pol III*
| Template base/dNTP | Km (μM) | Relative Vmax | Vmax/Km | finc |
|---|---|---|---|---|
| A/dTTP | 1.6 ± 0.4 | 2.9 ± 0.23 | 1.8 | 1 |
| A/8-oxo-dGTP | 2.5 ± 0.82 | 0.26 ± 0.03 | 0.1 | 0.056 |
| C/dGTP | 3.2 ± 0.72 | 5.6 ± 0.47 | 1.75 | 1 |
| C/8-oxo-dGTP | 212 ± 45.7 | 1.1 ± 0.1 | 0.005 | 0.0029 |
Excision of 8-oxo-dG at the end of the primer
The incorporated 8-oxo-dG opposite template A forms a mismatch, which is usually excised as an improper base by proofreading activities of pols. Thus, we examined whether incorporated 8-oxo-dGMP is excised by the proofreading activity of pol III* (Fig. 3A). Strikingly, 8-oxo-dGMP was not excised from the primer DNA regardless of the pairing template base of C or A efficiently. In contrast, pol III* effectively excised terminal normal dGMP incorrectly pairing with template A, and even correctly pairing with template C. When we plotted percentage of digestion of primer DNA periodically, it became evident that the order of primer most rapidly digested was primer having G/A mismatch at the termini > primer having G/C > primer having 8-oxo-G/A = primer having 8-oxo-G/C (Fig. 3B). Single-stranded DNA having 8-oxo-dGMP at the termini was rapidly degraded by pol III* as well as that having normal dGMP (Fig. 3C). For comparison, we conducted similar assays with other enzymes, i.e. T7 pol, E. coli pol I (KF) and exonuclease III. T7 pol displayed similar digesting patterns to those with pol III*. It could not effectively excise 8-oxo-dGMP at primer termini regardless of the template bases although it excised terminal normal dGMP incorrectly pairing with template A and less effectively correctly pairing normal dGMP with template C. It digested single-stranded DNA having 8-oxo-dGMP at the termini effectively. Pol I (KF) poorly excised 8-oxo-dGMP at primer termini as pol III* and T7 pol. In contrast to pol III* and T7 pol, pol I (KF) did not digest single-stranded DNA having 8-oxo-dGMP at the termini. Exonuclease III digested primer DNA having terminal 8-oxo-dGMP as well as normal dGMP. It did not digest single-stranded DNA regardless of the presence or the absence of 8-oxo-dGMP at the termini.
Fig. 3.
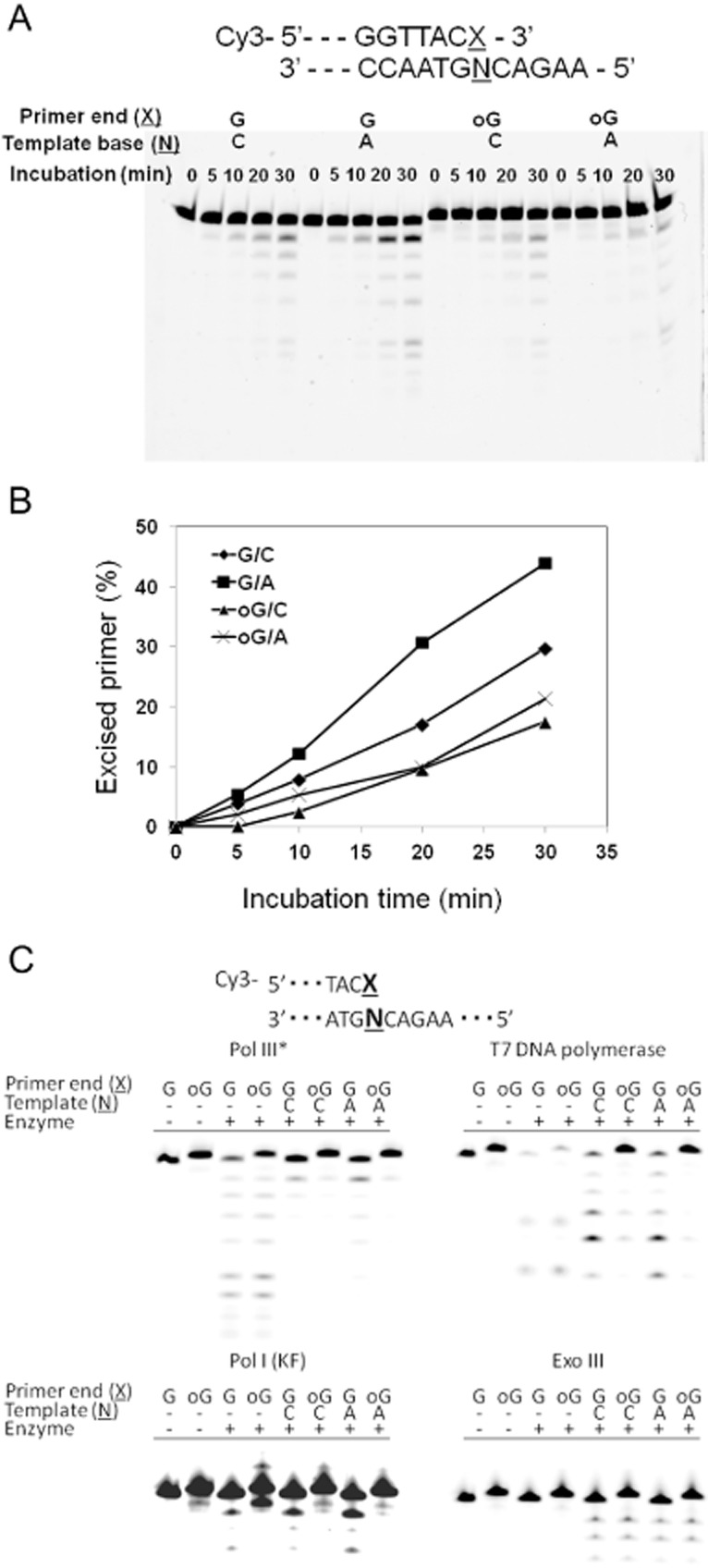
A. Exonuclease digestion of primers by pol III*. The Cy3-labelled 19-mer primer having G or 8-oxo-G at the 3′-termini/36-mer template having C or A at the position N (0.1 μM) were incubated with pol III* (1 nM) for 5, 10, 20 or 30 min at 25°C. The products were analysed by denaturing polyacrylamide gel electrophoresis and visualized by the Molecular Imager. oG represents 8-oxo-G.
B. Time course of digestion of primers by pol III*. Four types of primer/template DNA having G/C, G/A, 8-oxo-G/C or 8-oxo-G/A at the termini were incubated with pol III* (1 nM) for 5, 10, 20 or 30 min at 25°C and the percentage of the digested primer DNA was plotted.
C. Excision of 8-oxo-dGMP at the end of the primer by pol III*, T7 pol, pol I (KF) and exo III. The 19-mer primer/36-mer template DNA (0.1 μM) or the 19-mer primer DNA alone (0.1 μM) was incubated with pol III* (1 nM) at room temperature, T7 pol (0.0001 unit μl−1), pol I (KF)(0.001 unit μl−1) or exo III (0.0001 unit μl−1) at 37°C for 10 min without dNTP. The products were analysed as described in the legend to A.
Extension from dG or 8-oxo-dG at the end of a primer
Primer DNA having incorporated 8-oxo-dGMP at the termini has to be extended. Otherwise, it will induce DNA strand breaks. Next, we examined whether primer DNA having 8-oxo-dGMP at the termini was extended by pol III* in vitro. We also examined the effects of addition of the β subunit to pol III* on the extension reactions. For this purpose, we prepared template DNA having biotin/streptavidin at both ends (Fig. S2). The terminal streptavidin prevents the β subunit from falling off from the template/primer DNA. In standing start experiments, pol III* extended primer DNA having 8-oxo-dG at the termini and the extension was substantially enhanced by the addition of the β subunit (Fig. 4). The primer/template DNA having 8-oxo-dGMP pairing with template A appeared to be slightly more effectively extended compared with the primer/template DNA having 8-oxo-dGMP pairing with template C. In running start experiments where 8-oxo-dGMP was incorporated into DNA during DNA synthesis, pol III* extended primer DNA upon incorporation of 8-oxo-dGMP opposite template A (Fig. 5). In these reactions, addition of the β subunit substantially enhanced the extension reactions. It should be noted, however, that the extension reactions upon incorporation of 8-oxo-dGMP opposite template A were less efficient compared with the reactions upon incorporation of dTMP opposite template A even in the presence of the β subunit. As controls, we also examined the extension activity of pol I (KF) and pol IV with primers having 8-oxo-dG at the termini (Fig. S3). Pol I (KF) displayed potent extension activity with primers having terminal 8-oxo-dGMP pairing with template A or C. The primer DNA having 8-oxo-dGMP pairing with template A was more effectively extended by pol I (KF) than primer DNA having normal G pairing with template A. Pol IV exhibited moderate extension activity with primer DNA having 8-oxo-dGMP pairing with template A or C.
Fig. 4.

Standing start extension of primers having 8-oxo-G at the termini by pol III* with or without β clamp. Cy3-labelled 35-mer primer (X = T, G or 8-oxo-G) with 100-mer template DNA (N = A or C) having biotin/streptavidin at both ends (20 nM) was incubated with pol III* (10 nM) and β clamp (0, 10 or 100 nM) in the presence of 100 μM dNTPs. The reaction mixtures were incubated at room temperature for 1 min. The samples were analysed by denaturing polyacrylamide gel electrophoresis and visualized by the Molecular Imager. The alphabets shown in the figure represent: X, primer terminal base; N, template base; A, adenine; C, cytosine; G, guanine; T, thymine; oG, 8-oxo-G. The arrow indicates the position of primer. Although we have purified the primer DNA, there appears shorter primer DNAs, which were present below the position of the primer.
Fig. 5.
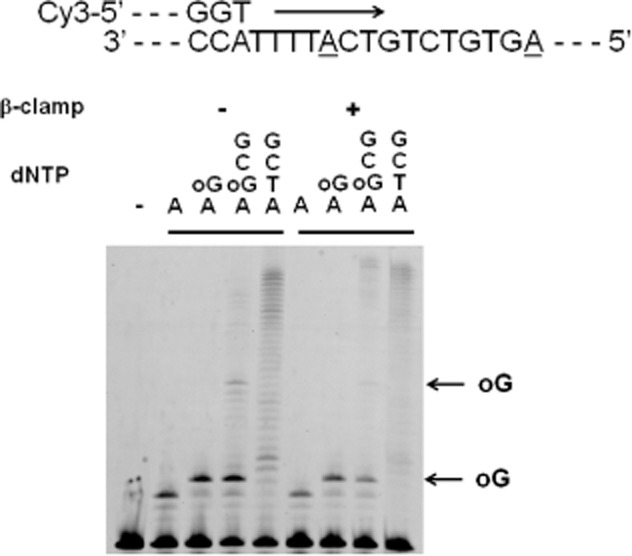
Incorporation of 8-oxo-dGTP and extension by pol III* with or without β clamp under running start conditions. The 30-mer primer/100-mer streptavidin bound template (sequences 2, 20 nM) were incubated with pol III* (10 nM) with or without β clamp (100 nM) in the presence of indicated dNTPs (100 μM each) for 1 min at 25°C. The samples were analysed by denaturing polyacrylamide gel electrophoresis and visualized by the Molecular Imager. The alphabets shown in the figure represent: A, dATP; C, dCTP; G, dGTP; T, dTTP; oG, 8-oxo-dGTP.
Discussion
dNTP pool and DNA are continuously exposed to a variety of exogenous and endogenous genotoxic agents, including ROS, and incorporation of oxidized dNTPs into DNA is a source of spontaneous mutagenesis and carcinogenesis (Ames and Gold, 1991). Here, we provided genetic and biochemical evidence that a replicative pol of E. coli, i.e. pol III HE, may be involved in oxidative mutagenesis through misincorporation of an oxidized nucleotide, i.e. 8-oxo-dGTP, during DNA synthesis in the mutT background. Although deletion of the genes encoding pol I, pol II, pol IV and pol V reduced the mutation frequency by more than 50%, the resulting strain YG6376 having pol III alone exhibited more than 50 times higher spontaneous mutation frequency in the mutT background than the wild-type strain (Fig. 1 and Table S1). Pol III* incorporated 8-oxo-dGTP opposite template A about 20 times more effectively than opposite template C in vitro (Fig. 2 and Table 1). Genetic analyses also suggest that 8-oxo-dGTP is preferentially incorporated opposite template A in the mutT background in vivo (Fowler et al., 2003). This biased specificity incorporating 8-oxo-dGTP opposite template A is reminiscent of that of X-family or Y-family pols involved in DNA repair and translesion DNA synthesis (Katafuchi and Nohmi, 2010). Mammalian pol β, a representative of X-family pols, incorporates 8-oxo-dGTP opposite template A and opposite template C at a ratio of 24:1 (Miller et al., 2000). Human pol η, a Y-family pol, preferentially incorporates 8-oxo-dGTP opposite template A at 60% efficiency of normal dTTP incorporation (Shimizu et al., 2007). Interestingly, α subunit, the catalytic subunit of pol III HE of E. coli and Thermus aquaticus, is structurally related to mammalian pol β, but not to mammalian replicative pols such as pol δ or pol ε, which are B-family enzymes (Bailey et al., 2006; Lamers et al., 2006). Therefore, we speculate that the structural similarity of the α subunit of pol III HE to mammalian pol β might be the structural basis for the preferential incorporation of 8-oxo-dGTP opposite A in template DNA.
In general, 8-oxo-dGTP is a poor substrate for most of pols (Nohmi et al., 2005; Katafuchi and Nohmi, 2010). For example, the efficiency of incorporation of 8-oxo-dGTP by pol δ involved in the chromosome replication in mammalian cells is more than 104-fold lower than that of incorporation of normal dGTP, and the enzyme prefers to incorporate 8-oxo-dGTP opposite template C (Einolf and Guengerich, 2001). 8-oxo-dGTP is poorly incorporated into DNA by T7 pol exo−, HIV reverse transcriptase, E. coli pol II and pol I (KF) exo− as well (Einolf et al., 1998). In contrast, pol III* incorporates 8-oxo-dGTP opposite template A at about 5% efficiency of normal dTTP incorporation (Table 1). We suggest that the erroneous and efficient incorporation of 8-oxo-dGTP opposite template A by pol III HE may account for the extremely high spontaneous mutations in the mutT mutants of E. coli.
At first, we expected that pol IV and pol V might be responsible for the misincorporation of 8-oxo-dGTP into DNA during DNA replication in the mutT background. This is because pol IV and pol V are involved in mutagenesis through incorporation of oxidized dNTPs into DNA in a ΔsodΔfur background of E. coli (Yamada et al., 2006). In the mutants, intracellular ROS levels are extremely elevated, and SOS responses are fully induced (Nunoshiba et al., 1999; 2002). Therefore, expression levels of pol IV and pol V are highly elevated. In contrast, in the mutT mutants, no SOS responses are induced (Janion et al., 2003). Thus, it is expected that the expression of pol IV and pol V is constitutive levels. The different status of SOS induction might explain the different contribution of SOS-inducible Y-family pols to oxidative mutagenesis in ΔsodΔfur and mutT strains although oxidized dNTPs are deeply involved in the mutagenic processes.
It appears, however, that pols other than pol III play roles in the high spontaneous mutations in the mutT background. This is because the mutant lacking all four DNA polymerases (YG6376) exhibited a 70% reduction in mutability compared with mutT (Fig. 1 and Table S1). Single deletions of each of the genes encoding the auxiliary pols did not reduce the mutation frequency. Only in the cases where three or four pols are absent, there was a significant reduction of the mutT mutator phenotype. Thus, pol I, pol II, pol IV and pol V might have redundant roles in the mutagenesis. One possible explanation for the redundant role is that they might play roles in extension of primer DNA upon incorporation of 8-oxo-dGMP into DNA by pol III HE (Fig. 6). This speculation is based on the results that extension of primer upon incorporation of 8-oxo-dGMP opposite template A by pol III HE is less efficient compared with the incorporation of normal dTMP opposite template A (Fig. 5). In addition, pol I and pol IV possess ability to extend primer DNA having 8-oxo-dGMP opposite template A at the termini (Fig. S3). Hence, the auxiliary pols might play roles in the extension step, thereby enhancing the mutagenesis in the mutT background. Another, but not exclusive, alternative would be that the presence of pol I, pol II, pol IV and pol V might affect the efficiency of pol III HE incorporating 8-oxo-dGTP into DNA during the chromosome replication. It is well known that all five pols in E. coli interact with the β subunit and compete to take over the primer termini (Lopez de Saro et al., 2003; Burnouf et al., 2004). Thus, it seems possible that the absence of the auxiliary pols might affect the interactions of pol III with the β subunit, which in turn affects the efficiency of pol III HE incorporating 8-oxo-dGTP during DNA synthesis. It is reported that processivity factors such as eukaryotic PCNA and β subunit of E. coli affect the processivity and specificity of pols (Bloom et al., 1997; Maga et al., 2007).
Fig. 6.
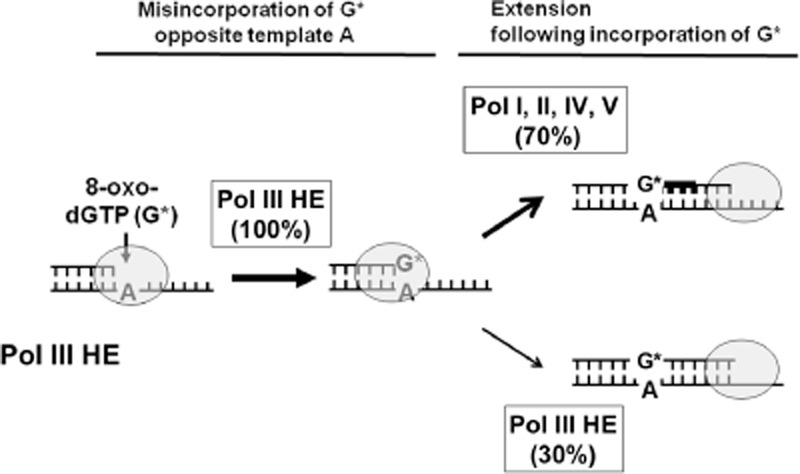
MutT mutator phenotype: a multi-polymerase affair. Pol III HE (oval) incorporates 8-oxo-dGTP (G*) opposite template A during the chromosomal replication. We suggest that while pol III accounts for nearly all 8-oxo-dGTP incorporation opposite template A, it only extends ≍ 30% of them, the remaining 70% being extended by the combined action of pol I, pol II, pol IV or pol V (based on data from Fig. 1). We also speculate that the roles of the auxiliary pols might be redundant because the mutation frequency was significantly reduced only when three or four auxiliary pols are deleted (Fig. 1). Following a short patch (a thick line) of DNA synthesis by the auxiliary pols, pol III HE will resume chromosomal replication.
Our biochemical results indicated that pol III*, T7 pol and pol I (KF) could not excise incorporated 8-oxo-dGMP from the primer effectively even when the oxidized dGMP was paired with template A (Fig. 3). Previous genetic analyses also suggest that the proofreading activity of pol III HE has little effect on the mutT mutator phenotype (Fowler et al., 1992). In pol III HE, the proofreading 3′ to 5′ exonuclease exists as a separate subunit ε, which binds to the catalytic subunit (McHenry, 2011). In contrast, exonucleases and polymerases exist in separate domains in single polypeptides of T7 pol and pol I (KF) (Beese et al., 1993; Doublie et al., 1998). Because pol III* and T7 pol can digest the single-stranded DNA having 8-oxo-dGMP and exonuclease III digested double-stranded DNA having 8-oxo-dGMP at the 3′-termini (Fig. 3), the exonucleases in the pols and exonucleases III appear to have the ability to hydrolyse phosphodiester bonds between the terminal 8-oxo-dGMP and the second terminal dNMP in the primer strands. Therefore, we speculate that the reason for the poor excision of 8-oxo-dGMP from the 3′-termini of primer strands by pol III* and T7 pol might be inefficient transfer of the primer strands having 8-oxo-dGMP from the polymerase domain (or subunit) to the exonuclease domain (or subunit) even when 8-oxo-dGMP pairs with template A. In contrast, pol I (KF) did not digest single-stranded DNA having 8-oxo-dGMP at the termini. Therefore, in the case of pol I (KF), the poor proofreading against 8-oxo-dGMP paired with template A or C may be due to its weak or defective nuclease activity against single-stranded DNA having 8-oxo-dGMP at the terminus. It might be interesting to investigate the structural and biochemical reasons for the poor excision of 8-oxo-dGMP at the 3′-termini of primers by the pols.
It may be counterintuitive that pol III HE, the replicative pol, which is supposed to be very accurate, positively produces errors during the chromosome replication if mutT is inactivated. One plausible explanation is that pol III HE incorporates the oxidized dGTP into DNA, thereby enhancing mutagenesis to generate progenies that can adapt to the stressful environments, when the bacteria are exposed to oxidative stress and the mutT gene is inactivated. The original habitat of E. coli is intestine, which is strictly anaerobic. Therefore, aerobic culture conditions may be somewhat stressful to E. coli. A precedent for such enhanced mutagenesis in stressful conditions is the SOS-induced mutagenesis where E. coli induces error-prone pol IV and pol V and enhances mutagenesis when DNA is damaged by ultraviolet light or other DNA-damaging agents (Echols, 1981; Friedberg et al., 2002). High mutation rates may be detrimental to individual bacterium but may be beneficial for the whole population because the high mutation rates may lead to generation of mutant progenies that can survive under the stressful conditions. Other C-family pols from organisms originally living in anaerobic conditions might have evolved in a manner to incorporate 8-oxo-dGTP into DNA effectively as in the case of E. coli pol III HE.
In summary, we presented genetic and biochemical evidence that suggests that the replicative pol of E. coli, i.e. pol III HE, effectively and incorrectly incorporates oxidized dGTP opposite template A during the chromosome replication in vivo. The auxiliary pols appear to help the erroneous DNA replication by pol III HE in the mutT background. The specificity and efficiency of incorporation of 8-oxo-dGTP into DNA by pol III HE are marked contrast with those of replicative pols in mammals, e.g. pol δ, which incorporate 8-oxo-dGTP into DNA very poorly. The erroneous nature of pol III HE, which is uniquely present in eubacteria, might explain the extremely high spontaneous mutations of the mutT background in E. coli compared with the moderate mutator effects of mammalian counterparts lacking enzymes with 8-oxo-dGTP hydrolysing activities such as MTH1.
Experimental procedures
Materials
Pol III* was purified as described (Fujii and Fuchs, 2004). Deoxyribonucleoside triphosphates (ultrapure grade) and 8-oxo-dGTP were purchased from Amersham Pharmacia Biotech (Buckinghamshire, UK) and TriLink BioTechnologies (San Diego, CA, USA) respectively. Oligonucleotides were synthesized and purified twice by high-performance liquid chromatography (BEX, Tokyo, Japan). Primers were annealed to templates at a 1:1 molar ratio. For primer extension and kinetic studies of nucleotide incorporation, two types of DNA substrates were used. The set of the 18-mer primer/36-mer template named sequences 1 was as follows: 5′-CGCGCGAAGACCGGTTAC-3′ (18-mer primer) and 3′-GCGCGCTTCTGGCCAATGNCAGAATTCCTAGGGAAG-5′ (36-mer template, where N = A, C, G or T). The set of the 30-mer primer/100-mer template named as sequences 2 used for measuring the incorporation frequencies was as follows: 5′-GTACCGCCACCCTTAGAACCGGTGTTTGGT-3′ (primer; 30-mer) and 3′-GGCCTTATCCACATAGTGGCATGAGTCCTCCAAATCATGGCGGTGGGAATCTTGGCCACAAACCATTTTXCTGTCTGTGACTCGTTCAGGCTATTACTGA-5′ [template; 100-mer, X (= A or C) is the target site]. The same 100-mer oligonucleotide with biotin was synthesized and purified twice by high-performance liquid chromatography (Tsukuba Oligo Service, Tsukuba, Japan).
Strain construction
All the strains and plasmids used in this study are listed in Table 2. P1 transduction was conducted at 37°C. When strains lacking pol I Klenow fragment (ΔKF) were used as either recipients or donors, the transduction was conducted at 30°C.
Table 2.
Strains and plasmids used in this study
| Strains | Genetic characteristics | Sources |
|---|---|---|
| AB1157 | F− thr1 ara14 leuB6 proA2 lacG1 tsx33 supE44 galK2 hisG4 rfbD1 mgl51 rpsL31 xyl5 mtl1 argE3 thi1 λ− rac− | Laboratory stock |
| V355 | F− lac-3350 galK2(Oc) galT22 λ− recD1014(Nuc−) IN(rrnD-rrnE)1 rpsL179(strR) | Shevell et al. (1988) |
| JW0059 | F− ΔpolB770::kan Δ(araD–araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD–rhaB)568 hsdR514; KmR | Keio Collection, NBRP |
| YG2004 | The same as V355, but deficient in mutT with KmR cassette insertion; KmR, ΔmutT | This study |
| YG6156 | The same as AB1157, but deficient in mutT with KmR cassette insertion; KmR, ΔmutT | This study |
| HRS7052 | Deficient in a part of polA gene, thus lacking both 3′–5′ exonuclease and polymerase activities; CmR, ΔKF | Wagner and Nohmi (2000) |
| YG6162 | The same as AB1157, but deficient in dinB with in frame deletion; ΔdinB | Salem et al. (2009) |
| YG6167 | The same as YG6162, but deficient in mutT with KmR cassette insertion; KmR, ΔdinB ΔmutT | This study |
| YG6168 | The same as AB1157, but deficient in umuDC with ermGT insertion, ΔumuDC | Salem et al. (2009) |
| YG6174 | The same as YG6168, but deficient in mutT with KmR cassette insertion; KmR, ΔumuDC ΔmutT | This study |
| YG6171 | The same as YG6168, but deficient in dinB with in frame deletion, ΔumuDC ΔdinB | Salem et al. (2009) |
| YG6172 | The same as YG6171, but deficient in mutT with KmR cassette insertion; KmR, ΔumuDC ΔdinB ΔmutT | This study |
| YG6343 | The same as AB1157, but deficient in a part of polA gene, thus lacking both 3′–5′ exonuclease and polymerase activities with CmR cassette insertion; CmR, ΔKF | This study |
| YG6347 | The same as YG6343, but deficient in mutT with KmR cassette insertion; CmR; KmR, ΔKF ΔmutT | This study |
| YG6371 | The same as YG6162, but deficient in a part of polA gene, thus lacking both 3′–5′ exonuclease and polymerase activities with CmR cassette insertion; CmR, ΔdinB ΔKF | This study |
| YG6372 | The same as YG6371, but deficient in mutT with KmR cassette insertion; CmR; KmR, ΔdinB ΔKF ΔmutT | This study |
| YG6375 | The same as YG6171, but deficient in a part of polA gene with CmR cassette insertion as HRS7052 and deficient in polB gene with in frame deletion, CmR; ΔumuDC ΔdinB ΔKF ΔpolB | This study |
| YG6376 | The same as YG6375, but deficient in mutT with KmR cassette insertion; CmR; KmR, ΔumuDC ΔdinB ΔpolB ΔKF ΔmutT | This study |
| YG6377 | The same as AB1157, but deficient in polB with in frame deletion; ΔpolB | This study |
| YG6378 | The same as YG6377, but deficient in mutT with KmR cassette insertion; KmR, ΔpolB ΔmutT | This study |
| YG6381 | The same as YG6168, but deficient in polB with in frame deletion; ΔumuDC ΔpolB | This study |
| YG6382 | The same as YG6171, but deficient in polB with in frame deletion; ΔumuDC ΔdinB ΔpolB | This study |
| YG6385 | The same as YG6382, but deficient in mutT with KmR cassette insertion; KmR, ΔumuDC ΔdinB ΔpolB ΔmutT | This study |
| YG6383 | The same as YG6171, but deficient in a part of polA gene with CmR cassette insertion as HRS7052; CmR, ΔumuDC ΔdinB ΔKF | This study |
| YG6386 | The same as YG6383, but deficient in mutT with KmR cassette insertion; CmR, KmR, ΔumuDC ΔdinB ΔKF ΔmutT | This study |
| YG6384 | The same as YG6381, but deficient in a part of polA gene with CmR cassette insertion as HRS7052, CmR; ΔumuDC ΔpolB ΔKF | This study |
| YG6387 | The same as YG6384, but deficient in mutT with KmR cassette insertion; CmR; KmR, ΔumuDC ΔpolB ΔKF ΔmutT | This study |
| YG6379 | The same as YG6371, but deficient in polB with in frame deletion; CmR, ΔdinB ΔKF ΔpolB | This study |
| YG6380 | The same as YG6379, but deficient in mutT with KmR cassette insertion; CmR, KmR, ΔdinB ΔKF ΔpolB ΔmutT | This study |
| Plasmids | ||
|---|---|---|
| pYG703 | Plasmid pUC19, but has the mutT gene | This study |
| pYG704 | The same as pYG703, but has mutT::KmR | This study |
| pCP20 | pCP20 has the yeast Flp recombinase gene, FLP, chloramphenicol and ampicillin resistance genes, and temperature-sensitive replication; ApR, CmR | CGSC |
To construct polA− derivative, the Δklenow::chloramphenicol resistance (CmR) gene (cat) allele of strain HRS7052 (Wagner and Nohmi, 2000), which was constructed by Dr H. Iwasaki (Tokyo Institute of Technology, Japan), was transferred to AB1157 by P1 transduction. The resultant strain was named as YG6343, which lacked pol and 3′ to 5′ exonuclease of pol I but retained 5′ to 3′ exonuclease activity. YG6371 (ΔKFΔdinB) was constructed by P1 transduction using HRS7052 as a donor and YG6162 (ΔdinB, see below) as a recipient. Similarly, YG6383 (ΔKFΔdinBΔumuDC), YG6384 (ΔKFΔpolBΔumuDC) and YG6375 (ΔKFΔpolBΔdinBΔumuDC) were constructed by P1 transduction with YG6171 (ΔdinBΔumuDC), YG6381 (ΔpolBΔumuDC, see below) and YG6382 (ΔpolBΔdinBΔumuDC, see below) respectively.
To construct polB− derivative, polB::kanamycin resistance (KmR) gene (kan) was transferred from JW0059 into a recipient AB1157, YG6171 (ΔdinBΔumuDC; Salem et al., 2009), YG6371 (ΔKFΔdinB) or YG6168 (ΔumuDC; Salem et al., 2009) by P1 transduction, and the KmR colonies were selected. The KmR strains were transformed with pCP20 [ampicillin resistance (ApR), CmR], which encodes a yeast FLP recombinase. The CmR transformants were selected at 30°C since the replication origin of the plasmid was temperature-sensitive. In the case of YG6371 (ΔKFΔdinB), ApR transformants were selected. A few colonies were picked up and streaked on a fresh plate, and then incubated at 43°C. An expression of the FLP was induced, which resulted in removal of the KmR cassette whose both ends had recognition sites for the FLP recombinase. The high temperature also makes pCP20 cured from the cells. The obtained colonies were confirmed for their sensitivity to Cm (except for ΔKF strain), Ap and Km and also the size of the bands by PCR analysis (Table S2). The resultant strains were designated as YG6377 (ΔpolB), YG6382 (ΔpolBΔdinBΔumuDC), YG6379 (ΔKFΔpolBΔdinB) and YG6381 (ΔpolBΔumuDC) respectively. They had no drug resistance maker for the polB deletion.
To construct mutT-deficient strains, we cloned the mutT gene from Kohara library (Kohara et al., 1987) into pUC19, then designated as pYG703. Inserting 1.3 kb EcoRI cassette carrying the KmR gene into EcoRI site of pYG703, the clone in which transcription direction of mutT is opposite to the kan gene was selected and named as pYG704. After digestion of pYG704 with KpnI, the linear fragment was isolated and introduced into strain V355, which lacks recD, to obtain the clone whose mutT was replaced with ΔmutT::kan. The resultant strain was designated as YG2004, which was used to transfer ΔmutT::kan to strain AB1157 and the pol-defective derivatives by P1 transduction. The mutT::kan derivative of AB1157 was designated as YG6156 (AB1157ΔmutT). Other host strains for the P1 transduction were YG6162 (ΔdinB; Salem et al., 2009), YG6168 (ΔumuDC), YG6171 (ΔdinBΔumuDC), YG6377 (ΔpolB) and YG6382 (ΔpolBΔdinBΔumuDC). The resultant strains were designated as YG6167 (ΔdinBΔmutT), YG6174 (ΔumuDCΔmutT), YG6172 (ΔdinBΔumuDCΔmutT), YG6378 (ΔpolBΔmutT), YG6385 (ΔpolBΔdinBΔumuDCΔmutT) respectively. In addition, the ΔmutT::kan allele was transferred from YG2004 to strains YG6343 (ΔKF), YG6371 (ΔKFΔdinB), YG6383 (ΔKFΔdinBΔumuDC), YG6384 (ΔKFΔpolBΔumuDC), YG6375 (ΔKFΔpolBΔdinBΔumuDC) and YG6379 (ΔKFΔpolBΔdinB) at 30°C, and the resultant strains were designated as YG6347 (ΔKFΔmutT), YG6372 (ΔKFΔdinBΔmutT), YG6386 (ΔKFΔdinBΔumuDCΔmutT), YG6387 (ΔKFΔpolBΔumuDCΔmutT), YG6376 (ΔKFΔpolBΔdinBΔumuDCΔmutT) and YG6380 (ΔKFΔpolBΔdinBΔmutT) respectively.
To confirm proper replacements in the chromosome, each constructed strain was subject to polymerase chain reactions with primers designed to display different amplification sizes when gene replacements successfully occur (Table S2).
Mutation assay
To determine mutation frequency, acquisition of resistance to rifampicin was used in fluctuation analysis. A fresh single colony was picked from LB agar and grown overnight in a tube at 37°C with aeration in LB medium and 10 tubes were prepared for each strain. The culture was diluted 106 times in fresh LB medium to achieve no mutants in one culture. The cultures were grown at 37°C with aeration (on a roller drum) to saturation (16 h), and 50 μl of aliquot for each tube was plated on a LB agar plate supplemented with 100 μg ml−1 rifampicin (Sigma, OH, USA). For viable cell count, three randomly selected cultures were serially diluted in cold saline and plated in LB agar without antibiotics. All plates were incubated at 37°C, except for the Klenow-deficient strains for which 30°C was used, for 24 h before counting colonies. We repeated the experiments three times for the determination of mutation frequency, which was calculated with median for 10 cultures divided by a mean value of viable cell count (Hasegawa et al., 2008).
Primer extension assay
The enzyme reaction buffer containing 20 mM Tris-HCl (pH 7.5), 4% glycerol, 8 mM DTT, 80 µg ml−1 BSA, 2.5 mM ATP, 8 mM MgCl2, 100 μM 8-oxo-dGTP, 1 nM pol III* was incubated with 0.1 μM 5′-Cy3-primer/template (sequences 1) at room temperature (20 to 25°C) for 1 min. Reactions were terminated by adding 10 μl of stop solution (98% formamide, 10 mM EDTA, 10 mg ml−1 Blue Dextran). Samples were denatured at 100°C for 10 min, then applied to 15% denaturing polyacrylamide gel for electrophoresis and the patterns were visualized by the Molecular Imager FX Pro System (Bio-Rad, CA, USA).
Kinetics analysis
For incorporation kinetics, 1 nM pol III*, 0.1 μM substrate (sequences 2) and 0.05–500 μM dNTPs were incubated at room temperature for 1 to 3 min in the reaction buffer written above. The reaction samples were subjected to 15% denaturing polyacrylamide gel. The gel band intensities were measured using the Molecular Imager FX Pro System and Quantity One software (Bio-Rad). The nucleotide incorporation efficiency opposite the target site was obtained by measuring ITΣ/IT−1, where ITΣ represents the integrated gel band intensities of primers extended to the target site and beyond, and IT-1 is the integrated gel band intensity of primers extended to the site just prior to the target site (Creighton and Goodman, 1995; Bloom et al., 1997). For each DNA substrate, the rate of incorporation was plotted as a function of dNTP concentration, and the relative Vmax and apparent Km values were determined by nonlinear regression fitting using the SigmaPlot software (SigmaPlot Software Sciences, IL, USA). The relative Vmax value is equal to the maximum value of ITΣ/IT−1. The frequency of incorporation (finc) was calculated using the equation finc = (Vmax/Km)incorrect / (Vmax/Km)correct. All values are means (± standard error) of three experiments.
Assay for excision of 8-oxo-G-ended primer
The set of the 19-mer primer/36-mer template was basically the same as sequences 1 (see above). It was as follows: 5′-CGCGCGAAGACCGGTTACX-3′ (19-mer primer, where X = G or 8-oxo-G) and 3′-GCGCGCTTCTGGCCAATGNCAGAATTCCTAGGGAAG-5′ (36-mer template, where N = A or C). The primer whose 3′-terminus is 8-oxo-G was purchased from Tsukuba Oligo Service (Tsukuba, Japan). For the assay for excision of 8-oxo-G-ended primer, these primer/templates (0.1 μM) were incubated with 1 nM pol III* at 25°C for 5, 10, 20 or 30 min in the reaction buffer described above. The products were analysed by denaturing polyacrylamide gel electrophoresis and visualized by the Molecular Imager. Percentage of the amount of digested primers compared with that of the original primers was calculated. For comparison, the primer/template DNA or the primer DNA alone was incubated pol III* (1 nM) at room temperature, T7 pol (0.0001 unit μl−1), pol I (KF) (0.001 unit μl−1) or exo III (0.0001 unit μl−1, New England BioLabs) at 37°C for 10 min in a reaction mixture for each enzyme without dNTP. The products were analysed as described above.
Assay for primer extension with β clamp
Primer extension with the β subunit was examined under two different conditions, i.e. standing start and running start experiments. In the standing start experiments, the primer was the same as that of sequences 2, but had extra five bases at the 3′-end, i.e. 5′-GTACCGCCACCCTTAGAACCGGTGTTTGGTAAAAX-3′ (35-mer, where X = T, G or 8-oxo-G). The template was the same as that of sequences 2 (100-mer), but had biotin/streptavidin at both ends (see Fig. S2). The primer/template DNA (20 nM), pol III* (10 nM) and four normal dNTPs (100 μM) were incubated for 1 min at 25°C. When the β subunit (10 or 100 nM) was included, the reaction mixture without dNTPs was pre-incubated for 10 min at 25°C and the reaction was started by addition of dNTPs. The reaction buffer and the methods to analyse the reaction products were the same as those described in the primer extension assay. In running start experiments, the primer (30-mer)/template (100-mer) DNA was the same as those of sequences 2, but the template DNA had A at the position of N and biotin/streptavidin at both ends. The primer/template DNA (20 nM), pol III* (10 nM) and dNTP(s) (100 μM) were incubated in the presence or the absence of the β subunit (100 nM) for 1 min at 25°C. In these experiments, four types of dNTP solution were used. Each contained dATP alone, dATP and 8-oxo-dGTP, dATP, 8-oxo-dGTP, dCTP and dGTP, or four normal dNTPs.
Acknowledgments
We thank Dr Su-Ryang Kim for construction of strain YG2004 and Ms Makiko Takamune for technical assistance. All authors have no conflict of interest. This work was supported by grants in aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT, 18201010; MEXT, 22241016), the Ministry of Health, Labour and Welfare, Japan (MHLW, H21-Food-General-009) and the Japan Health Science Foundation (KHB1007); for cancer research from MHLW (20 designated-8); and the Food Safety Commission.
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Akiyama M, Maki H, Sekiguchi M, Horiuchi T. A specific role of MutT protein: to prevent dG.dA mispairing in DNA replication. Proc Natl Acad Sci USA. 1989;86:3949–3952. doi: 10.1073/pnas.86.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science. 1983;221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutat Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or ‘Nudix’ hydrolases, a family of versatile, widely distributed, ‘housecleaning’ enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Bloom LB, Chen X, Fygenson DK, Turner J, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of b, g complex processivity proteins and e proofreading exonuclease on nucleotide misincorporation efficiencies. J Biol Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- Burnouf DY, Olieric V, Wagner J, Fujii S, Reinbolt J, Fuchs RP, Dumas P. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J Mol Biol. 2004;335:1187–1197. doi: 10.1016/j.jmb.2003.11.049. [DOI] [PubMed] [Google Scholar]
- Cai JP, Kakuma T, Tsuzuki T, Sekiguchi M. cDNA and genomic sequences for rat 8-oxo-dGTPase that prevents occurrence of spontaneous mutations due to oxidation of guanine nucleotides. Carcinogenesis. 1995;16:2343–2350. doi: 10.1093/carcin/16.10.2343. [DOI] [PubMed] [Google Scholar]
- Cai JP, Ishibashi T, Takagi Y, Hayakawa H, Sekiguchi M. Mouse MTH2 protein which prevents mutations caused by 8-oxoguanine nucleotides. Biochem Biophys Res Commun. 2003;305:1073–1077. doi: 10.1016/s0006-291x(03)00864-7. [DOI] [PubMed] [Google Scholar]
- Creighton S, Goodman MF. Gel kinetic analysis of DNA polymerase fidelity in the presence of proofreading using bacteriophage T4 DNA polymerase. J Biol Chem. 1995;270:4759–4774. doi: 10.1074/jbc.270.9.4759. [DOI] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Echols H. SOS functions, cancer and inducible evolution. Cell. 1981;25:1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Egashira A, Yamauchi K, Yoshiyama K, Kawate H, Katsuki M, Sekiguchi M, et al. Mutational specificity of mice defective in the MTH1 and/or the MSH2 genes. DNA Repair (Amst) 2002;1:881–893. doi: 10.1016/s1568-7864(02)00113-1. [DOI] [PubMed] [Google Scholar]
- Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase d. Steady-state and pre-steady-state kinetic analysis. J Biol Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- Einolf HJ, Schnetz-Boutaud N, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry. 1998;37:13300–13312. doi: 10.1021/bi981346d. [DOI] [PubMed] [Google Scholar]
- Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- Fowler RG, Amutan MV, Isbell RJ. The interaction of the Escherichia coli mutD and mutT pathways in the prevention of A:T→C:G transversions. Mutat Res. 1992;284:307–319. doi: 10.1016/0027-5107(92)90015-t. [DOI] [PubMed] [Google Scholar]
- Fowler RG, Erickson JA, Isbell RJ. Activity of the Escherichia coli mutT mutator allele in an anaerobic environment. J Bacteriol. 1994;176:7727–7729. doi: 10.1128/jb.176.24.7727-7729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler RG, White SJ, Koyama C, Moore SC, Dunn RL, Schaaper RM. Interactions among the Escherichia coli mutTmutM, and mutY damage prevention pathways. DNA Repair (Amst) 2003;2:159–173. doi: 10.1016/s1568-7864(02)00193-3. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi M, Yoshida MC, Oda H, Tajiri T, Nakabeppu Y, Tsuzuki T, Sekiguchi M. Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics. 1994;24:485–490. doi: 10.1006/geno.1994.1657. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Yoshiyama K, Maki H. Spontaneous mutagenesis associated with nucleotide excision repair in Escherichia coli. Genes Cells. 2008;13:459–469. doi: 10.1111/j.1365-2443.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Hayakawa H, Sekiguchi M. A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 2003;4:479–483. doi: 10.1038/sj.embor.embor838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- Janion C, Sikora A, Nowosielska A, Grzesiuk E. E. coli BW535, a triple mutant for the DNA repair genes xthnth, and nfo, chronically induces the SOS response. Environ Mol Mutagen. 2003;41:237–242. doi: 10.1002/em.10154. [DOI] [PubMed] [Google Scholar]
- Kakuma T, Nishida J, Tsuzuki T, Sekiguchi M. Mouse MTH1 protein with 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphatase activity that prevents transversion mutation. cDNA cloning and tissue distribution. J Biol Chem. 1995;270:25942–25948. doi: 10.1074/jbc.270.43.25942. [DOI] [PubMed] [Google Scholar]
- Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H. Chemistry-based studies on oxidative DNA damage: formation, repair, and mutagenesis. Free Radic Biol Med. 2002;33:450–456. [PubMed] [Google Scholar]
- Katafuchi A, Nohmi T. DNA polymerases involved in the incorporation of oxidized nucleotides into DNA: their efficiency and template base preference. Mutat Res. 2010;703:24–31. doi: 10.1016/j.mrgentox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J. Crystal structure of the catalytic a subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Lopez de Saro FJ, Georgescu RE, Goodman MF, O'Donnell M. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 2003;22:6408–6418. doi: 10.1093/emboj/cdg603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- McHenry CS. DNA replicases from a bacterial perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H, Prasad R, Wilson SH, Johnson F, Grollman AP. 8-oxodGTP incorporation by DNA polymerase b is modified by active-site residue Asn279. Biochemistry. 2000;39:1029–1033. doi: 10.1021/bi991789x. [DOI] [PubMed] [Google Scholar]
- Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc Natl Acad Sci USA. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y, Sakumi K, Sakamoto K, Tsuchimoto D, Tsuzuki T, Nakatsu Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol Chem. 2006;387:373–379. doi: 10.1515/BC.2006.050. [DOI] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T, Obata F, Boss AC, Oikawa S, Mori T, Kawanishi S, Yamamoto K. Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J Biol Chem. 1999;274:34832–34837. doi: 10.1074/jbc.274.49.34832. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T, Watanabe T, Nakabeppu Y, Yamamoto K. Mutagenic target for hydroxyl radicals generated in Escherichia coli mutant deficient in Mn− and Fe− superoxide dismutases and Fur, a repressor for iron-uptake systems. DNA Repair (Amst) 2002;1:411–418. doi: 10.1016/s1568-7864(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Sakai A, Nakanishi M, Yoshiyama K, Maki H. Impact of reactive oxygen species on spontaneous mutagenesis in Escherichia coli. Genes Cells. 2006;11:767–778. doi: 10.1111/j.1365-2443.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- Salem AM, Nakano T, Takuwa M, Matoba N, Tsuboi T, Terato H, et al. Genetic analysis of repair and damage tolerance mechanisms for DNA-protein cross-links in Escherichia coli. J Bacteriol. 2009;191:5657–5668. doi: 10.1128/JB.00417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi M, Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21:8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- Setoyama D, Ito R, Takagi Y, Sekiguchi M. Molecular actions of Escherichia coli MutT for control of spontaneous mutagenesis. Mutat Res. 2011;707:9–14. doi: 10.1016/j.mrfmmm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Shevell DE, Abou-Zamzam AM, Demple B, Walker GC. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J Bacteriol. 1988;170:3294–3296. doi: 10.1128/jb.170.7.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Gruz P, Kamiya H, Masutani C, Xu Y, Usui Y, et al. Efficient and erroneous incorporation of oxidized DNA precursors by human DNA polymerase η. Biochemistry. 2007;46:5515–5522. doi: 10.1021/bi062238r. [DOI] [PubMed] [Google Scholar]
- Tajiri T, Maki H, Sekiguchi M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat Res. 1995;336:257–267. doi: 10.1016/0921-8777(94)00062-b. [DOI] [PubMed] [Google Scholar]
- Treffers HP, Spinelli V, Belser NO. A factor (or mutator gene) influencing mutation rates in Escherichia coli. Proc Natl Acad Sci USA. 1954;40:1064–1071. doi: 10.1073/pnas.40.11.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Egashira A, Igarashi H, Iwakuma T, Nakatsuru Y, Tominaga Y, et al. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc Natl Acad Sci USA. 2001;98:11456–11461. doi: 10.1073/pnas.191086798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Nunoshiba T, Shimizu M, Gruz P, Kamiya H, Harashima H, Nohmi T. Involvement of Y-family DNA polymerases in mutagenesis caused by oxidized nucleotides in Escherichia coli. J Bacteriol. 2006;188:4992–4995. doi: 10.1128/JB.00281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C, Cox EC, Horn V. The unusual mutagenic specificity of an E. coli mutator gene. Proc Natl Acad Sci USA. 1966;55:274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


