Abstract
The Abl family nonreceptor tyrosine kinase Arg/Abl2 interacts with cortactin, an Arp2/3 complex activator, to promote actin-driven cell edge protrusion. Both Arg and cortactin bind directly to filamentous actin (F-actin). While protein– protein interactions between Arg and cortactin have well-characterized downstream effects on the actin cytoskeleton, it is unclear whether and how Arg and cortactin affect each other’s actin binding properties. We employ actin cosedimentation assays to show that Arg increases the stoichiometry of binding of cortactin to F-actin at saturation. Using a series of Arg deletion mutants and fragments, we demonstrate that the Arg Cterminal calponin homology domain is necessary and sufficient to increase the stoichiometry of binding of cortactin to F-actin. We also show that interactions between Arg and cortactin are required for optimal affinity between cortactin and the actin filament. Our data suggest a mechanism for Arg-dependent stimulation of binding of cortactin to F-actin, which may facilitate the recruitment of cortactin to sites of local actin network assembly.
The Abl2/Arg (Abelson-related gene) nonreceptor tyrosine kinase regulates cytoskeletal rearrangement, in part through binding and phosphorylating cortactin, a nucleation-promoting factor of the Arp2/3 complex.1–7 Binding between Arg and cortactin and phosphorylation of cortactin by Arg are important for actin-driven cell edge protrusion.3,8 The cortactin SH3 domain binds a unique PYLPRLP motif within the proline-rich domain of Arg.9 Integrin-mediated adhesion and growth factor stimulation activate Arg to phosphorylate cortactin, and these phosphorylated tyrosines serve as a binding site for SH2 domain-containing proteins, including Arg itself.2,3,9 Disruption of these interactions significantly reduces the efficiency of actin-driven cellular processes, including cell edge protrusion and dorsal ruffle formation in fibroblasts and invadopodia formation and maturation in breast cancer cells.2,3,8
Arg and cortactin both also bind filamentous actin (F-actin). The Arg C-terminal half has two distinct F-actin-binding domains separated by a microtubule-binding domain (Figure 1A), with which it can bundle F-actin and cross-link it to microtubules.10,11 The internal I/LWEQ actin-binding motif binds actin subdomain 1 or 4, while the C-terminal calponin homology (CH) domain binds subdomain 1 and induces a 30° tilt in the actin filament.12 Both of these F-actin-binding domains are required for Arg to promote formation of F-actin-rich structures at the fibroblast periphery.10 Cortactin has six “cortactin repeats”, but studies in which each repeat was deleted indicate that only loss of the fourth repeat compromises F-actin binding (Figure 1A).7,13,14 While the fourth repeat alone is not sufficient to bind F-actin, a polypeptide containing the 6.5- repeat region of cortactin binds F-actin between actin subdomains 1 and 3.7,14 Cortactin requires both its F-actin-binding and the N-terminal acid (NTA) domains to localize to the cell edge and activate the Arp2/3 complex.1,7 While several studies have investigated the functions of protein—protein interactions between Arg and cortactin, it is unclear whether and how Arg and cortactin each affect the other’s F-actin binding activity.
Figure 1.
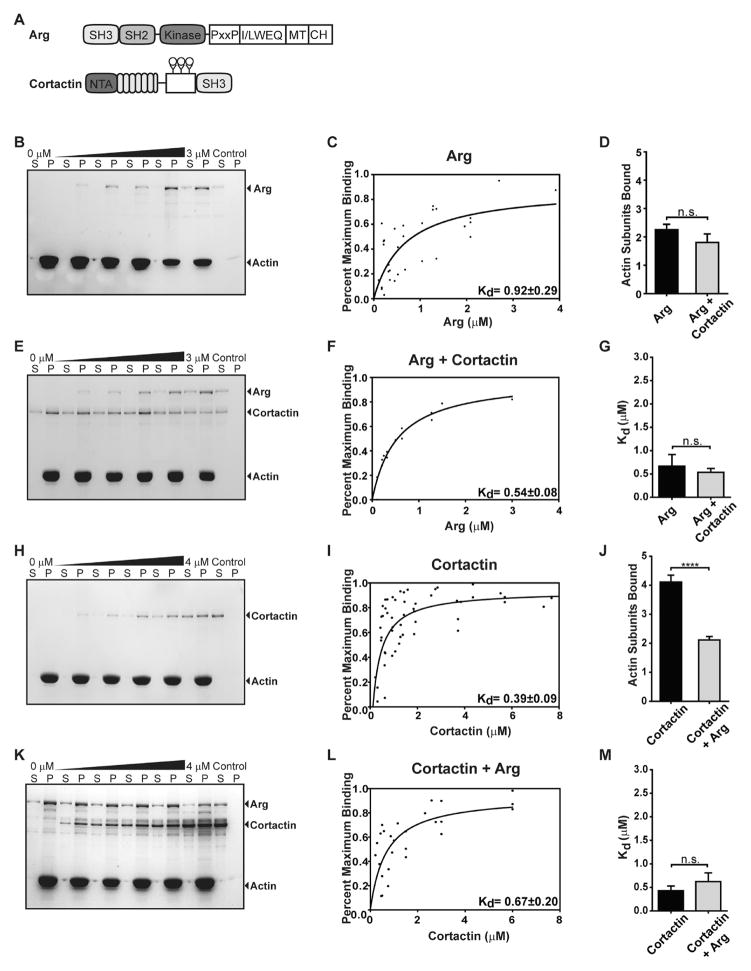
Arg increases the stoichiometry between cortactin and F-actin. (A) Arg and cortactin domain structures. (B) Representative cosedimentation assay gel showing increasing concentrations of Arg binding to F-actin (quantified in panel C). Kd values are means ± the standard error of the mean. (E) Representative cosedimentation assay gel showing increasing concentrations of Arg binding to F-actin preincubated with saturating cortactin (quantified in panel F). (D) Quantification of binding stoichiometry and (G) affinity from panels C and F. (H) Representative cosedimentation assay gel of increasing concentrations of cortactin binding to F-actin (quantified in panel I). (K) Representative cosedimentation assay gel of increasing concentrations of cortactin binding to F-actin preincubated with saturating Arg (quantified in panel L). (J and M) Quantification of (J) binding stoichiometry and (M) affinity from panels I and L. S and P denote the supernatant and pellet, respectively; controls are proteins centrifuged in the absence of F-actin. Values in panels D, G, J, and M are means ± the standard error of the mean. One-way ANOVA with a post hoc unpaired two-tailed t test. ***p < 0.001; n.s., not significant.
We show here that Arg increases the stoichiometry of binding between cortactin and F-actin at saturation, while cortactin does not impact Arg—actin binding. We show that this augmentation of cortactin binding stoichiometry does not require protein—protein interactions between Arg and cortactin. Rather, we find that the Arg C-terminal CH domain is both necessary and sufficient to induce this increase in cortactin binding stoichiometry. Saturating Arg concentrations do not affect the binding affinity of cortactin for F-actin. However, Arg mutants with disruptions in key cortactin interaction interfaces reduce the affinity of cortactin for F-actin, suggesting that Arg—cortactin binding interfaces may influence the exposure of cortactin binding sites on the actin filament. Together, these data suggest that Arg binding to Factin may recruit and concentrate cortactin to sites where it is needed to stimulate F-actin network formation and/or stabilization.
EXPERIMENTAL PROCEDURES
Molecular Cloning and Purification of Recombinant Proteins
Murine Arg and cortactin cDNAs were cloned with an engineered His6 tag into the pFastbac1 vector (Invitrogen). Arg deletion constructs and fragments as well as cortactin and W525A mutants were generated using PCR-based mutagenesis. 3,10 Recombinant baculoviruses expressing these proteins were generated using the Bac-to-Bac baculovirus expression system as previously described.15 Cells were lysed in lysis buffer [50 mM Tris (pH 7.5), 500 mM NaCl, 0.01% Triton X-100, 5% glycerol, 20 mM imidazole, 1 mM DTT, 1 mM EDTA, and protease inhibitors]. His6-tagged proteins were purified over a Ni2+ affinity column and eluted with 250 mM imidazole, followed by an SP-Sepharose ion exchange column, using a step salt gradient to elute. Proteins were dialyzed into storage buffer (SB) [20 mM Hepes (pH 7.25), 500 mM NaCl, 0.01% Triton X-100, 5% glycerol, 1 mM DTT, and 1 mM EDTA].
The Arg calponin homology (CH) domain (residues 1058— 1182) was generated using PCR-based amplification followed by cloning of the fragment into pGEX-6P-1 (GE Healthcare Life Sciences). Recombinant GST-Arg CH domain fusion protein (GST-ArgCH) was expressed in Escherichia coli using 0.1 mM IPTG induction and overnight expression at 37 °C. Bacteria were lysed at 4 °C in lysis buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM DTT, 1 mM EDTA, and protease inhibitors] using a French press. The lysate was clarified by centrifugation at 100000g for 30 min and bound to a glutathione agarose column. GST-ArgCH was washed and eluted from the glutathione column with 50 mM glutathione. Purified protein was dialyzed into SB as described above.
Dephosphorylation of Cortactin W525A and Immunoblotting
Purified recombinant His-cortactin W525A was incubated with 1.0 unit of calf intestinal phosphatase (CIP) (New England Biolabs, Inc.) for each microgram of protein for 4 h at room temperature (27 °C). His-cortactin W525A was repurified as described above to remove all CIP. Dephosphorylation was confirmed by immunoblotting. Protein (20 μg) was run on a 12% SDS—PAGE gel and transferred to a nitrocellulose membrane. The membrane was immunoblotted with 4G10 α-phosphotyrosine (Upstate) and 4F11 α-cortactin (Upstate), followed by HRP-conjugated secondary antibodies, and detected by chemiluminescence.
Actin Cosedimentation and Bundling Assays
Native monomeric actin was purified from chicken skeletal muscle as described previously.16–18 The bovine Arp2/3 complex was purified from bovine calf thymus as previously described.16 Actin was polymerized by dilution with an equal volume of F-actin polymerization buffer (FAPB) (150 mM KCl, 5 mM MgCl2, 2 mM EGTA, 20 mM imidazole, and 4 mM DTT) and stabilized by the addition of equimolar phalloidin (Sigma, St. Louis, MO). Cosedimentation assays were performed as previously described.10 Equal amounts of supernatant and pellet were separated on an SDS—PAGE gel and stained with Coomassie blue. The stoichiometry was determined by measuring Arg or cortactin and F-actin at saturating concentrations using densitometry, normalizing density to molecular weight, and calculating the ratio between Arg/cortactin and F-actin. The binding affinity (Kd) was quantified by plotting densitometry values of F-actin-bound Arg/cortactin against the concentration of titrated Arg/cortactin and fit using a one-site specific binding curve (GraphPad Prism). The equation y = Bmax × x/(Kd + x) was used to fit the curve, where y is specific binding, x is the concentration of the ligand, Bmax is the maximal binding in the same units as y, and Kd is the binding affinity in the same units as x.
Total Internal Reflection Microscopy (TIRFM) Experiments
Open-ended glass flow chambers were prepared and washed with skeletal muscle myosin as previously described. 19–21 Native monomeric actin was purified from chicken skeletal muscle as described previously16–18 and labeled with Alexa 488 actin conjugate (Invitrogen). Prior to the reaction, the chamber was washed with KMEI buffer [50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole (pH 7.0)]; 20% Alexa 488-labeled Ca2+-ATP actin was converted to Mg2+-ATP actin by addition of 0.05 mM MgCl2 and 0.2 mM EGTA followed by a 5 min incubation period. Equal parts 2× microscopy buffer [70 mM KCl, 1.4 mM MgCl2, 1.4 mM EGTA, 14 mM imidazole (pH 7.0), 1% (4000 cP) methylcellulose, 30 mM glucose, 400 μM ATP, 100 mM DTT, 40 μg/mL catalase, and 200 μg/mL glucose oxidase] and SB (see above) were mixed and added to the actin to induce polymerization. Samples then flowed into the chamber for imaging.
Images were collected every 20 s using prism-style total internal reflection microscopy on an Olympus IX-70 inverted microscope and Hamamatsu C4747-95 CCD (Orca-ER) camera with MetaMorph (Molecular Devices, Union City, CA). Changes in filament length were measured using ImageJ for approximately five filaments of each condition over 10—20 frames, which covered a span of at least 200 s. The rate of subunit growth or shrinkage was calculated using values of Huxley and Brown.22
RESULTS
Arg Increases the Stoichiometry of Binding of Cortactin to F-Actin
We measured whether the presence of cortactin affects the binding of Arg to F-actin using an actin cosedimentation assay (Figure 1). Saturating concentrations of cortactin (4 μM) were preincubated with a fixed amount of F-actin (1 μM), and increasing concentrations of Arg (0—3 μM) were then added to reach binding saturation. After high-speed centrifugation at 120000g, the amount of Arg cosedimenting with actin was measured. Consistent with previously published results, we found that binding of Arg to F-actin saturates at a stoichiometry of one Arg molecule to two actin monomers (1:2) (Figure 1B—D).10 This Arg:actin stoichiometry was unchanged when F-actin was preincubated with saturating concentrations of cortactin (Figure 1D—F).
We also performed the reciprocal experiment to test if Arg affects cortactin:actin binding stoichiometry. After preincubating F-actin with saturating amounts of Arg (3 μM), we added increasing concentrations of cortactin (0—4 μM) to reach saturation and quantified the stoichiometry of binding of cortactin to F-actin as described above (Figure 1K,L). Cortactin binding saturated at a ratio of one cortactin to four actin monomers (1:4) (Figure 1H—J). Interestingly, we found that inclusion of saturating concentrations of Arg in the reaction mixture increased cortactin binding stoichiometry by 2-fold to one cortactin to two actin monomers (1:2) (Figure 1J—L).
The Arg-Mediated Increase in Cortactin:F-Actin Stoichiometry Is Not Affected by Nucleating Concentrations of the Arp2/3 Complex
Cortactin is a weak activator of the Arp2/3 complex in actin polymerization.6,7 To determine if the Arp2/3 complex impacts the stoichiometry of cortactin—F-actin binding, we preincubated F-actin with 100 nM Arp2/3 complex, the concentration previously used in cortactin-activated nucleation promoting assays.6 The Arp2/3 complex does not affect the binding stiochiometry of cortactin, which maintained a saturation binding stoichimetry of 1:4 in the presence of the Arp2/3 complex (Figure 2A,B,E). The Arp2/3 complex decreased the affinity of cortactin for F-actin (Figure 2F). The Arp2/3 complex and cortactin both bind actin subdomain 1,14,23 and competition or steric hindrance by Arp2/3 complex binding may explain this result. We also tested the effects of the Arp2/3 complex on the ability of Arg to increase the binding stiochiometry of cortactin. We also found that the presence of 100 nM Arp2/3 complex did not affect the 1:2 cortactin:F-actin binding ratio observed in the presence of a saturating level of Arg (3 μM) (Figure 2C—E).
Figure 2.
Arg-dependent increase in cortactin:F-actin stoichiometry is not changed by the Arp2/3 complex. (A) Representative cosedimentation assay gel showing increasing concentrations of cortactin binding to F-actin preincubated with 100 nM Arp2/3 complex (quantified in panel B). Asterisks indicate Arp2/3 complex subunits. Kd values are means ± the standard error of the mean. (C) Representative cosedimentation assay gel showing increasing concentrations of cortactin binding to F-actin preincubated with saturating Arg and 100 nM Arp2/3 complex (quantified in panel D). (E and F) Quantification of (E) binding stoichiometry and (F) affinity from panels B and D. S and P denote the supernatant and pellet, respectively; controls are proteins centrifuged in the absence of F-actin. Values in panels E and F are means ± the standard error of the mean. One-way ANOVA with a post hoc unpaired two-tailed t test. **p < 0.01; ***p < 0.001.
Arg Stabilizes Actin Filaments
One possible explanation for the Arg-induced increase in stoichiometry between cortactin and F-actin would be destabilization or severing of the actin filaments by Arg. This would decrease the length of filaments and increase the number of free ends. If cortactin binds solely to the ends of filaments, it would increase the number of binding sites. To test this, we performed total internal reflection microscopy (TIRFM) to visualize actin filaments in the absence and presence of 2 μM Arg (Figure 3). We find that Arg does not sever filaments and, in fact, stabilizes filaments, leading to an approximatey 4-fold decrease in the rate of depolymerization (Figure 3B,C).
Figure 3.

Arg stabilizes actin filaments. (A) Representative TIRFM images of Alexa 488-labeled actin filaments alone (top) and filaments with 2 μM Arg (bottom). (B) Quantification of filament growth from time-lapse images. (C) Quantification of growth rates from panel B. Two-tailed t test. ***p < 0.001.
Arg—Cortactin Interactions Do Not Mediate the Increase in Cortactin—Actin Binding Stoichiometry
Arg and cortactin have two known interactions: the cortactin SH3 domain binds to a PYLPRLP motif in Arg, while the Arg SH2 domain can bind to tyrosine-phosphorylated cortactin.3,9 Arg increases the binding stoichiometry between cortactin and Factin from 1:4 to 1:2. On the basis of our finding that binding of Arg to F-actin saturates with an identical 1:2 binding ratio, we first hypothesized that F-actin-bound Arg may employ protein—protein interactions with cortactin to increase the cortactin binding stoichiometry. To test this hypothesis, we measured the F-actin binding stoichiometry of cortactin mutants in which these interaction interfaces were disrupted (Figures 1A and 4A), either alone or in the presence of saturating Arg concentrations. We mutated the conserved tryptophan residue in the cortactin SH3 domain to alanine (W525A), which disrupts its binding to the Arg PYLPRLP motif.3 We also mutated the three phosphorylatable tyrosine residues in cortactin (Y421, Y466, and Y482) to phenylalanine (3YF) to abrogate phosphorylation in insect cells and prevent Arg SH2 domain binding. Both cortactin W525A and cortactin 3YF exhibited saturating binding stoichiometries of 1:4 with F-actin (Figure 4C—F,O), identical to that of wild-type cortactin.
Figure 4.
Arg—cortactin interactions are not required for increased stoichiometry between cortactin and F-actin. (A) Domain structure of cortactin mutants. (B) Twenty micrograms of insect cell-purified WT cortactin, 3YF, and W525A (untreated or dephosphorylated with CIP) was immunoblotted with anti-phosphotyrosine antibody (pY, top) or anti-cortactin antibody (cortactin, bottom). WT and W525A exhibit tyrosine phosphorylation, while 3YF and CIP-treated samples do not. (C, E, and G) Representative cosedimentation assay gels showing increasing concentrations of 3YF, W525A, and W525A(-pY) binding to F-actin, respectively (quantified in panels D, F, and H, respectively). (I, K, and M) Representative cosedimentation assay gels showing increasing concentrations of 3YF, W525A, and W525A(-pY) binding to F-actin preincubated with saturating Arg, respectively (quantified in panels J, L, and N, respectively). Kd values are means ± the standard error of the mean. (O and P) Quantification of (O) binding stoichiometry and (P) affinity from panels D, F, H, J, L, and N. S and P denote the supernatant and pellet, respectively; controls are proteins centrifuged in the absence of F-actin. Values in panels O and P are means ± the standard error of the mean. One-way ANOVA with a post hoc two-tailed unpaired t test. *p < 0.05; n.s., not significant.
We also tested whether Arg affected the stoichiometry of binding of these cortactin mutants to F-actin. We found that both cortactin W525A and 3YF bound Arg-coated F-actin at the same stoichiometry (1:2) that was observed for WT cortactin, indicating that neither of these interaction interfaces is required to induce the observed increase in binding stoichiometry (Figure 4I—L,O). It remained possible that the presence of either of the two interaction interfaces was sufficient for the Arg-induced increase in cortactin:actin stoichiometry. However, we found that the Arg-enhanced binding ratio of 1:2 persisted even when we enzymatically dephosphorylated cortactin W525A [W525A(-pY)], abolishing both interaction interfaces (Figure 4G,H,M—O). These data indicate that neither of the defined Arg—cortactin binding interfaces mediates the Arg-induced increase in cortactin:actin binding stoichiometry.
The Arg CH Domain Is Necessary and Sufficient To Increase the Binding Stoichiometry between Cortactin and F-Actin
Arg binds actin via two distinct F-actin-binding domains, an internal I/LWEQ motif and a C-terminal calponin homology (CH) domain.10 Interestingly, the Arg CH domain induces a tilt in F-actin, which may affect how other proteins bind the filament.12 To test if either of the Arg F-actin-binding domains are required to increase cortactin:actin stoichiometry, we preincubated saturating amounts of various Arg deletion constructs (Figure 5A) with 1 μM F-actin and titrated cortactin to saturation (in 0—4 μM) (Figure 5). An Arg mutant lacking the entire cytoskeletal binding C-terminus of Arg (ArgΔC), and therefore both F-actin-binding domains, did not increase the cortactin:actin binding stoichiometry over that observed for cortactin alone (Figure 5B,C,N). This observation indicates that binding of F-actin by Arg is essential for its stimulation of cortactin:actin binding stoichiometry. Indeed, we found that that an Arg C-terminal fragment containing both F-actin-binding domains (Arg688-C) was sufficient to induce a 1:2 cortactin:actin binding stoichiometry (Figure 5D,E,N). Interestingly, an Arg deletion mutant lacking the internal I/LWEQ motif (ArgΔI/LWEQ) also promoted increased 1:2 binding stoichiometry to F-actin as well as wild-type Arg (Figure 5F,G,N), indicating that this internal F-actin-binding domain is not required for an increased cortactin:actin binding stoichiometry. In contrast, an Arg mutant lacking the C-terminal CH domain (ArgΔCH) did not affect cortactin:actin binding stoichiometry, showing that the Arg CH domain is necessary for augmentation of binding of cortactin to F-actin (Figure 5H,I,N). Furthermore, a glutathione S-transferase (GST) fusion to the Arg CH (GST-ArgCH) domain by itself was sufficient to induce a 1:2 binding stoichiometry between cortactin and F-actin (Figure 5J,K,N). GST alone had no effect on the cortactin:actin binding stoichiometry (Figure 5L—N).
Figure 5.
Arg CH domain is necessary and sufficient to increase stoichiometry between cortactin and F-actin. (A) Arg deletion mutant domain structures. Representative cosedimentation assay gels showing increasing concentrations of cortactin binding to F-actin preincubated with saturating ArgΔC (B), Arg688-C (D), ArgΔI/LWEQ (F), ArgΔCH (H), GST-ArgCH (J), or GST (L) (quantified in panels C, E, G, I, K, and M, respectively). Kd values are means ± the standard error of the mean. S and P denote the supernatant and pellet, respectively; controls are proteins centrifuged in the absence of F-actin. Quantification of binding stoichiometry (N) and affinity (O) from panels C, E, G, I, K, and M. Values in panels N and O are means ± the standard error of the mean. One-way ANOVA with a post hoc unpaired two-tailed t test. **p < 0.01; ***p < 0.001.
Previous studies have shown that the Arg CH domain acts cooperatively to induce a twist in the actin filament.12 We reasoned that if this helical twist played a role in the observed increased stoichiometry of cortactin, subsaturating concentrations of Arg might be sufficient to achieve this effect. We preincubated 0.3 μM Arg with 1 μM F-actin and titrated in cortactin to saturating concentrations (0—4 μM) (Figure 6A,B). We found that even at subsaturating concentrations of Arg, cortactin still bound F-actin at a saturation binding density of one cortactin per two actin monomers (Figure 6C), supporting the hypothesis that this cooperatively induced twist in the actin filament promotes enhanced binding of cortactin.
Figure 6.

Subsaturating amounts of Arg are sufficient to stimulate an increased cortactin:actin binding stoichiometry. (A) Representative cosedimentation assay gels showing increasing concentrations of cortactin binding to F-actin preincubated with subsaturating (0.3 μM) Arg (quantified in panel B). Kd values are means ± the standard error of the mean. S and P denote the supernatant and pellet, respectively; controls are proteins centrifuged in the absence of F-actin. (C and D) Quantification of binding stoichiometry (C) and affinity (D) from panel B. Values in panels C and D are means ± the standard error of the mean. One-way ANOVA with a post hoc unpaired two-tailed t test. **p < 0.001; n.s., not significant.
Disruption of Arg—Cortactin Interactions Affects the Affinity of Cortactin for F-Actin
To determine if Arg affects the affinity of cortactin for F-actin and vice versa, we quantified binding affinities from each cosedimentation assay performed. The presence of cortactin at saturating concentrations did not change the affinity of Arg for F-actin (Figure 1G). Similarly, saturating and subsaturating concentrations of wild-type Arg do not affect the affinity of cortactin for F-actin (Figures 1M and 6D).
We also measured the binding affinity of cortactin 3YF, W525A, and dephosphorylated W525A, which disrupt interactions with Arg. In the absence of Arg, these mutants bind F-actin with an affinity similar to that of wild-type cortactin. However, in the presence of saturating concentrations of Arg, all mutants showed an approximately 3-fold decrease in their affinity for F-actin (Figure 4P). This suggests that interactions with Arg help to increase the affinity of cortactin for F-actin, possibly by stabilizing binding of cortactin to F-actin in the presence of Arg. We also measured the affinity of cortactin for F-actin in the presence of saturating concentrations of Arg deletion mutants described above. We found that Arg688-C and GST-ArgCH cause a similar decrease in the affinity of cortactin for F-actin (Figure 5O). These two Arg deletion mutants lack domains necessary for interaction with cortactin, supporting the idea that these interactions stabilize cortactin on the filament, which translates into an increase in affinity.
DISCUSSION
Together, our studies reveal a potentially important mechanism by which Arg recruits cortactin to F-actin by increasing its binding stoichiometry with the filament. Importantly, this process does not rely on interactions between Arg and cortactin but rather occurs indirectly by the binding of the Arg CH domain to F-actin, which is necessary and sufficient to induce an increased cortactin:actin binding stoichiometry. Together with previously published work,12 our experiments show that increased stoichiometry may be due to structural changes in F-actin induced by the Arg CH domain, allowing cortactin to bind at a higher density.
We show that Arg increases the stoichiometry between cortactin and F-actin from one cortactin per four actin subunits to one cortactin per two actin subunits. The cortactin binding stoichiometry we observed was significantly greater than the 1:14 GST-cortactin:actin monomer ratio previously reported.4 It is possible that the GST moiety constrained cortactin binding or caused steric hindrance.4 Even when the two known Arg— cortactin interactions were mutationally disrupted, we found that Arg still increases the stoichiometry of binding of cortactin to F-actin, indicating that these interactions are not required for this stimulatory effect.
The observation that Arg—cortactin interactions are not required for this effect led to our hypothesis that one or both of the Arg F-actin-binding domains were allowing the filament to bind more molecules of cortactin. We used Arg deletion mutants and fragments to show that the C-terminal CH domain is both necessary and sufficient to induce an increased cortactin:actin stoichiometry. The Arg CH domain is approximately 27% identical to the CH domains of spectrin and fimbrin and binds to the same site on the SD1 actin subdomain as fimbrin, utrophin, and α-actinin.12,24–26 Binding by the Arg CH domain cooperatively induces a tilt in F-actin, such that the conformational change is able to propagate throughout the filament.12 We propose that this twist in F-actin alters the cortactin binding site to allow enhanced binding. Interestingly, the conformational tilt induced by the Arg CH domain is similar to that observed in ADF/cofilin binding, although no severing activity was observed for Arg (Figure 3).12,27 Studies have shown that a force-induced twist in the actin filament via application of force can increase the affinity of proteins such as myosin II for the filament.28 To the best of our knowledge, this is the first example of an actin-binding protein increasing the binding stoichiometry of another protein for the actin filament.
This novel actin recruitment mechanism may also be reflected by the unique nature of the actin-binding cortactin repeat itself, which shares little homology with other known actin-binding domains.4,5 Binding of Arg via its CH domain to F-actin also increases the level of disorder in actin subdomain 2,12 which could potentially increase the accessibility of the cortactin binding site (subdomains 1 and 3)14 on the adjacent actin monomer, allowing more molecules to bind as suggested in our model (Figure 7).
Figure 7.
Model for increased binding stoichiometry between cortactin and Arg-bound F-actin. In the absence of Arg, cortactin binds subdomains 1 and 3 of the actin subunit within an actin filament,14 at a binding stoichiometry of one cortactin molecule per four actin monomers (1:4). In the presence of the Arg CH domain, cortactin binds every two actin monomers (1:2). The Arg CH domain propagates a twist throughout the F-actin filament and leads to an increased level of dissociation in actin subdomain 2,12 which may modify the cortactin binding site on the filament or eliminate steric hindrance, allowing cortactin to bind with increased density to F-actin.
The Arg-stimulated increase in cortactin:actin stoichiometry may help recruit cortactin to sites of concentrated actin assembly or stabilization. The estimated concentration of Arg in dendritic spines, which contain concentrated, dynamic pools of F-actin, is approximately 0.5 μM.10,29 We show here that this Arg concentration is sufficient to increase the stoichiometry between cortactin and F-actin. Furthermore, the concentration of cortactin in MDA-MB-231 breast cancer cells has been found to be approximately 1 μM but may be nearly 4 times this in invadopodia, which would allow for recruitment of high concentrations of cortactin as proposed in our model (Figure 7).30 Arg and cortactin both localize to invadopodia where both are required for actin polymerization-driven maturation of invadopodia, and Arg-stimulated recruitment of cortactin to F-actin may be important for this process.8,30–32 Additionally, previous studies have shown that fibroblasts expressing Arg mutants lacking the F-actin-binding domains are unable to form F-actin-rich structures at the cell edge, and this failure may possibly be due to inefficient recruitment of cortactin to actin filaments.10
Finally, we also show that Arg—cortactin interactions are important for optimal cortactin affinity for F-actin, supporting our previous observations that these interactions are critical for actin-driven processes in cells.2,3 In the context of these previous studies, our work suggests a novel mechanism for increasing the concentration of cortactin on actin filaments, which may serve as an important recruitment mechanism at sites of actin polymerization.
Acknowledgments
Funding
This work was supported by U.S. Public Health Service Grants NS39475 and CA133346 to A.J.K. S.M.M. is supported by an American Heart Association Founders Affiliate Predoctoral Fellowship (11PRE7450039).
We thank Xianyun Ye for technical assistance; N. Courtemanche and T. Pollard for expert assistance with TIRFM experiments; C. Mader for experimental advice; E. De La Cruz, B. Rosenberg, M. Simpson, and D. Wetzel for critical comments on the manuscript; and members of the Koleske lab for helpful discussion and feedback.
ABBREVIATIONS
- Arg
Abl-related gene
- Arp
actin-related protein
- CH
calponin homology
- DTT
dithiothreitol
- EB
elution buffer
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- F-actin
filamentous actin
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- Kd
dissociation constant
- NTA
Nterminal acidic region
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- SDS
sodium dodecyl sulfate
- SH2
Src-homology 2
- SH3
Src-homology 3
- TIRFM
total internal reflection microscopy
- WT
wild type
Footnotes
The authors declare no competing financial interest.
References
- 1.Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- 2.Boyle SN, Michaud GA, Schweitzer B, Predki PF, Koleske AJ. A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr Biol. 2007;17:445–451. doi: 10.1016/j.cub.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 3.Lapetina S, Mader CC, Machida K, Mayer BJ, Koleske AJ. Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J Cell Biol. 2009;185:503–519. doi: 10.1083/jcb.200809085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Reynolds AB, Kanner SB, Vines RR, Parsons JT. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991;11:5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 7.Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mader CC, Oser M, Magalhaes MA, Bravo-Cordero JJ, Condeelis J, Koleske AJ, Gil-Henn H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, MacGrath SM, Koleske AJ, Boggon TJ. Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex. Acta Crystallogr. 2012;F68:154–158. doi: 10.1107/S1744309111056132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Miller AL, Mooseker MS, Koleske AJ. The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc Natl Acad Sci USA. 2001;98:14865–14870. doi: 10.1073/pnas.251249298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller AL, Wang Y, Mooseker MS, Koleske AJ. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J Cell Biol. 2004;165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galkin VE, Orlova A, Koleske AJ, Egelman EH. The Arg non-receptor tyrosine kinase modifies F-actin structure. J Mol Biol. 2005;346:565–575. doi: 10.1016/j.jmb.2004.11.078. [DOI] [PubMed] [Google Scholar]
- 13.Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: Systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 14.Pant K, Chereau D, Hatch V, Dominguez R, Lehman W. Cortactin binding to F-actin revealed by electron microscopy and 3D reconstruction. J Mol Biol. 2006;359:840–847. doi: 10.1016/j.jmb.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 15.Tanis KQ, Veach D, Duewel HS, Bornmann WG, Koleske AJ. Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol Cell Biol. 2003;23:3884–3896. doi: 10.1128/MCB.23.11.3884-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard TD. Polymerization of ADP-actin. J Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 19.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courtemanche N, Pollard TD. Determinants of Formin Homology 1 (FH1) Domain Function in Actin Filament Elongation by Formins. J Biol Chem. 2012;287:7812–7820. doi: 10.1074/jbc.M111.322958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul A, Pollard T. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huxley HE, Brown W. Low-Angle X-Ray Diagram of Vertebrate Striated Muscle and Its Behaviour during Contraction and Rigor. J Mol Biol. 1967;30:383. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 23.Goley ED, Rammohan A, Znameroski EA, Firat-Karalar EN, Sept D, Welch MD. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci USA. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galkin VE, Orlova A, VanLoock MS, Rybakova IN, Ervasti JM, Egelman EH. The utrophin actin-binding domain binds F-actin in two different modes: Implications for the spectrin superfamily of proteins. J Cell Biol. 2002;157:243–251. doi: 10.1083/jcb.200111097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanein D, Matsudaira P, DeRosier DJ. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J Cell Biol. 1997;139:387–396. doi: 10.1083/jcb.139.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGough A, Way M, DeRosier D. Determination of the α-actinin-binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uyeda TQ, Iwadate Y, Umeki N, Nagasaki A, Yumura S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS One. 2011;6:e26200. doi: 10.1371/journal.pone.0026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- 30.Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, Chen X, Koleske AJ, Condeelis J. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. 2011;195:903–920. doi: 10.1083/jcb.201103045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oser M, Mader CC, Gil-Henn H, Magalhaes M, Bravo- Cordero JJ, Koleske AJ, Condeelis J. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J Cell Sci. 2010;123:3662–3673. doi: 10.1242/jcs.068163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J, Koleske AJ, Condeelis J. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]