Abstract
Friedreich ataxia (FRDA) is an autosomal recessive neurodegenerative disorder caused by a dynamic GAA repeat expansion mutation within intron 1 of the FXN gene. Studies of mouse models for other trinucleotide repeat (TNR) disorders have revealed an important role of mismatch repair (MMR) proteins in TNR instability. To explore the potential role of MMR proteins on intergenerational GAA repeat instability in FRDA, we have analyzed the transmission of unstable GAA repeat expansions from FXN transgenic mice which have been crossed with mice that are deficient for Msh2, Msh3, Msh6 or Pms2. We find in all cases that absence of parental MMR protein not only maintains transmission of GAA expansions and contractions, but also increases GAA repeat mutability (expansions and/or contractions) in the offspring. This indicates that Msh2, Msh3, Msh6 and Pms2 proteins are not the cause of intergenerational GAA expansions or contractions, but act in their canonical MMR capacity to protect against GAA repeat instability. We further identified differential modes of action for the four MMR proteins. Thus, Msh2 and Msh3 protect against GAA repeat contractions, while Msh6 protects against both GAA repeat expansions and contractions, and Pms2 protects against GAA repeat expansions and also promotes contractions. Furthermore, we detected enhanced occupancy of Msh2 and Msh3 proteins downstream of the FXN expanded GAA repeat, suggesting a model in which Msh2/3 dimers are recruited to this region to repair mismatches that would otherwise produce intergenerational GAA contractions. These findings reveal substantial differences in the intergenerational dynamics of expanded GAA repeat sequences compared with expanded CAG/CTG repeats, where Msh2 and Msh3 are thought to actively promote repeat expansions.
Keywords: Friedreich ataxia, FRDA, frataxin, GAA trinucleotide repeat, transgenic mouse model, mismatch repair, MMR, Msh2, Msh3, Msh6, Pms2
Introduction
Friedreich ataxia (FRDA) is an autosomal recessive neurodegenerative disorder caused by homozygous GAA repeat expansion within intron 1 of the FXN gene (Campuzano et al., 1996), which induces heterochromatin formation, possibly due to abnormal DNA or DNA•RNA hybrid triplex structures, resulting in FXN gene silencing and hence reduced expression of the essential mitochondrial protein frataxin (Campuzano et al., 1997). Frataxin insufficiency leads to oxidative stress, mitochondrial iron accumulation and resultant cell death, with the primary site of pathology being in the large sensory neurons of the DRG and the dentate nucleus of the cerebellum (Koeppen, 2011). The outcome is progressive spinocerebellar neurodegeneration and cardiomyopathy, with death commonly in early adulthood (Pandolfo, 2009). At present there is no effective treatment for FRDA. Unaffected individuals have FXN alleles containing 5–32 GAA repeats, there is a premutation range of 33–65 GAA repeats, and affected individuals have alleles of 66–1700 GAA repeats. The GAA repeats are dynamic, exhibiting both intergenerational and somatic instability. Thus, nonpathogenic parental premutations can be transmitted to offspring as expanded pathogenic GAA repeats (Montermini et al., 1997). Further expansion and contraction of pathological GAA repeat expansions are detected equally during maternal transmission, while there is a preference towards contraction during paternal transmissions (De Michele et al., 1998; Delatycki et al., 1998; Monros et al., 1997; Pianese et al., 1997). Progressive somatic GAA repeat expansion occurs in a subset of tissues throughout life, being particularly prominent in the disease-relevant cerebellum and dorsal root ganglia (DRG) (De Biase et al., 2007a; De Biase et al., 2007b). Therefore, GAA repeat dynamics may play an important role in disease progression, and identifying ways to prevent GAA repeat expansion or to induce GAA repeat contractions would be very useful therapeutic strategies. However, we first need to understand much more about the molecular mechanisms underlying FXN GAA repeat expansion dynamics. With this in mind, precedents from mouse model studies of the other trinucleotide repeat (TNR) expansion diseases, Huntington disease (HD) and myotonic dystrophy type 1 (DM1), have revealed an important role of mismatch repair (MMR) proteins in the dynamics of CAG and CTG repeats, respectively (Lopez Castel et al., 2010). Thus, Msh2 and Msh3 proteins have been shown to promote both intergenerational and somatic TNR expansions, perhaps by unusual Msh2–3 complex stabilization of insertion/deletion loops or else by an alternative role for Msh2 which does not involve the usual Msh2–3 and Msh2–6 heterodimer functioning of MMR (Dragileva et al., 2009; Foiry et al., 2006; Kovtun and McMurray, 2001; Savouret et al., 2003; Wheeler et al., 2003). The role, if any, of the Msh6 protein is less clear, with some studies showing parental gender-specific intergenerational effects to promote TNR expansion and protect against TNR contraction (Dragileva et al., 2009; Foiry et al., 2006), while other studies show either no effect (CAG repeats) or protection against expansion (CTG repeats) at the somatic instability level (Dragileva et al., 2009; van den Broek et al., 2002). Pms2 has not previously been studied at an intergenerational level, but has been shown to play a role in enhancing somatic expansion of CTG repeats in a DM1 mouse model (Gomes-Pereira et al., 2004). An important outcome of these previous studies has been the proposal to target either MSH3 or MSH6 as a form of TNR-reducing therapy in HD and DM1 (Dragileva et al., 2009; Foiry et al., 2006).
To enable similar studies of GAA repeat expansion dynamics for FRDA, we have previously generated two lines of GAA repeat expansion-containing human FXN transgenic mice, YG8 (90 and 190 GAA repeats) and YG22 (190 GAA repeats), that exhibit intergenerational and somatic instability of the GAA repeat expansion (Al-Mahdawi et al., 2004). In particular, we have detected specific age-related expansion of the GAA repeat in YG8 and YG22 transgenic mouse cerebellum and DRG tissues (Clark et al., 2007), as seen in FRDA patient autopsy tissues. To now explore the potential role of MMR proteins on intergenerational GAA repeat instability, we have crossed YG8 and YG22 transgenic mice with knockout mice that are deficient for Msh2, Msh3, Msh6 or Pms2 (Baker et al., 1995; de Wind et al., 1995; de Wind et al., 1999), and analyzed the GAA repeat sizes in offspring from mice that were either wild type (+/+), heterozygous (+/−) or homozygous (−/−) for MMR genotypes. We find in all cases that absence of the MMR protein in the parent results in increased GAA repeat mutability (expansions or contractions) in the offspring. Thus we conclude that Msh2, Msh3, Msh6 and Pms2 proteins each act in a typical MMR fashion to protect against GAA repeat instability. However, we also identified some distinct differences between the inferred actions of the four MMR proteins. Thus, Msh2 and Msh3 protect against GAA contractions, while Msh6 protects against both expansions and contractions, and Pms2 protects against expansions and promotes contractions.
Materials and methods
Animal procedures
Mice were housed in conventional open cages with Litaspen Premium 8/20 bedding, paper wool nesting and standard fun tunnel environmental enrichment, with 13 hours light, 11 hours dark, 20–23°C and 45–60% humidity. The mice were given a diet of SDS RM3 Expanded food pellets and standard drinking water. All procedures were carried out in accordance with the UK Home Office ‘Animals (Scientific Procedures) Act 1986’. YG8 and YG22 FXN GAA repeat expansion-containing transgenic mice (Al-Mahdawi et al., 2004) with GAA repeat sizes ranging from 90–230 repeats were crossed with Msh2, Msh3, Msh6 or Pms2 heterozygous knockout mice (Baker et al., 1995; de Wind et al., 1995; de Wind et al., 1999). All mice were maintained in a predominant C57BL/6J genetic background. Double genetically modified mice containing the FXN GAA transgene together with wild type, heterozygous or homozygous MMR knockout alleles were then crossed with non-GAA transgenic mice to obtain the necessary offspring for subsequent analysis.
Trinucleotide repeat analysis
Genomic DNA was isolated from tail biopsies of the parents and offspring by phenol/chloroform extraction. GAA PCR amplification was carried out with a conventional PCR kit (Qiagen) using 200–500ng DNA, GAA-F and GAA-R primers and PCR conditions as previously described (Campuzano et al., 1996). GAA PCR products were resolved in 20-cm long 1.5% agarose 1 X TBE gels by electrophoresis at 50V for 16–20h and band sizes were determined by comparison with 100bp DNA size markers (Invitrogen). The number of GAA repeats were then determined by subtracting 451bp (flanking non-repeat DNA) from the PCR product size, followed by division of the remaining base pair repeat size by 3. The mouse chromosome 19 CGG repeat was amplified as a 386bp PCR product using primers CGG-m-F: ATGTCCTAATTCGTATGACAGCTGG and CGG-m-R: TGCTACAGGAATCACAGGCTTCTA, and the mouse chromosome 13 CAG repeat was amplified as a 605bp PCR product using primers CAG-m-F: TGTCGCCTCAGCCCTTGTGG and CAG-m-R: CTGCTCTGCCGTTTGGGGCA.
MMR chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out using snap-frozen mouse brain tissue from Y47 (Pook et al., 2001), YG8 and YG22 transgenic mice (Al-Mahdawi et al., 2004) as previously described (Al-Mahdawi et al., 2008), using antibodies against mouse Msh2 (Abcam ab16833), Msh3 (Abcam ab74607), Msh6 (Abcam ab92471) and Pms2 (Abcam ab13900). Quantitative PCR amplification of ChIP DNA was performed for 5 regions of the FXN locus: −1254, promoter (Pro), GAA upstream (Up), GAA downstream (Down) and intron 2 (+11482), using previously described human FXN primer pairs (Herman et al., 2006; Ku et al., 2010).
Statistical analysis
Significant differences of the frequency distributions of transmitted ‘GAA repeat expansions’, ‘no changes’ and ‘GAA repeat contractions’ between groups of wild type, heterozygous or homozygous MMR knockout parental genotypes were determined using χ2 analysis. Differences between MMR ChIP values were determined using the Student’s t test. A P value of 0.05 was chosen as the significance threshold.
Results
MMR deficits produce increased GAA repeat mutability
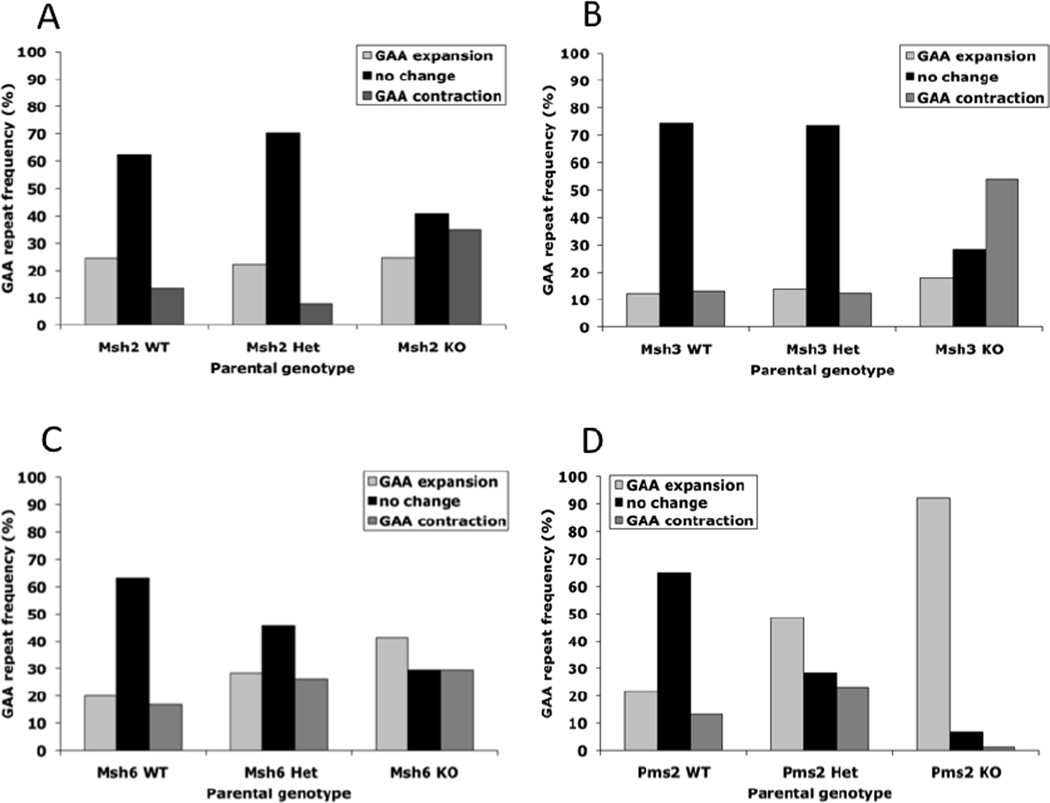
The effects of Msh2, Msh3, Msh6 and Pms2 MMR proteins on the intergenerational transmission of YG8 and YG22 FXN transgenic GAA repeats were assessed by comparing the GAA repeat sizes of offspring with those of the parent (Fig. 1). In each case, three parental MMR genotypes were employed: FXNGAA/MMR+/+, FXNGAA/MMR+/− and FXNGAA/MMR−/−. Furthermore, both YG8 and YG22 parents and both male and female parents were used (except for Pms2−/− males, which are sterile). For each subset, the transmitted GAA repeat size differences were collated into ‘GAA expansion’, ‘no change’ or ‘GAA contraction’ groups and the percentage frequencies and mean size changes of each group were determined to give an overall ‘transmission profile’.
Fig. 1.
Representative example of the ethidium bromide-stained agarose gels used to determine GAA repeat sizes, showing an inverted image of PCR products obtained from a YG22 GAA+/Msh3−/− female parent (P) and 12 GAA+ offspring. M=100bp DNA size markers.
Initial analysis of data based only upon parental MMR genotype (Fig 2) reveals a similar level of mutability (combined frequencies of expansions and contractions) of 25–37% for each of the four MMR WT (FXNGAA/MMR+/+) transmission profiles. This is consistent with our previously reported findings for YG8 and YG22 transgenic mice (Al-Mahdawi et al., 2004). The level of mutability then notably increases for all four MMR KO (FXNGAA/MMR−/−) groups: Msh2−/− is 59% (24% expansions, 35% contractions); Msh3−/− is 72% (18% expansions, 54% contractions); Msh6−/− is 71% (42% expansions, 29% contractions), and Pms2−/− is 93% (92% expansions, 1% contractions). These results indicate that Msh2, Msh3, Msh6 and Pms2 are not the cause of intergenerational GAA expansions or contractions, but are all involved in MMR processes to protect against intergenerational GAA repeat instability. In the case of Msh2 and Msh3, increased mutability only occurs in the absence of both MMR alleles, as revealed by identifying a significant difference between Msh2+/+ and Msh2−/− or Msh2+/− and Msh2−/− transmission profiles, but finding no difference between Msh2+/+ and Msh2+/− transmission profiles (Fig. 2A, Table 1), and similarly there is a significant difference between Msh3+/+ and Msh3−/− or Msh3+/− and Msh3−/− transmission profiles, but no difference between Msh3+/+ and Msh3+/− transmission profiles (Fig. 2B, Table 1). These findings suggest that single alleles of Msh2 and Msh3 are sufficient to produce adequate functional levels of MMR proteins. In contrast, Msh6 and Pms2 show a degree of increased mutability with the loss of only one allele, but an even greater increase in mutability with the loss of both alleles, as demonstrated by the identification of significant differences between all three pair-wise comparisons of transmission profiles (Figs 2C and 2D, Table 1). This suggests that both Msh6 and Pms2 alleles are required to fully maintain adequate MMR protection against GAA repeat instability, but a single allele can afford partial protection.
Fig. 2.
Intergenerational GAA repeat expansion analysis based upon MMR parental genotype (WT = GAA+/MMR+/+, Het = GAA+/MMR+/−, KO = GAA+/MMR−/−). Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions. (A) Msh2: WT n=164, Het n=198, KO n=98. (B) Msh3: WT n=114, Het n=137, KO n=186. (C) Msh6: WT n=65, Het n=241, KO n=181. (D) Pms2: WT n=156, Het n=457, KO n=88.
Table 1.
χ2 analysis of GAA repeat transmissions.
| MMR parental genotype 1 |
MMR parental genotype 2 |
χ2 value | df | P value |
|---|---|---|---|---|
| Msh2+/+ | Msh2+/− | 4.04 | 2 | 0.132 |
| Msh2+/+ | Msh2−/− | 18.17 | 2 | <0.001 |
| Msh2+/− | Msh2−/− | 38.63 | 2 | <0.001 |
| Msh3+/+ | Msh3+/− | 0.39 | 2 | 0.822 |
| Msh3+/+ | Msh3−/− | 64.40 | 2 | <0.001 |
| Msh3+/− | Msh3−/− | 67.50 | 2 | <0.001 |
| Msh6+/+ | Msh6+/− | 6.26 | 2 | 0.044 |
| Msh6+/+ | Msh6−/− | 23.25 | 2 | <0.001 |
| Msh6+/− | Msh6−/− | 12.87 | 2 | <0.01 |
| Pms2+/+ | Pms2+/− | 66.63 | 2 | <0.001 |
| Pms2+/+ | Pms2−/− | 111.00 | 2 | <0.001 |
| Pms2+/− | Pms2−/− | 57.00 | 2 | <0.001 |
MMR deficits produce different intergenerational GAA repeat instability effects
Although Msh2, Msh3, Msh6 and Pms2 deficits each produce an overall effect of increased levels of GAA repeat mutability, there are differences as to whether this mutability is due to increased GAA repeat expansion or increased GAA repeat contraction. Thus, both Msh2−/− and Msh3−/− mice show a significantly increased frequency of transmitting GAA repeat contractions to their offspring, while the frequency of GAA expansions remains unchanged (Figs. 2A and 2B, Table 1). Furthermore, the mean GAA repeat size variations (i.e. the summation of all GAA size changes divided by the total number of GAA repeat transmissions) for both Msh2−/− and Msh3−/− show a bias towards greater contraction size, compared with both the wild-type and heterozygous states (Table 2). In contrast, both Pms2+/− and Pms2−/− mice transmit significantly increased frequencies of GAA repeat expansions and increased mean size variations of GAA repeat expansions to their offspring, which appear to occur in an allele-dose dependent manner (Fig. 2D, Tables 1–2). Pms2−/− mice also transmit fewer GAA repeat contractions (Fig. 2D). On the other hand, Msh6+/− and Msh6−/− mice transmit marginally increased frequencies of both GAA repeat expansions and contractions to their offspring, with a small trend towards an increased mean GAA repeat size variation (Fig. 2C, Tables 1–2).
Table 2.
Mean transmitted GAA repeat size variations
| Parental genotype | Mean GAA repeat size increase of expansions |
Mean GAA repeat size decrease of contractions |
Mean GAA repeat size variation of all transmissions |
|---|---|---|---|
| Msh2+/+ | +2.6 | −3.3 | +0.19 |
| Msh2+/− | +8.0 | −17.9 | +0.42 |
| Msh2−/− | +2.6 | −9.1 | −2.54 |
| Msh3+/+ | +2.2 | −3.5 | −0.19 |
| Msh3+/− | +2.7 | −2.9 | +0.03 |
| Msh3−/− | +3.7 | −4.7 | −1.88 |
| Msh6+/+ | +1.8 | −3.6 | −0.24 |
| Msh6+/− | +6.2 | −5.5 | +0.31 |
| Msh6−/− | +3.7 | −3.5 | +0.53 |
| Pms2+/+ | +6.4 | −8.2 | +0.28 |
| Pms2+/− | +5.7 | −4.0 | +1.93 |
| Pms2−/− | +7.6 | −1.0 | +7.00 |
The effects of MMR deficits on GAA repeat expansion transmissions are independent of both FXN transgene type and parental gender
To determine if the two GAA repeat expansion-containing FXN transgenes showed any differences in intergenerational GAA repeat instability, data from YG8 and YG22 parental genotypes were analyzed separately (Figs. S1 and S2). Individual YG8 and YG22 results were found to be consistent with our previous combined YG8/YG22 analysis (Fig. 2). Thus, we observed increased GAA repeat mutability in the offspring from all YG8 or YG22 MMR−/− mice, together with an increased frequency of GAA repeat contractions from Msh2−/− and Msh3−/− mice, increased expansions and contractions from Msh6−/− mice and a significantly increased frequency of GAA repeat expansions from Pms2−/− mice. Furthermore, to determine if parental gender had any effect on the resultant intergenerational GAA repeat instability, we further analyzed the data on the basis of male and female parents separately (Figs. S3 and S4). Once again, the results of GAA repeat expansion and contraction profiles from separate paternal and maternal transmissions were each in general accord with our previous combined parental analysis. Thus, there is increased GAA repeat mutability from both paternal and maternal MMR−/− mice, increased GAA repeat contractions from Msh2−/− and Msh3−/− mice, increased expansions and contractions from Msh6−/− mice and increased expansions from Pms2+/− mice (n.b. there are no values for Pms2 KO males as these are sterile). We conclude that the main effects of MMR parental genotype on intergenerational GAA repeat instability are not significantly modified by parental transgenic GAA sequence or parental gender.
MMR deficits have different effects on expanded GAA repeats compared with tandem CGG and CAG repeats
To determine if the increased mutability induced by MMR deficits is specific for the expanded GAA repeat or a more general effect for TNR sequences, we similarly investigated the parent-to-offspring transmission of a 12 repeat CGG sequence and a 21 repeat CAG sequence, which were located on mouse chromosomes 19 and 13, respectively (Fig. 3). Loss of Msh2 increased mutability for both CGG and CAG repeats and loss of Pms2 increased mutability for CGG repeats. However, Msh3 and Msh6 loss resulted in either decreased mutability or no change in mutability for CGG and CAG repeats, while loss of Pms2 produced no change in mutability for CAG repeats. Therefore, we conclude that the overall effects of MMR deficits on expanded GAA repeats compared with CGG or CAG repeats are different. MMR actions are likely to be specific for expanded GAA repeats within the context of the FXN transgene sequence.
Fig. 3.
Mouse CGG and CAG tandem repeat analysis. (A) Representative example of mouse chromosome 19 CGG tandem repeat PCR products, showing unstable transmission of repeats from Msh3 WT parents, compared with stable transmission of repeats from an Msh3 KO parent. (B) Representative example of mouse chromosome 13 CGG tandem repeat PCR products, showing stable transmission of repeats from both Msh6 WT and Msh6 KO parents. P = parent sample, M = 100bp MW markers. The tables shown beneath indicate the percentages of transmitted expansions (↑), no changes (−) or contractions (↓), n = 9–12.
Enrichment of Msh2 and Msh3 proteins within expanded GAA repeat-containing FXN transgenic loci
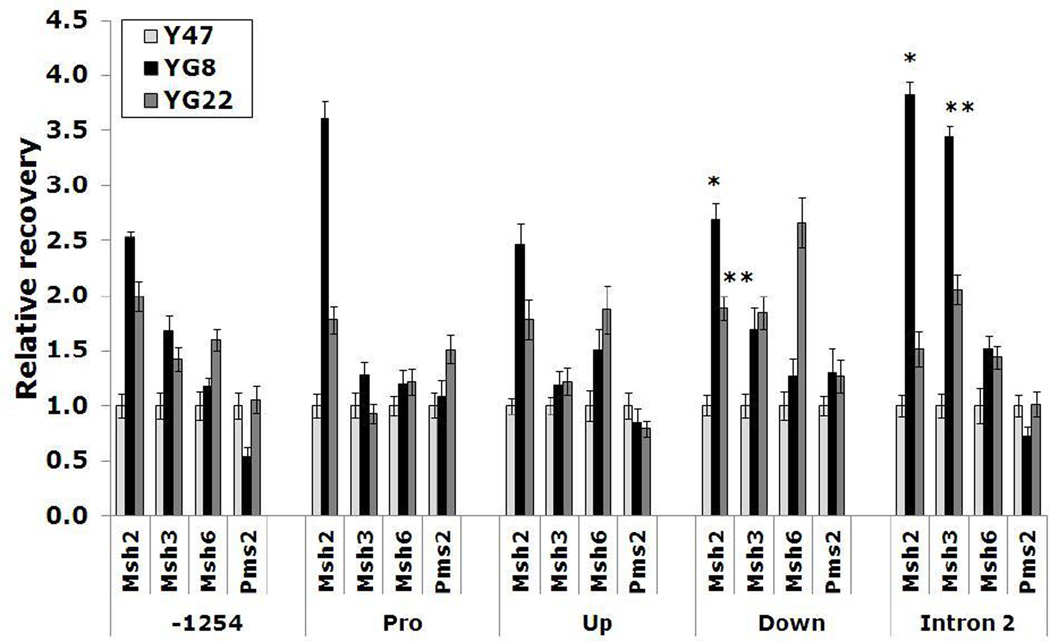
Recently, it has been reported that MSH2 and MSH3 proteins are enriched just downstream of the FXN expanded GAA repeat tract in human induced pluripotent stem cells (iPSCs) (Ku et al., 2010). To determine if this is also the case for mouse cells, ChIP experiments were performed to investigate occupancy of mouse Msh2, Msh3, Msh6 and Pms2 proteins at five regions of the transgenic FXN locus (-1254, promoter, upstream GAA, downstream GAA and intron 2) in unstable expanded GAA repeat-containing YG8 and YG22 FXN transgenic mice (Al-Mahdawi et al., 2004), compared with stable normal-sized GAA repeat-containing Y47 FXN transgenic mice (Pook et al., 2001). Consistent enrichment of Msh2 occupancy was detected at all 5 regions of the FXN locus in both YG8 and YG22 mice compared with Y47 mice (Fig. 4). However, the most significant levels of Msh2 enrichment were found at the downstream GAA region and the intron 2 region of the FXN locus. Msh3 also showed enriched occupancy of the downstream GAA and intron 2 regions (Fig. 4). In contrast, Msh6 and Pms2 did not show changes of occupancy at any region of the FXN locus, with the one exception of Msh6 enrichment at the downstream GAA region in YG22 mice. Overall, these results concur with the previous findings in human cells, indicating a role for Msh2 and Msh3 proteins at the FXN region immediately downstream of expanded GAA repeat sequences.
Fig. 4.
ChIP data showing Msh2, Msh3, Msh6 and Pms2 occupancy at 5 regions of the FXN locus: 1254bp upstream of the transcription start site (−1254), promoter (Pro), upstream GAA (Up), downstream GAA (Down) and 11482bp downstream of the transcription start site (intron 2). Columns represent quantitative PCR data from IP samples relative to input. YG8 and YG22 values are shown as fold changes relative to Y47 set at 1.0. * = P<0.05, ** = P<0.01. Error bars = s.e.m. of independent experiments, n = 4.
Discussion
MMR proteins play a role in intergenerational GAA repeat instability in FRDA
To investigate potential molecular mechanisms involved in the intergenerational GAA repeat expansion dynamics of the inherited TNR disorder FRDA, we have analyzed the role of the MMR proteins Msh2, Msh3, Msh6 and Pms2 in the transmission of the GAA repeats from YG8 and YG22 FXN transgenic mice. We report that the deficit of each MMR gene in parental mice results in continued transmission of GAA expansions and contractions, but increases overall mutability of the GAA repeat expansion in offspring. Therefore, we conclude that during intergenerational transmission of the GAA repeat, Msh2, Msh3, Msh6 and Pms2 proteins are not the cause of GAA expansions or contractions, but rather they act in a repair capacity to protect against the adverse effects of GAA repeat instability. This would agree with the generally accepted view of the function of the MMR system, which is to repair DNA sequence errors following replication or recombination, with Msh2–3 heterodimers identifying small insertion/deletion loops (IDLs), Msh2–6 heterodimers identifying single base pair mismatches and single base IDLs, and Mlh1-Pms2 heterodimers subsequently interacting with both Msh2–3 and Msh2–6 heterodimers to continue the repair process (Schofield and Hsieh, 2003). However, we also found distinct differences between the effects of Msh2 and Msh3 proteins compared with Pms2 and, to a lesser extent, with Msh6. Thus, Msh2 and Msh3 were found to protect against GAA repeat contractions, whereas Pms2 was seen to significantly protect against GAA repeat expansions, while also contributing to the promotion of contractions. Msh6 had only a minor protective effect against both GAA expansions and contractions. Therefore, rather than there being a single MMR system in operation, there are likely to be at least two distinct mechanisms involving MMR proteins acting on FRDA intergenerational GAA repeat instability: one mechanism predominantly involving Msh2 and Msh3 proteins that acts to limit GAA repeat contractions, and another mechanism predominantly involving the Pms2 protein that acts to limit GAA repeat expansions, with the Msh6 protein perhaps playing a minor role in one or other system.
Comparison with CAG/CTG transgenic mouse intergenerational instability studies
Our findings of increased frequencies of GAA repeat contractions in the offspring of Msh2−/− and Msh3−/− mice, and to a lesser extent of Msh6−/− mice, are in agreement with previous studies of HD and DM1 transgenic mice, which identified similar effects of Msh2−/−, Msh3−/− and Msh6−/− deficits on CAG repeats and CTG repeats, respectively (Dragileva et al., 2009; Foiry et al., 2006; Savouret et al., 2003; Wheeler et al., 2003). This suggests that there may be a common repair component of the MMR system that is operating on all TNR expansion sequences to suppress intergenerational repeat contraction. However, other findings highlight differences in the way that the MMR system acts on expanded GAA versus CAG/CTG repeats. Thus, an important difference between our GAA repeat results and those for CAG/CTG repeats is the fact that we do not observe any loss of TNR expansions transmitted from Msh2−/− and Msh3−/− mice as seen with HD and DM1 transgenic mice; our GAA repeat expansion frequencies remain unaffected by parental Msh2 or Msh3 genotype. Therefore, Msh2 and Msh3 cannot be considered to be essential for intergenerational GAA repeat expansions in FRDA as has been proposed for the CAG/CTG repeats of HD and DM1 (Dragileva et al., 2009; Foiry et al., 2006). This is most likely due to differences in the secondary structure of GAA repeat expansion compared with CAG/CTG repeat expansions. For CAG repeats, one proposal, described as ‘hijacking’ of the MMR system, is where the normal MMR function of Msh2–3 heterodimers to detect and repair IDLs no longer takes place when encountering CAG hairpin loops. Instead there is non-canonical Msh2–3 binding that prevents repair and preserves the expansion (McMurray, 2008). Another proposal is that CAG expansions are induced by an alternative, as yet unknown, Msh2-dependent mechanism that is independent of Msh3 or Msh6 (Dragileva et al., 2009). Such divergent MMR mechanisms for Msh2 and Msh3 proteins do not appear to operate for intergenerational transmission of GAA repeat expansions, since neither Msh2 nor Msh3 deficits were found to have any effect on GAA expansion frequencies.
However, we did identify Msh6 and Pms2 effects on GAA repeat expansion frequencies. Thus, our results show that absence of either Msh6 or Pms2 proteins produces an effect that increases the frequency and size of GAA expansions, and in both cases the effects are allele-dose dependent. Therefore, transmission of GAA repeat expansions may occur when there is insufficient expression of either of these MMR proteins to perform their normal functions in DNA repair. Our study is, to the best of our knowledge, the first to investigate the role of Pms2 in intergenerational TNR instability. Therefore, it would be interesting to now see if Pms2 has a similar effect to protect against expansions during the genetic transmission of CAG/CTG repeats in HD and DM1, especially as this would differ from the expansion-enhancing role of Pms2 identified in somatic tissues (Gomes-Pereira et al., 2004). In contrast to our findings with GAA repeats, no increase in intergenerational CAG/CTG expansion frequency has previously been observed in Msh6-deficient HD or DM1 transgenic mice. However, interestingly, increased somatic CTG expansions have been reported in some tissues of Msh6-deficient DM1 mice (van den Broek et al., 2002). Finally, we did not identify any of the parental gender-specific effects that have previously been reported for both HD and DM1 transgenic mice (Dragileva et al., 2009; Foiry et al., 2006; Savouret et al., 2003; Wheeler et al., 2003). Overall, we conclude that the differences of MMR effects on GAA and CAG/CTG repeat dynamics in transgenic mice outweigh their similarities, perhaps reflecting an underlying difference in how the MMR system recognizes and acts upon the TNR secondary structures. Interactions of the MMR system with other molecular mechanisms may also affect repeat dynamics in the context of each specific TNR sequence. Thus, in human cell culture systems, transcription has been shown to increase the frequency of CAG repeat contractions by a mechanism that is dependent on MSH2 and MSH3, but not MSH6 (Lin et al., 2006). In contrast, transcription has been shown to induce GAA repeat expansions (Ditch et al., 2009), possibly by the interaction of transcription-coupled nucleotide excision repair (TC-NER) with the MSH2–3 complex (Zhao et al., 2009). Furthermore, base excision repair (BER) has been implicated in the formation of CAG, but not GAA, repeat expansions (Goula et al., 2009; Kovtun et al., 2007), and BER has been shown to interact with the MSH2–6 complex (Gu et al., 2002; Kovtun and McMurray, 2007).
Comparison with other MMR GAA repeat instability studies
Our findings support the general notion that the MMR system has an important role to play in GAA repeat instability, as first reported by a study that described MMR-dependent chromosome fragility of GAA repeat expansions in yeast (Kim et al., 2008), and more recently by a study of GAA repeats in human FRDA iPSCs (Ku et al., 2010). In this latter study, the GAA expansion instability of iPSCs correlated with increased MSH2 expression and increased occupancy of MSH2 and MSH3 proteins in the downstream GAA region of the FXN gene. Knockdown of MSH2 expression in human iPSCs produced attenuation of GAA repeat expansion, thus resembling the Msh2-dependent mechanism of somatic CAG/CTG expansion in HD and DM1 transgenic mice (Savouret et al., 2003; Wheeler et al., 2003). We now report that Msh2 and Msh3 proteins also show enriched occupancy of the FXN downstream GAA region in transgenic mice that contain unstable expanded GAA repeats compared with transgenic mice that contain stable normal-sized GAA repeats. However, somewhat in contrast to previous studies, we found that Msh2 protects against GAA contraction, but is not necessary for GAA expansion, during intergenerational transmission in mice. This suggests that differences in GAA repeat dynamics may exist between somatic cell division and intergenerational germline transmission, which now warrant further investigation.
Conclusions
Our studies highlight the importance of the MMR system in the intergenerational GAA repeat expansion dynamics of FRDA, as previously described for the CAG/CTG repeat expansions of HD and DM1 (Lopez Castel et al., 2010). However, it is apparent that intergenerational GAA repeat expansion dynamics differ from those of CAG/CTG repeats. Within our proposed model, neither Msh2 nor Msh3 are involved in causing GAA expansions or contractions, since both GAA expansions and contractions still occur to some degree in the absence of either Msh2 or Msh3. Therefore, additional, as yet unknown, mechanisms of GAA instability that do not involve the MMR system must be in operation. We propose that Msh2/3 dimers are recruited to the FXN downstream GAA region to repair mismatches that occur here, most likely within the 3’ segment of the GAA repeat itself. Msh2/3 repair processes then act to prevent the occurrence of GAA contractions. This model contrasts with that for CAG/CTG repeats, where Msh2 and Msh3 appear to promote intergenerational repeat expansion. Within our model of MMR effects on GAA repeat instability, Pms2 acts by a different mechanism to Msh2/3, and at an undefined location of the GAA repeat, to prevent the occurrence of GAA expansions and even to promote contractions. As with CAG/CTG intergenerational instability, Msh6 has a less obvious role to play in intergenerational GAA repeat dynamics, but appears to involve some protection against both expansions and contractions. Ultimately, both GAA and CAG/CTG intergenerational repeat dynamics are likely to arise from a complicated interplay between several different molecular mechanisms, including transcription and DNA repair systems, of which MMR is only one. Thus, further investigations, are now needed to uncover precise mechanisms of GAA repeat instability, which may then lead to the identification of potential targets for FRDA therapy.
Supplementary Material
Fig. S1. GAA repeat transmissions from YG8 transgenic mice. Intergenerational GAA repeat analysis based upon YG8 MMR parental genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=109, Msh2 Het n=56, Msh2 KO n=48, Msh3 WT n=51, Msh3 Het n=33, Msh3 KO n=15, Msh6 WT n=27, Msh6 Het n=45, Msh6 KO n=43, Pms2 WT n=102, Pms2 Het n=256, Pms2 KO n=18).
Fig. S2. GAA repeat transmissions from YG22 transgenic mice. Intergenerational GAA repeat analysis based upon YG22 MMR parental genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=55, Msh2 Het n=142, Msh2 KO n=50, Msh3 WT n=63, Msh3 Het n=104, Msh3 KO n=171, Msh6 WT n=38, Msh6 Het n=196, Msh6 KO n=138, Pms2 WT n=54, Pms2 Het n=201, Pms2 KO n=70).
Fig. S3. Paternal GAA repeat transmissions. Intergenerational GAA repeat analysis based upon paternal MMR genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=32, Msh2 Het n=114, Msh2 KO n=32, Msh3 WT n=23, Msh3 Het n=94, Msh3 KO n=130, Msh6 WT n=44, Msh6 Het n=194, Msh6 KO n=93, Pms2 WT n=102, Pms2 Het n=296, Pms2 KO not determined - males are strelie).
Fig. S4. Maternal GAA repeat transmissions. Intergenerational GAA repeat analysis based upon maternal MMR genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=132, Msh2 Het n=84, Msh2 KO n=66, Msh3 WT n=91, Msh3 Het n=43, Msh3 KO n=56, Msh6 WT n=21, Msh6 Het n=47, Msh6 KO n=88, Pms2 WT n=54, Pms2 Het n=161, Pms2 KO n=88).
Highlights.
> GAA repeat FRDA mice were crossed with Msh2, Msh3, Msh6 and Pms2 knockout mice. > FXN GAA repeat sizes were measured in parents and offspring. > Transmitted changes in GAA repeat sizes were determined. > MMR knockout parents produced increased intergenerational GAA repeat instability. > MMR status affects intergenerational GAA dynamics different to CAG/CTGs.
Acknowledgements
We thank Sanjay Bidichandani (University of Oklahoma Health Sciences Centre) for initiating collaborative studies, Michael Liskay (Oregon Health & Science University) and Darren Monckton (University of Glasgow) for providing the Pms2 knockout mice, and Steve Pash for assistance with animal husbandry. This work was supported by the National Institutes of Health (NIH), USA; Muscular Dystrophy Association (MDA) USA; Ataxia UK; Friedreich’s Ataxia Research Alliance (FARA); GoFAR; and the Wellcome Trust [089757]. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement number 242193/EFACTS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement.
None to declare.
References
- Al-Mahdawi S, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- Al-Mahdawi S, et al. GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics. 2004;84:301–310. doi: 10.1016/j.ygeno.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Baker SM, et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- Campuzano V, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- Campuzano V, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Clark RM, et al. The GAA triplet-repeat is unstable in the context of the human FXN locus and displays age-dependent expansions in cerebellum and DRG in a transgenic mouse model. Hum Genet. 2007;120:633–640. doi: 10.1007/s00439-006-0249-3. [DOI] [PubMed] [Google Scholar]
- De Biase I, et al. Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann Neurol. 2007a;61:55–60. doi: 10.1002/ana.21052. [DOI] [PubMed] [Google Scholar]
- De Biase I, et al. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007b;90:1–5. doi: 10.1016/j.ygeno.2007.04.001. [DOI] [PubMed] [Google Scholar]
- De Michele G, et al. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum Mol Genet. 1998;7:1901–1906. doi: 10.1093/hmg/7.12.1901. [DOI] [PubMed] [Google Scholar]
- de Wind N, et al. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- de Wind N, et al. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- Delatycki MB, et al. Sperm DNA analysis in a Friedreich ataxia premutation carrier suggests both meiotic and mitotic expansion in the FRDA gene. J Med Genet. 1998;35:713–716. doi: 10.1136/jmg.35.9.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditch S, et al. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragileva E, et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiry L, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- Gomes-Pereira M, et al. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum Mol Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- Goula AV, et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, et al. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J Biol Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- Herman D, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- Kim HM, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen AH. Friedreich's ataxia: pathology, pathogenesis, and molecular genetics. J Neurol Sci. 2011;303:1–12. doi: 10.1016/j.jns.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair (Amst) 2007;6:517–529. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Ku S, et al. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, et al. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- Lopez Castel A, et al. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair (Amst) 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monros E, et al. Phenotype correlation and intergenerational dynamics of the Friedreich ataxia GAA trinucleotide repeat. Am J Hum Genet. 1997;61:101–110. doi: 10.1086/513887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montermini L, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- Pandolfo M. Friedreich ataxia: the clinical picture. J Neurol. 2009;256(Suppl 1):3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]
- Pianese L, et al. The effect of parental gender on the GAA dynamic mutation in the FRDA gene. Am J Hum Genet. 1997;60:460–463. [PMC free article] [PubMed] [Google Scholar]
- Pook MA, et al. Rescue of the Friedreich's ataxia knockout mouse by human YAC transgenesis. Neurogenetics. 2001;3:185–193. doi: 10.1007/s100480100118. [DOI] [PubMed] [Google Scholar]
- Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- van den Broek WJ, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, et al. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- Zhao J, et al. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37:4420–4429. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. GAA repeat transmissions from YG8 transgenic mice. Intergenerational GAA repeat analysis based upon YG8 MMR parental genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=109, Msh2 Het n=56, Msh2 KO n=48, Msh3 WT n=51, Msh3 Het n=33, Msh3 KO n=15, Msh6 WT n=27, Msh6 Het n=45, Msh6 KO n=43, Pms2 WT n=102, Pms2 Het n=256, Pms2 KO n=18).
Fig. S2. GAA repeat transmissions from YG22 transgenic mice. Intergenerational GAA repeat analysis based upon YG22 MMR parental genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=55, Msh2 Het n=142, Msh2 KO n=50, Msh3 WT n=63, Msh3 Het n=104, Msh3 KO n=171, Msh6 WT n=38, Msh6 Het n=196, Msh6 KO n=138, Pms2 WT n=54, Pms2 Het n=201, Pms2 KO n=70).
Fig. S3. Paternal GAA repeat transmissions. Intergenerational GAA repeat analysis based upon paternal MMR genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=32, Msh2 Het n=114, Msh2 KO n=32, Msh3 WT n=23, Msh3 Het n=94, Msh3 KO n=130, Msh6 WT n=44, Msh6 Het n=194, Msh6 KO n=93, Pms2 WT n=102, Pms2 Het n=296, Pms2 KO not determined - males are strelie).
Fig. S4. Maternal GAA repeat transmissions. Intergenerational GAA repeat analysis based upon maternal MMR genotypes: (A) Msh2, (B) Msh3, (C) Msh6 and (D) Pms2. Frequencies of GAA expansions, no changes and GAA contractions transmitted to offspring are represented as percentages of total GAA repeat transmissions (Msh2 WT n=132, Msh2 Het n=84, Msh2 KO n=66, Msh3 WT n=91, Msh3 Het n=43, Msh3 KO n=56, Msh6 WT n=21, Msh6 Het n=47, Msh6 KO n=88, Pms2 WT n=54, Pms2 Het n=161, Pms2 KO n=88).