INTRODUCTION
Conventional flow cytometry uses dichroic mirrors and band pass filters to select specific bands of the optical spectrum for detection using point detectors such as photomultiplier tubes (PMTs). This approach has been very effective for detecting the signal from specific fluorophores, but its low resolution sampling of the optical spectrum limits its usefulness for the more sophisticated optical analyses. Over the years there have been many efforts to develop flow cytometers that performed more similarly to a spectrofluorimeter to collect a continuous, high resolution optical spectrum from individual particles. A key challenge for such efforts was the very short signal integration times available for single particle measurements in flow. Recent advances in optics and detectors have made it feasible to make full spectral measurements on the sub-millisecond time scales in which flow cytometry measurements typically occur. In this Unit, we review the general considerations for flow-based spectral measurements, discuss the options for hardware, considerations for software, and current and future applications of spectral flow cytometry.
HISTORY
Interest in measuring the complete fluorescence spectra of cells in flow can be traced (Table 1) to the early days of flow cytometry, with many notable instrument development efforts employing state-or-the-art (for the time) detectors, electronics, and software. In general, these early efforts used dispersive optics such as gratings and prisms to disperse the light over a detector array, which was often the limiting factor in performance. The earliest report used this approach to measure the average spectra of many particles(Wade et al., 1979), while later detectors enabled the measurement of spectra of single particles(Dubelaar et al., 1999; Fuller and Sweedler, 1996; Gauci et al., 1996). Alternative approaches included the use of a scanning monochromometer and a PMT to make successive measurements at different wavelengths, which enabled the measurement of population average spectra with relatively high resolution(Asbury et al., 1996; Steen and Stokke, 1986), and an interferometric approach that enabled single cell measurements but with relatively low spectral resolution(Buican, 1990; Marrone et al., 1991). The trade-offs between speed, sensitivity, and spectral resolution limited the impact of these early systems but in recent years improvements in optics, detectors, and data systems have enabled the development of spectral flow cytometers that can routinely make fast and sensitive high resolution measurements of cell and other particles.
Table 1.
Development of Spectral Flow Cytometry
| Reference | Approach | Comments |
|---|---|---|
| Wade et al, 1979(Wade et al., 1979) | Grating spectrograph and silicon intensified detector | Measured average spectra of many cells |
| Steen and Stokke, 1986(Steen and Stokke, 1986) | Grating monochromometer and PMT | Measured average intensity at 10 nm intervals |
| Buican, 1990(Buican, 1990) | Fourier transform FC | |
| Gauci et al, 1996(Gauci et al., 1996) | Prism and intensified photodiode array | Measured spectra of single cells and microspheres |
| Fuller and Sweedler, 1996(Fuller and Sweedler, 1996) | Grating spectrograph and CCD | Measured spectra of single microspheres an liposomes |
| Asbury et al, 1996(Asbury et al., 1996) | Scanning monochromometer and PMT | Constructed population average spectra from many single cells, single wavelength measurements |
| Dubelaar et al, 1999(Dubelaar et al., 1999) | Grating spectrograph and a 7 pixel hybrid PMT | Used 3 of 7 detector elements to measure light scatter and two colors of fluorescence |
| Isailovic et al, 2005(Isailovic et al., 2005) | Grating and ICCD camera | Measured spectra of bacterial cells in a capillary flow system |
| Robinson et al, 2005(Robinson et al., 2005);Gregori et al 2012(Grégori et al., 2011) | Prism or grating and multianode PMT | Measured spectra of fluorescence-stained cells, PCA analysis |
| Goddard et al, 2006(Goddard et al., 2006) | Grating and CCD | Measured spectra from single beads and cells |
| Watson et al, 2008(Watson et al., 2008); Nolan et al, 2012(Nolan and Sebba, 2011) | Imaging spectrograph and CCD | Measured fluorescence and SERS spectra from individual beads and cells, PCA and linear unmixing analysis |
GENERAL CONSIDERATIONS
Modern spectral flow cytometry presents several options in instrument design and data analysis, the choice of which is ultimately determined by the needs of the biological applications. As discussed above, analysis speed, sensitivity, and spectral resolution are often competing considerations that must be balanced against the needs of the biological application. As for conventional flow cytometry, high speed measurement implies shorter measurement times with less light collected. In spectral flow cytometry, higher resolution results in the same amount of light being distributed over more detector elements, resulting in fewer photons per measurement. The spectral data analysis methods employed will depend on whether the spectra of the components measured are known and constant or if there are unknown and/or changing contributions to the measured spectra, factors that are also defined by the experimental design and aims of the biological application. Finally, the need to physically sort cells based on their spectral features presents additional constraints on both the data acquisition and analysis that will impact instrument design. In the following sections we provide an overview of spectral flow cytometry instrument design and performance, data analysis and software, and survey potential applications.
INSTRUMENT DESIGN
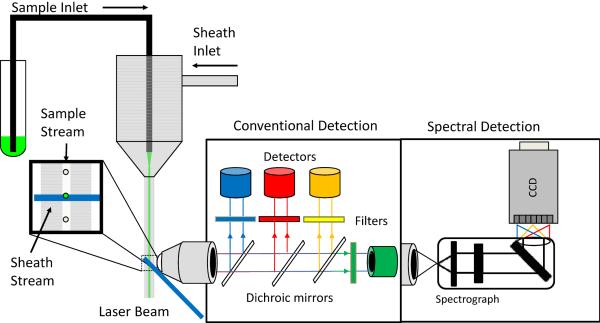
In terms of instrumentation, spectral flow cytometry differs from conventional flow cytometry in the optics and detectors required to obtain high resolution spectra (Figure 1). Currently available commercial flow cytometers use dichroic mirrors and band pass filters to serially isolate specific wavelength ranges for detection. Lerner and colleagues have prepared an excellent reviews of spectral imaging principles and hardware (Lerner, 2006; Lerner et al., 2010), much of which applies directly to spectral flow cytometry, although the short measurement times involved in flow preclude several approaches that can be applied in imaging, where the time constraints are not so stringent.
Figure 1. Schematic comparison of a conventional and spectral detection in flow cytometry.
Conventional detection uses dichroic mirrors and bandpass filters to select colors of light for detection on PMTs. Spectral detection uses gratings or prisms to disperse light across a detector array.
To a large degree, considerations of excitation and illumination in a flow cell and/or sheath stream are the same for conventional and spectral flow cytometry. Tight hydrodynamic focusing of the sample stream within the measurement probe volume allows uniform illumination and a low coefficient of variation (CV). Optimized collection optics that maximize collection efficiency from the probe volume while rejecting out of focus scattering or autofluorescence maximize sensitivity. In most cases, the optical bench of a conventional commercial flow cytometer can be adapted to make spectral flow cytometry measurements.
The first important difference between a conventional flow cytometer and a spectral flow cytometer, from the point of view of a collected photon, is dispersion. While conventional flow cytometry optics block, reflect, or transmit a photon based on its wavelength, spectral flow cytometers disperse photons according to wavelength. The two most common types of dispersive optics are prisms and gratings. Prisms are the classic dispersive optic, refracting the light in a wavelength-dependent manner. The throughput of a prism is typically very high (>90%) and dispersion is efficient over a wide wavelength range, however the dispersion is non-linear and the spectral resolution changes as a function of wavelength, typically by a few nanometers in the visible spectrum. Ruled gratings can offer linear dispersion with higher resolution when used at their intended wavelength, but the presence of higher order diffraction can decrease efficiency and throughput and increase stray light. Volume phase holographic gratings can also offer high efficiency and resolution when used at their optimum wavelength, but with lower levels of stray light. A full discussion of dispersive optics is beyond our scope, but a number of instructive resources are available on line. Beyond issues of dispersion, other optical effects such as spherical aberration, can also affect performance. Most commercial spectrographs are designed to minimize these issues to provide maximum optical performance.
The second major difference between conventional flow cytometry and spectral flow cytometry is the detector. Measurement of the continuous spectrum produced by the prism or grating requires a linear array of detectors. The density and spacing of the detectors in the array, together with the performance of the dispersive optics will determine resolution. Detector technology has been developing rapidly in recent years, and the detectors of choice for spectral flow cytometry at the moment are CCDs and multianode PMTs.
Rectangular format CCDs designed for spectroscopy typically have pixel dimensions on the order of 15–30 um in arrays between 128 × 1024 pixels to 512 × 2048 pixels (height × width) that, when combined with a highly efficiency grating, can provide sub-nanometer spectral resolution. Such CCD chips can have very high quantum efficiencies (>95%) across the visible spectrum in back illuminated mode, very low dark counts with appropriate cooling, and are available in a number of commercial packages with features such as electron multiplication, which provides up to a 1000× gain and on-chip binning. Binning enables spectral resolution to be exchanged for increased signal, and doing this on chip before CCD readout minimizes read noise, and major background source in CCD measurements. In flow cytometry applications, the maximum event rate of the CCD detector is limited by the number of pixels used (after binning) and the readout speed, with mid-range detectors supporting hundreds of events per second and state of the art detectors capable of frame rates of thousands of events per second. CCD-based spectral flow cytometers have been used for visible (Goddard et al., 2006; Nolan et al., 2012a) and NIR fluorescence measurements (Nolan et al., 2012a), as well as for measurements of nanoparticle surface enhanced Raman scattering (SERS) tags (Nolan et al., 2012b; Sebba et al., 2009; Watson et al., 2008; Watson et al., 2009).
Multianode PMTs have smaller arrays of larger detector elements (32 anodes each ~800 um wide in the Hammamatsu H7260), which limits spectral resolution. For example, 300 nm of the optical spectrum dispersed over 32 detector elements would allow a maximum resolution of 10 nm per channel, sufficient for many fluorescence applications where emission spectra tend to be broad. Multianode PMTs also have lower quantum efficiency (< 20%) with a steep drop off in the red characteristic of PMTs. On the other hand, PMTs have much higher gain and faster speed compared to CCDs. At sheath volumetric flow rates that gave 1–2 microsecond transit times through the laser beam, Gregori et al reported measurements between 1000 and 3000 events per second on a 32 channel multianode PMT-based system (Grégori et al., 2011).
EXPERIMENT DESIGN AND DATA ANALYSIS
Beyond the general considerations for the design and execution of conventional flow cytometry experiments, including titrating reagents and including appropriate calibration and control samples, spectral flow cytometry involves several additional considerations including spectral calibration. Similarly, while most of the principles of data analysis for conventional flow cytometry are relevant, spectral data analysis provides additional challenges and opportunities for extracting biological information form samples.
Spectral Calibration
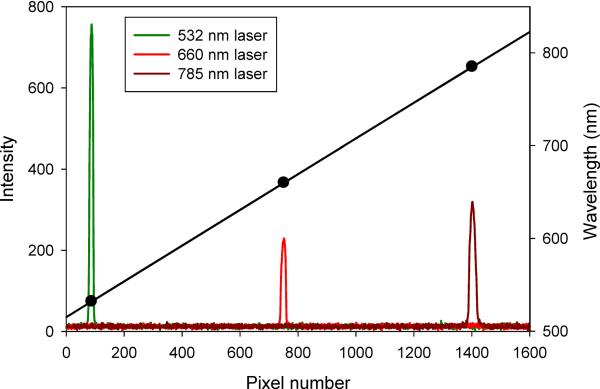
Imaging spectrographs interfaced to array detectors such as CCDs are generally calibrated using low pressure mercury, argon, and/or xenon lamps with distinctive and well characterized spectra. Alternatively, scattered laser light from different sources can be used to define the spectral range of a system. In either case, spectra are acquired, peaks assigned, and a plot of pixel vs wavelength is fitted to interpolate the spectral range of the detector (Figure 1). The general principles of this approach have been discussed in the context of spectral imaging (Lerner, 2006), and these apply to spectral flow cytometry as well.
Spectral data formats and display
A first challenge for spectral flow cytometry is how to store and display the data in a manner that is compatible with conventional flow cytometry measurements. One approach is to store each spectral data point as a flow cytometry parameter, such that a spectra acquired with a 32 channel multianode PMT would be stored at 32 discrete parameters in and FCS format data file. This approach has the advantage of being compatible with the current FCS file format standard, allowing it to be read, in principle, by any third party software. The disadvantage of this approach is that it sacrifices the continuous nature of the optical spectrum, including the ability to display and analyze the data as spectra. Moreover, this is an inefficient way to store the data, resulting in large data files and slow processing, especially in the case of spectra collected with a CCD array producing hundreds or thousands of spectral data points (Watson et al., 2008).
An alternative approach is to store the spectra as spectra, allowing it to be displayed and analyzed as a continuous string of data. This has the advantage of making display and sophisticated spectral analysis fairly straightforward, but has the disadvantage of being incompatible with the existing FCS file format and not supported by the flow cytometry data analysis software designed to read this data format. This has necessitated the development of custom analysis software to support spectral flow cytometry. For example, Robinson's group has developed a software package call Cytospec for the analysis of spectral data from the 32 channel multianode PMT-based spectral flow cytometer (Grégori et al., 2011). This package can display full spectra, as well as a number of different analyses including principle components analysis (PCA). The limitations of the FCS file format are well known, and the ISAC Flow Cytometry Data Standards Task Forces has been developing an alternative format that packages the FCS format data along with other data types into a zip format container file. We have taken a similar approach for spectral flow cytometry data by developing a zip format container that contains the FCS format data, text format spectral data, and text format analysis data resulting from spectral analysis (Nolan et al., 2012b).
A reader plug-in for this file format was developed for a commercial flow cytometry software package (FCS Express 4, de Novo Software) allowing the zip container to be opened, the different data types read, and a customized version of this software was developed to facilitate spectral display and some data analysis. With this approach it is possible to display single cell spectra, or the average spectra of many cells, to display only the spectra of cells with a gate set on a conventional flow cytometry parameter, apply virtual band pass filters to the spectral data to create new parameters, and to display and gate on these parameters.
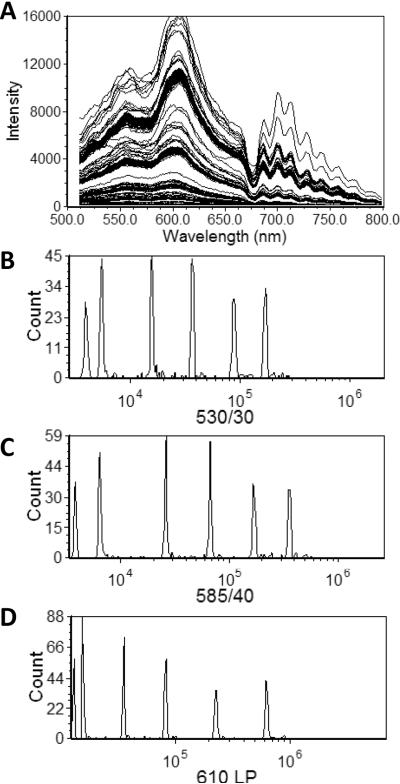
As an example, presented in Figure 3A are spectra from multifluorophore beads (UltraRainbow, Spherotech). We can define a number of virtual band pass filters in which we integrate the signal over a defined wavelength range to generate new parameters for display as histograms (Figure 3B–D). Once the spectral data has been reduced to a new set of parameters, those parameters can be handled as any other conventional data parameter.
Figure 3. Single particle spectra and virtual band pass filters.
A) Spectral flow cytometry data from multifluorphore beads (UltraRainbow, Spherotech). Intensity histograms generated by applying virtual bandpass filters: B) 530/30 C) 585/40, D) 610 LP.
Intensity Calibration
Intensity calibration is conceptually similar to conventional flow cytometry in that multifluorophore reference beads can be used to assess the system stability over time and qualitatively assess the resolution of dimly stained beads. Calibrated single fluorophore particles can be used to quantify detector response in absolute units of mean equivalent soluble fluorophores (MESF). These same principles can be applied to spectral flow cytometry.
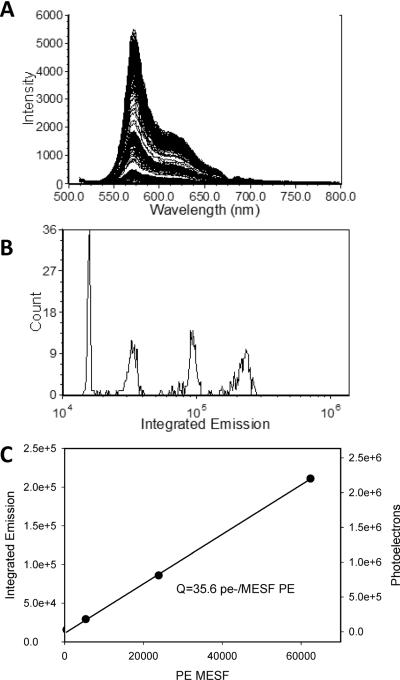
Presented in Figure 4A and B are spectra and intensity histograms from calibrated fluorophore-specific beads measured on a spectral flow cytometer. The fluorophore-specific beads can be used to calibrate the multifluorophore beads for use over a defined wavelength range for a particular instrument (Hoffman et al., 2012) and allow the intensities of unknown samples to be expressed in absolute units of mean equivalent soluble fluorophores. In the case of CCD detectors, often the digital output of the detector has been calibrated by the manufacturer in terms of photoelectrons/ADC unit, allowing the detection efficiency (Q) to be estimated. In principle, multianode PMT-based systems should be able to use the variance in CV as a function of bead intensity to estimate the fundamental measurement performance parameters Q and B (Chase and Hoffman, 1998; Hoffman and Wood, 2007; Wood and Hoffman, 1998), although this has not been reported yet.
Figure 4. Intensity calibration using single fluorophore calibration beads.
A) Spectral flow cytometry data of PE calibration beads (PE Quantibrite, BD Biosciences). B) Histogram of Integrated Emission intensity for the calibration bead set. C) plot of calibration bead MESF value vs Integrated Intensity and the number of photons detected, as calculated from the detector response.
Spectral analysis and spectral unmixing
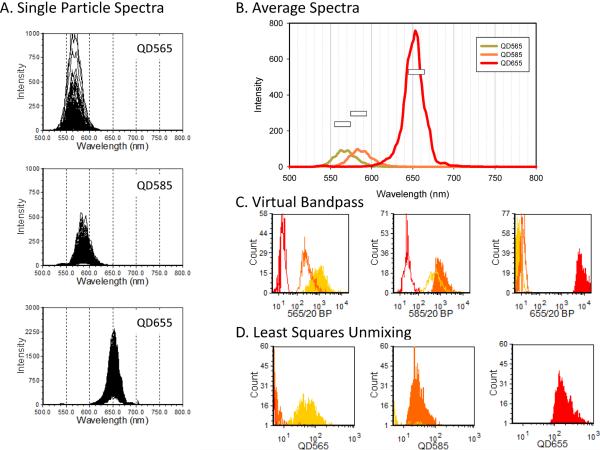
As described above, single cell spectra can be analyzed by applying virtual bandpass filters to measure the signal from specific wavelength bands. As another example of this approach, consider the analysis of quantum dot labeled microspheres measured by spectral flow cytometry using a CCD-based system (Nolan et al., 2012a). Presented in Figure 5A are spectra from individual microspheres labeled with one of three different quantum dots. When we examine the average spectra of each of the three QDots (Figure 5B) we can appreciate the spectral overlap and differences in brightness of the three fluorescent labels. We defined a virtual bandpass for each Qdot centered on the peak emission wavelength, and calculated the integrated emission for each to create a set of three new parameters that can be displayed as histograms (Figure 5C).
Figure 5. QDot-labeled particles: virtual band pass filters and spectral unmixing.
Biotinylated beads labeled with different streptavidin QDots were analyze by spectral flow cytometry. A) Spectra of individual beads B) average particle spectra for each QDot. C) Contribution of each QDot as estimated using band pass filters. D) Contribution of each QDot as estimated using CLS spectral unmixing.
Like experiments using real band pass filters, the application of virtual band pass filters spectral data must confront spillover, or crosstalk, of the probe fluorescence into adjacent spectral bands. For multicolor experiments in conventional flow cytometry using bandpass filters, single stained samples are measured in all channels and the result used to calculate the compensation matrix that allows spillover from one channel to another to be accounted for. Compensation could also be applied to data using virtual bandpass filters such as that presented in Figure 5C, however, in spectral flow cytometry, we can use the full spectral data from singly stained samples to determine the contribution of each tag in a mixture to the total signal using least squares spectral unmixing. This is illustrated in Figure 5D, where classical least squares (CLS) unmixing, an approach often used in spectral imaging (Garini et al., 2006), allows labels with highly overlapping spectra to be resolved.
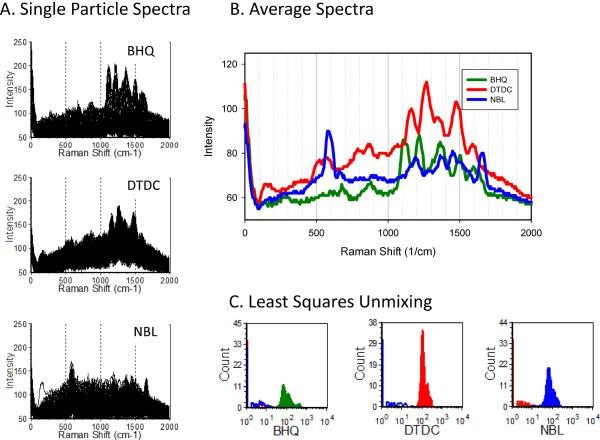
A more striking illustration is given by the ability of spectral unmixing to discriminate SERS tags that have highly complex and overlapping spectra. Presented in Figure 6 are the spectra of particles labeled with one each of three different nanoparticle SERS tags (with the spectral axis expressed as Raman shift, as is the convention for Raman scattering measurements). When the average spectra of particle labeled with each tag are examined, the fine structure and significant overlap of the Raman spectra can be appreciated. Yet, when these spectra are used as reference spectra with CLS-based spectral unmixing, the three tags are readily discriminated, allowing the contribution of each to be clearly resolved when presented in histogram form (Figure 6 C–D).
Figure 6. SERS tag-labeled particles: spectral unmixing.
Avidin-coated beads labeled with different biotinylated SERS tags were analyze by spectral flow cytometry. A) Spectra of individual beads B) average particle spectra for each SERS tag. C) Contribution of each SERS tag as estimated using CLS spectral unmixing.
CLS-based spectral unmixing assumes a linear contribution of unchanging reference spectra to the mixture spectra of an unknown sample, allowing the contribution of each stain to be estimated, in a manner that is mathematically similar to spillover compensation in conventional flow cytometry. Other spectral analysis approaches can be used when the individual spectra contributing to a mixture are changing or even unknown.
These different spectral analysis approaches can be divided in two main categories: supervised and unsupervised methods. The first group assumes that the spectra of the pure components are known a priori. In this case the data analysis translates into the solution of an overdetermined system of equations (as long as the number of channels exceeds the number of fluorophores) usually using a least squares approach. CLS unmixing is a widely used approach that belongs to this group.
In the second group of algorithms nothing is known before to start the data processing. The spectrum of the pure components and their respective concentration profiles are extracted simultaneously from the raw data minimizing an error or cost function chosen depending on the assumptions previously made. An example belonging to this family is Multivariate Curve Resolution. In the following sections, we will highlight the algorithms that have the most direct application to spectral flow cytometry.
Classic Least Squares Unmixing
Classic least squares unmixing uses a linear decomposition algorithm to solve the matrix equation:
where Y is the measured spectrum matrix, A is the pure component spectrum matrix, C is the concentration matrix, and E is the random measurement error (Garini et al., 2006). The result is the linear least squares estimate of the concentration matrix C, which consists of the contribution of each tag for each cell.
The CLS approach is very simple and versatile since it is possible to add almost any kind of constraint to the solution (i.e. non-negativity, equalty, unimodality, etc). As previously stated, the pure components matrix must be known a priori, so a separate measurement of single stained samples with same instrument conditions must be performed. This algorithm makes the assumption that the measured spectrum is a linear combination of the spectra of the pure components. This requires that the spectra of the end members are linearly independent and do not change, but this condition doesn't always apply. For example, if two probes exhibit fluorescence resonance energy transfer (FRET), or changed their spectra in response to the cell environment, this assumption would not be valid. Furthermore, it is important to pay particular attention to how the background is modeled and taken into account. The largest source of background is typically cell autofluorescence and it is often modeled by a polynomial that is included as component in the CLS unmixing. However, autofluorescence can vary between cell types and experimental treatment, and also with the excitation wavelength, so some care and judgment is required in deciding how to model autofluorescence in CLS unmixing.
Principal Component Analysis (PCA)
PCA is an unsupervised unmixing technique that reveals the internal structure of the data by extracting the components with most variance and reducing the dimensions of the data accordingly(Ringner, 2008). In a few words, after performing PCA, it is possible to navigate through the data using a new set of uncorrelated parameters, or principal components, that let the user identify the main contributions to the observed spectra.
In spectral flow cytometry, PCA can be used to find a small set of reference spectra that represents the whole dataset. The first spectrum in the set is a spectrum that contributes the most to the measured spectrum; the second one contributes less, and so on(Garini et al., 2006).
This type of statistical analysis is performed automatically and gives the user the possibility to determine the presence of different fluorophores/tags in every measured spectrum without any information known a priori. Watson et al. used PCA to distinguish particles stained with four different nanoparticle SERS tags measured on a CCD-based spectral flow cytometer (Watson et al., 2008). More recently, Gregori et al. (Grégori et al., 2011) showed how PCA and other data-reduction tools can be used to discriminate four different subsets of lymphocytes using a multianode PMT-based instrument.
PCA is a very powerful method able to distinguish different components within your dataset (noise, cell subsets, etc) but often some sort of statistical preprocessing of the data is required to get better separation between the subsets. These preprocessing steps can include smoothing, normalization, and other data transforms, and PCA results depend critically on preprocessing of the data and on the selection of variables(Ringner, 2008). Furthermore, non-negativity and other typical constraints used in CLS and other unmixing methods cannot be applied to standard linear PCA. Therefore, the results may be optimal in the mathematical sense but lack in physical significance and as consequence it is not possible do many types of quantitative analysis on the differentiated populations. For these reasons, PCA and other unmixing methods are often used together with the goal of exploiting the best characteristics of each (for example PCA for pre-classification and CLS-based approach for quantification).
Multivariate Curve Resolution – Alternating Least Squares ( MCR-ALS)
MCR-ALS is a powerful factor analysis technique that can be used in conjunction with PCA or other unsupervised extraction algorithms to analyze spectral imaging data(Tauler, 1995; Tauler et al., 1995). MCR-ALS has been used extensively in chemometric analysis (Tauler et al., 1993) and more recently in microscopic imaging (Duponchel et al., 2003), hyperspectral microarray scanning (Timlin et al., 2005) and in-vivo fluorescence imaging (Xu and Rice, 2009). In spectral imaging, the unmixing process usually involves a components extraction to estimate pure reference spectra and/or concentration profiles (usually done with PCA or singular value decomposition (SVD), another unsupervised analysis technique. Subsequently, the MCR-ALS algorithm is run on the dataset using this intermediate data and the desired algorithm constraints as input. This step can be thought as an optimization process that alternates the refining of the pure components spectra and the concentration matrices.
The benefits of MCR over other multivariate analysis methods (least squares, linear unmixing) include the ability to model unknown spectral contributions such as interferents, backgrounds, and instrument artifacts and discover the pure component spectra with little or no information a priori (((Timlin et al., 2005)). However, it must be mentioned that the unmixing results deriving from this method strongly depend on the initial estimates of the number of pure components and of their spectra(Duponchel et al., 2003).
EXAMPLES AND BIOLOGICAL APPLICATIONS
The development of biological applications of spectral flow cytometry is still at an early stage, with most published work to date taking the form of demonstration or validation studies. Accordingly, an important first test for an instrument is the analysis of reference or calibration beads. For example, polymer microspheres impregnated with different fluorochromes were used to demonstrate fluorescence detection on an early generation of high resolution spectral flow cytometer (Goddard et al., 2006). A later example used calibrated multi-intensity microspheres for a quantitative analysis of instrument performance and sensitivity, showing it to be comparable to conventional flow cytometry (Nolan et al., 2012a). Antibody capture beads are also widely used in conventional flow cytometry for both calibration (Zenger et al., 1998) and as compensation reference standards (Maecker and Trotter, 2006), and these have been used in a similar manner for spectral flow cytometry (Nolan et al., 2012a). Beads have also been used as calibration and reference particles for measurements using nanoparticle SERS tag-based reagents (Nolan and Sebba, 2011; Watson et al., 2008).
Immunofluorescence detection is probably the single largest class of flow cytometry applications, and the use of spectral flow cytometry for immunofluorescence of peripheral blood mononuclear cells (PBMCs) has been demonstrated using a high speed, low resolution PMT-based spectral flow cytometer (Grégori et al., 2011) as well as a slower speed, high resolution system (Nolan et al., 2012a). In the first case, the authors used principle components analysis to identify lymphocyte subsets, while the latter study used least squares spectral unmixing to determine the amount of each fluorescent antibody bound to each cell, and used conventional parameter-based gating to identify subsets. Immunodetection using SERS-tagged antibodies has also been demonstrated on cultured cells and PMBCs, also using least squares-based linear unmixing to determine the amount of tagged antibody bound to cells (Nolan and Sebba, 2011).
Many other conventional flow cytometry applications can be adapted to spectral flow cytometry. For example, measurement of DNA intercalating dyes in cells has been reported on several different spectral flow cytometers over the years (Asbury et al., 1996; Goddard et al., 2006; Steen and Stokke, 1986). For this and other potential applications, a key question will be does the availability of spectral information provide a significant advantage over the conventional optical filter/PMT approach? For many applications, the practical advantages may be limited. For applications where spectral information is important, for example environmentally sensitive probes and indicators, or systems involving fluorescence resonance energy transfer, the advantages may be compelling.
One potentially useful application may be in the analysis of the intrinsic fluorescence of cells. Autofluorescence is often a major source of background in flow cytometry measurements, and several compensation or subtraction-based approaches have been described to account or correct for its contribution to the total cell signal (Alberti et al., 1987; Roederer and Murphy, 1986; Steinkamp and Stewart, 1986). Autofluorescence intensity can vary between cells, especially in heterogeneous primary cell samples, but its spectra is generally distinct from commonly used fluorochromes. This presents the possibility to use spectral unmixing approaches to separate signal from intrinsic cell autofluorescence from that of extrinsic fluorescence probes. Spectral flow cytometry has been used to characterize the autofluorescence spectra of various cultured cell lines (Goddard et al., 2008), and meeting abstract presentations have described resolving autofluorescence from signal from that of fluorescence labels.
Another potentially very useful application of spectral flow cytometry is in multiplexed cell or bead-based assays. The approach of optically encoding particles (Nolan and Mandy, 2006; Nolan and Sklar, 2002), cells (Krutzik and Nolan, 2006), or ligands (Hadrup et al., 2009; Newell et al., 2009) for multiplexed analyses has been exploited for a range of applications, especially in large scale systems biology (Clutter et al., 2010; Krutzik et al., 2011; Krutzik and Nolan, 2006) or high throughput screening studies (Saunders et al., 2010; Simons et al., 2011; Young et al., 2009). For fluorescence labels such as quantum dots with narrower emission spectra, or Raman scattering labels with very narrow spectral features (Sebba et al., 2009), spectral flow cytometry combined with spectral unmixing or classification approaches has the potential to enable much higher levels of multiplexing using a smaller region of the optical spectrum.
SUMMARY AND PROSPECTS
Full spectral analysis of single cells, using the flow cytometer as a spectrometer, has been a goal of instrument developers since the earliest days of the field. In recent years, advances in optics, detectors, and electronics have made single cell flow spectroscopy a reality. With several research labs making such measurements routinely, capable commercial software, and rumors of impending releases of commercial instruments, it is likely that this capability will lead to the development of new applications and further adoption of a spectral flow cytometry approach.
Figure 2. Spectral calibration.
Spectra of three different laser lines taken on a grating spectrograph coupled to a CCD detector.
Acknowledgments
Supported by NIH EB003824
LITERATURE CITED
- Alberti S, Parks DR, Herzenberg LA. A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry. 1987;8:114–119. doi: 10.1002/cyto.990080203. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Esposito R, Farmer C, van den Engh G. Fluorescence spectra of DNA dyes measured in a flow cytometer. Cytometry. 1996;24:234–242. doi: 10.1002/(SICI)1097-0320(19960701)24:3<234::AID-CYTO6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Buican TN. Real-time Fourier transform spectrometry for fluorescence imaging and flow cytometry. 1990:126. [Google Scholar]
- Chase ES, Hoffman RA. Resolution of dimly fluorescent particles: A practical measure of fluorescence sensitivity. Cytometry. 1998;33:267–279. [PubMed] [Google Scholar]
- Clutter MR, Heffner GC, Krutzik PO, Sachen KL, Nolan GP. Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry. Cytometry Part A. 2010;77:1020–1031. doi: 10.1002/cyto.a.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubelaar GBJ, Gerritzen PL, Beeker AER, Jonker RR, Tangen K. Design and first results of CytoBuoy: A wireless flow cytometer for in situ analysis of marine and fresh waters. Cytometry. 1999;37:247–254. [PubMed] [Google Scholar]
- Duponchel L, Elmi-Rayaleh W, Ruckebusch C, Huvenne JP. Multivariate Curve Resolution Methods in Imaging Spectroscopy:⍰ Influence of Extraction Methods and Instrumental Perturbations. Journal of Chemical Information and Computer Sciences. 2003;43:2057–2067. doi: 10.1021/ci034097v. [DOI] [PubMed] [Google Scholar]
- Fuller RR, Sweedler JV. Characterizing submicron vesicles with wavelength-resolved fluorescence in flow cytometry. Cytometry. 1996;25:144–155. doi: 10.1002/(SICI)1097-0320(19961001)25:2<144::AID-CYTO3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Garini Y, Young IT, McNamara G. Spectral imaging: Principles and applications. Cytometry Part A. 2006;69A:735–747. doi: 10.1002/cyto.a.20311. [DOI] [PubMed] [Google Scholar]
- Gauci M, et al. Observation of single-cell fluorescence spectra in laser flow cytometry. Cytometry. 1996;25:388–393. doi: 10.1002/(SICI)1097-0320(19961201)25:4<388::AID-CYTO11>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Goddard G, et al. Single particle high resolution spectral analysis flow cytometry. Cytometry Part A. 2006;69:842–851. doi: 10.1002/cyto.a.20320. [DOI] [PubMed] [Google Scholar]
- Goddard GR, Houston J, Martin JC, Graves SW, Freyer JP. Cellular discrimination based on spectral analysis of instrinic fluorescence. 2008:685908–1. [Google Scholar]
- Grégori G, et al. Hyperspectral cytometry at the single-cell level using a 32-channel photodetector. Cytometry Part A. 2011;81A:35–44. doi: 10.1002/cyto.a.21120. [DOI] [PubMed] [Google Scholar]
- Hadrup SR, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nature methods. 2009;6:520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- Hoffman RA, Wang L, Bigos M, Nolan JP. NIST/ISAC standardization study: Variability in assignment of intensity values to fluorescence standard beads and in cross calibration of standard beads to hard dyed beads. Cytometry Part A:n/a-n/a. 2012 doi: 10.1002/cyto.a.22086. [DOI] [PubMed] [Google Scholar]
- Hoffman RA, Wood J. Characterization of flow cytometer instrument sensitivity. Current Protocols in Cytometry. 2007 doi: 10.1002/0471142956.cy0120s40. [DOI] [PubMed] [Google Scholar]
- Isailovic D, Li HW, Phillips GJ, Yeung ES. High-throughput single-cell fluorescence spectroscopy. Applied spectroscopy. 2005;59:221–226. doi: 10.1366/0003702053085124. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. Curr. Protocol Cytom. 2011 doi: 10.1002/0471142956.cy0631s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nature Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Lerner JM. Imaging spectrometer fundamentals for researchers in the biosciences—a tutorial. Cytometry Part A. 2006;69:712–734. doi: 10.1002/cyto.a.20242. [DOI] [PubMed] [Google Scholar]
- Lerner JM, Gat N, Wachman E. Approaches to spectral imaging hardware. Curr. Protoc. Cytom. 2010;53:1–12.20. doi: 10.1002/0471142956.cy1220s53. [DOI] [PubMed] [Google Scholar]
- Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry Part A. 2006;69:1037–1042. doi: 10.1002/cyto.a.20333. [DOI] [PubMed] [Google Scholar]
- Marrone BL, et al. Single cell endocrinology: analysis of P-450scc activity by fluorescence detection methods. Endocrinology. 1991;128:2654–2656. doi: 10.1210/endo-128-5-2654. [DOI] [PubMed] [Google Scholar]
- Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nature Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JP, Condello D, Duggen E, Naivar M. Visible and NIR Fluorescence Spectral Flow Cytometry. Cytometry Part A. 2012a doi: 10.1002/cyto.a.22241. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JP, et al. Single cell analysis using surface enhanced Raman scattering (SERS) tags. Methods. 2012b;57:272–9. doi: 10.1016/j.ymeth.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JP, Mandy F. Multiplexed and microparticle based analyses: Quantitative tools for the large scale analysis of biological systems. Cytometry Part A. 2006;69:318–325. doi: 10.1002/cyto.a.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JP, Sebba DS. Surface-Enhanced Raman Scattering (SERS) Cytometry. In: Paul M, editor. Methods in Cell Biology. Academic Press; 2011. pp. 515–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JP, Sklar LA. Suspension array technology: evolution of the flat-array paradigm. TRENDS in Biotechnology. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- Ringner M. What is principal component analysis? Nat Biotech. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- Robinson J, Patsekin V, Gregori G, Rajwa B, Jones J. Multispectral flow cytometry: Next generation tools for automated classification. Microscopy and Microanalysis. 2005;11:2–3. [Google Scholar]
- Roederer M, Murphy RF. Cell-by-cell autofluorescence correction for low signal-to-noise systems: Application to epidermal growth factor endocytosis by 3T3 fibroblasts. Cytometry. 1986;7:558–565. doi: 10.1002/cyto.990070610. [DOI] [PubMed] [Google Scholar]
- Saunders MJ, Graves SW, Sklar LA, Oprea TI, Edwards BS. High-throughput multiplex flow cytometry screening for botulinum neurotoxin type A light chain protease inhibitors. Assay and drug development technologies. 2010;8:37–46. doi: 10.1089/adt.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebba DS, Watson DA, Nolan JP. High throughput single nanoparticle spectroscopy. ACS Nano. 2009;3:1477–1484. doi: 10.1021/nn9003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons PC, et al. Simultaneous in vitro molecular screening of protein-peptide interactions by flow cytometry, using six Bcl-2 family proteins as examples. Nature Protocols. 2011;6:943–952. doi: 10.1038/nprot.2011.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen HB, Stokke T. Fluorescence spectra of cells stained with a DNA-specific dye, measured by flow cytometry. Cytometry. 1986;7:104–106. doi: 10.1002/cyto.990070117. [DOI] [PubMed] [Google Scholar]
- Steinkamp JA, Stewart CC. Dual-laser, differential fluorescence correction method for reducing cellular background autofluorescence. Cytometry. 1986;7:566–574. doi: 10.1002/cyto.990070611. [DOI] [PubMed] [Google Scholar]
- Tauler R. Multivariate curve resolution applied to second order data. Chemometrics and Intelligent Laboratory Systems. 1995;30:133–146. [Google Scholar]
- Tauler R, Kowalski B, Fleming S. Multivariate curve resolution applied to spectral data from multiple runs of an industrial process. Analytical Chemistry. 1993;65:2040–2047. [Google Scholar]
- Tauler R, Smilde A, Kowalski B. Selectivity, local rank, three-way data analysis and ambiguity in multivariate curve resolution. Journal of Chemometrics. 1995;9:31–58. [Google Scholar]
- Timlin J, et al. Hyperspectral microarray scanning: impact on the accuracy and reliability of gene expression data. Bmc Genomics. 2005;6:72. doi: 10.1186/1471-2164-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade C, Rhyne R, Woodruff W, Bloch D, Bartholomew J. Spectra of cells in flow cytometry using a vidicon detector. Journal of Histochemistry & Cytochemistry. 1979;27:1049. doi: 10.1177/27.6.110874. [DOI] [PubMed] [Google Scholar]
- Watson DA, et al. A flow cytometer for the measurement of Raman spectra. Cytometry Part A. 2008;73:119–128. doi: 10.1002/cyto.a.20520. [DOI] [PubMed] [Google Scholar]
- Watson DA, Gaskill DF, Brown LO, Doorn SK, Nolan JP. Spectral measurements of large particles by flow cytometry. Cytometry Part A. 2009;75:460–464. doi: 10.1002/cyto.a.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JCS, Hoffman RA. Evaluating fluorescence sensitivity on flow cytometers: An overview. Cytometry. 1998;33:256–259. [PubMed] [Google Scholar]
- Xu H, Rice BW. In-vivo fluorescence imaging with a multivariate curve resolution spectral unmixing technique. Journal of Biomedical Optics. 2009;14:064011–9. doi: 10.1117/1.3258838. [DOI] [PubMed] [Google Scholar]
- Young SM, et al. Duplex high-throughput flow cytometry screen identifies two novel formylpeptide receptor family probes. Cytometry Part A. 2009;75:253–263. doi: 10.1002/cyto.a.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenger VE, Vogt R, Mandy F, Schwartz A, Marti GE. Quantitative flow cytometry: Inter-laboratory variation. Cytometry. 1998;33:138–145. doi: 10.1002/(sici)1097-0320(19981001)33:2<138::aid-cyto8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]