Abstract
A series of polyaminohydroxamic acids (PAHAs) and polyaminobenzamides (PABAs) were synthesized and evaluated as isoform-selective histone deacetylase (HDAC) inhibitors. These analogues contain a polyamine chain to increase affinity for chromatin and facilitate cellular import. Seven PAHAs inhibited HDAC >50% (1 µM), and two PABAs inhibited HDAC >50% (5 µM). Compound 17 increased acetylated α-tubulin in HCT116 colon tumor cells 253-fold but only modestly increased p21waf1 and acetylated histones 3 and 4, suggesting that 17 selectively inhibits HDAC 6. PABA 22 alone minimally increased p21waf1 and acetylated histones 3 and 4 but caused dose-dependent increases in p21waf1 in combination with 0.1 µM 5-azadeoxycytidine. Finally, 22 appeared to be a substrate for the polyamine transport system. None of these compounds were cytotoxic at 100 µM. PAHAs and PABAs exhibit strikingly different cellular effects from SAHA and have the potential for use in combination antitumor therapies with reduced toxicity.
Introduction
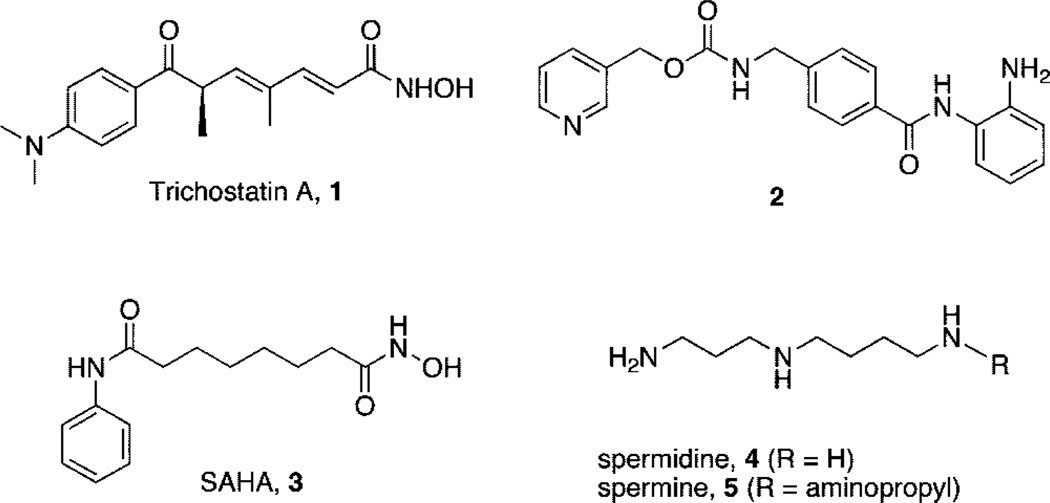
The dynamic status of acetylation on specific histone lysine residues, mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs),a plays a critical role in the regulation of gene expression.1,2 In some tumor cell types, hypoacetylation of histones caused by aberrant HDAC activity results in the underexpression of growth regulatory factors such as the cyclin dependent kinase inhibitor p21Waf1 (also known as CDKN1A and CIP1) and thus contributes to the development of cancer.1,2 Histone hyperacetylation caused by HDAC inhibitors such as trichostatin A (TSA, 1), N-(2-aminophenyl)-4-[N-(pyridin-3-ylmethoxycarbonyl)aminomethyl]benzamide 2 (MS-275),3 and SAHA (3) (Figure 1) can cause growth arrest in a wide range of transformed cells and can inhibit the growth of human tumor xenografts.1,2,4,5 Both 2 and 3 are effective in the clinic, especially when used in combination with DNA methyltransferase inhibitors such as 5-aza-2′-deoxycytidine (5-azadC). 6 Although they are effective both in vitro and in vivo, HDAC inhibitors typified by 1–3 inhibit multiple isoforms of HDAC and produce side effects through activity in noncancerous cells. Thus, it is desirable to identify potent HDAC inhibitors that restore the expression of normal tumor suppressor genes without producing significant dose-limiting toxicity.6 Current structure-activity studies involving analogues of 1–3 have focused largely on modifications to the aromatic ring moiety and the aliphatic linker region present in these molecules.5,7 We recently reported a series of polyaminohydroxamic acid (PAHA) derivatives that incorporate structural features of the polyamines spermidine and spermine (4 and 5, respectively, Figure 1) and the linker and hydroxamic acid moieties found in 1 and 3.8 This strategy is based on the observation that polyamine analogues have high affinities for DNA9–12 and enter cells using the cellular polyamine transport system.9,13 Thus, PAHAs could be selectively directed to DNA and the associated histones. It has been shown that HDAC isoforms have a high degree of sequence homology in the binding pocket but differ in primary sequence at residues in the rim region outside the lysine residue binding site.4 Thus, it may be possible to produce isoform specific inhibitors for individual HDACs by altering the polyamine chain composition and its associated terminal alkyl group, since these portions of the molecule would be expected to interact with the rim region. Using these design criteria, we successfully identified three lead compounds from a library consisting of only 16 analogues.8 We now report 15 additional analogues in the PAHA series (compounds 6–20) and have extended our studies to include polyaminobenzamides (PABAs) 21–23, which incorporate the benzamide moiety of 2 and also possess HDAC inhibitory activity in vitro.
Figure 1.
Structures of 1 (trichostatin A), 2, 3 (SAHA), 4 (spermidine), and 5 (spermine).
Chemistry
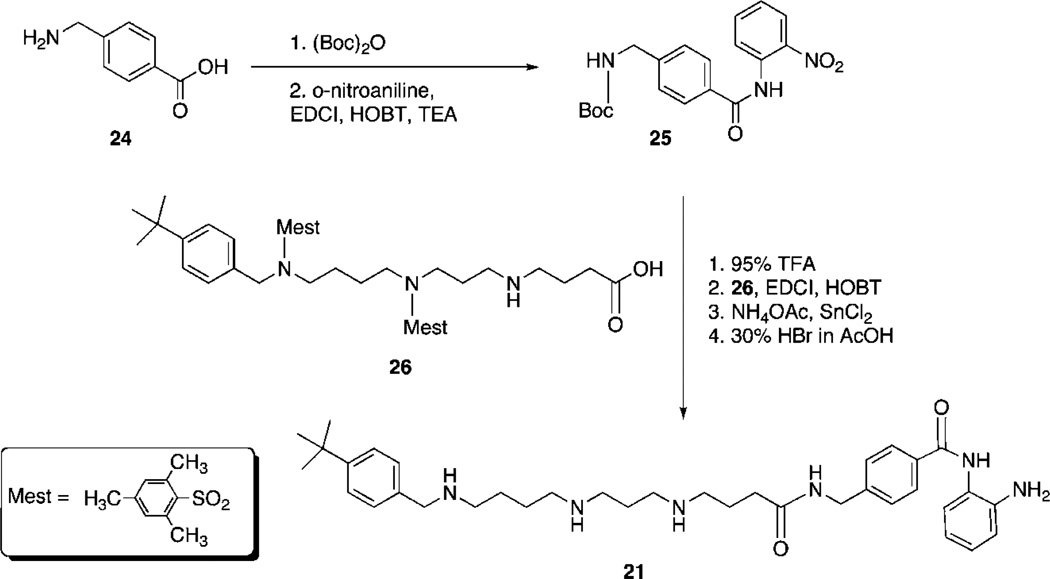
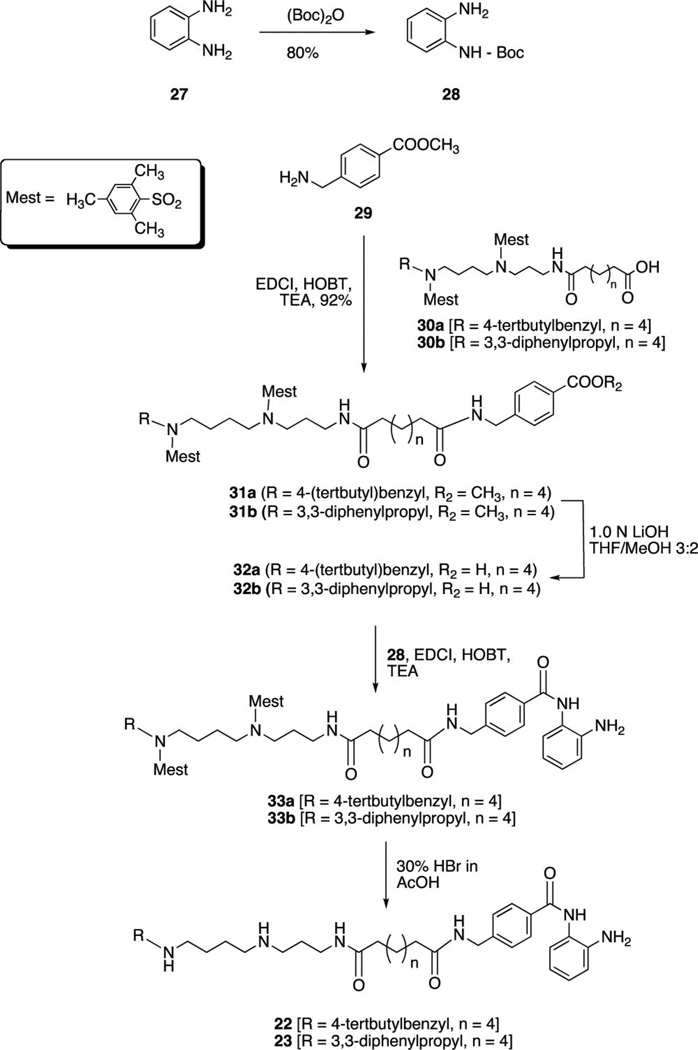
PAHA analogues 6–20 (Table 1) were synthesized using routes that were previously described.8 The synthetic routes to PABA analogues 21–23 are shown in Schemes 1 and 2. Our initial strategy involved N-Boc protection14 of 4-(amino)methylbenzoic acid 24, followed by coupling with o-nitroaniline (EDCI, HOBT, TEA) to form 25.15,16 Compound 25 was then appended to the protected carboxylate 268 as previously described8 followed by reduction of the nitro group (SnCl2)3,16 and deprotection (30% HBr in AcOH)8 to afford target benzamide 21. Although the EDCI coupling to form 25 proceeded in 65% yield, the reaction resulted in multiple side products that complicated purification. In addition, in our hands the reduction of the nitro group produced an unacceptable yield (32%). We thus adopted the synthetic strategy outlined in Scheme 2. o-Aminoaniline 27 was mono-N-Boc protected14,16 to form 28 in 80% yield. Commercial 4-(methoxycarbonyl)benzylamine 29 was then coupled to the previously described intermediates 30a and 30b8,17 to afford compounds 31a and 31b in 95.3% and 92.4% yield, respectively. Hydrolysis of the ester (1.0 N LiOH) yielded the corresponding carboxylates 32a and 32b, which were coupled to 288,17 to form 33a and 33b. Removal of the mesityl protecting groups (30% HBr in AcOH)18,19 then afforded to the desired benzamides 22 and 23 (average of 74% yield from 31).
Table 1.
Structures of PAHA Analogues 6–20 and Their Inhibitory Activity against HeLa Cell Extract Histone Deacetylase at 1.0 µM
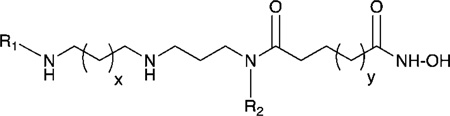 | |||||
|---|---|---|---|---|---|
| Compound | R1 | R2 | x | y | % inhibition (1.0 µM) |
| 6 | 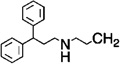 |
H | 1 | 4 | 55.2 |
| 7 | 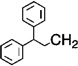 |
H | 2 | 3 | 46.3 |
| 8 | 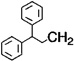 |
H | 2 | 5 | 54.6 |
| 9 | 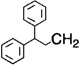 |
H | 2 | 6 | 73.9 |
| 10 | H | 2 | 4 | 46.7 | |
| 11 | H | 2 | 2 | 15.9 | |
| 12 | H | 2 | 4 | 41.4 | |
| 13 | H | 2 | 2 | 71.3 | |
| 14 | H | 2 | 4 | 38.6 | |
| 15 | H | 2 | 2 | 6.7 | |
| 16 | H | 2 | 4 | 16.5 | |
| 17 | H | 2 | 3 | 51.5 | |
| 18 | H | 2 | 5 | 88.7 | |
| 19 | 2 | 2 | 2.0 | ||
| 20 | H | 2 | 4 | 58.6 | |
Scheme 1.
Scheme 2.
Biological Evaluation
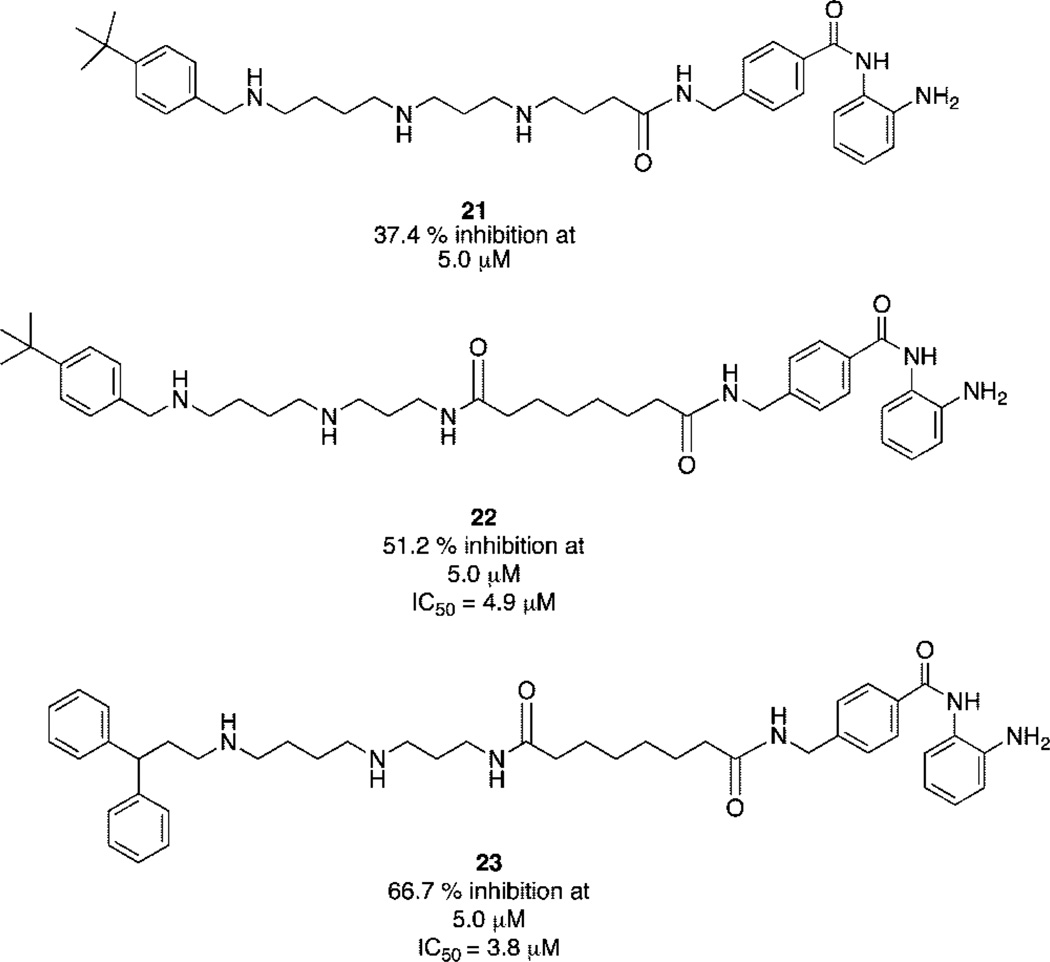
PAHAs 6–20 were evaluated for their ability to inhibit isolated HDAC activity at 1 µM in the Fluor de Lys assay system (Biomol International LP, Plymouth Meeting, PA), employing 1.0 µM TSA (1) as a positive control.8 The results of these studies are summarized in Table 1. PAHAs 6, 8, 9, 13, 17, 18, and 20 all produced greater than 50% inhibition, with compound 18 producing the greatest inhibition (88.7%). Under the same conditions, TSA produced 100% inhibition of the enzyme. PABA analogues 21–23 were similarly evaluated at concentrations of 5.0 µM, using 5.0 µM 2 as a positive control. Compounds 21–23 produced 37.4%, 51.2%, and 66.7% inhibition, respectively (Figure 2), compared to 51.6% inhibition by 2 at the same concentration. Dose–response experiments (not shown) revealed that 22 and 23 possess IC50 values of 4.9 and 3.8 µM, respectively, which compares favorably to the IC50 value of 4.8 µM reported for 2.3
Figure 2.
Structures of PABA analogues 21–23 and their inhibitory activity against histone deacetylase at 5.0 μM. Percent inhibition values reported are the average of three determinations that in all cases differed by 5% or less.
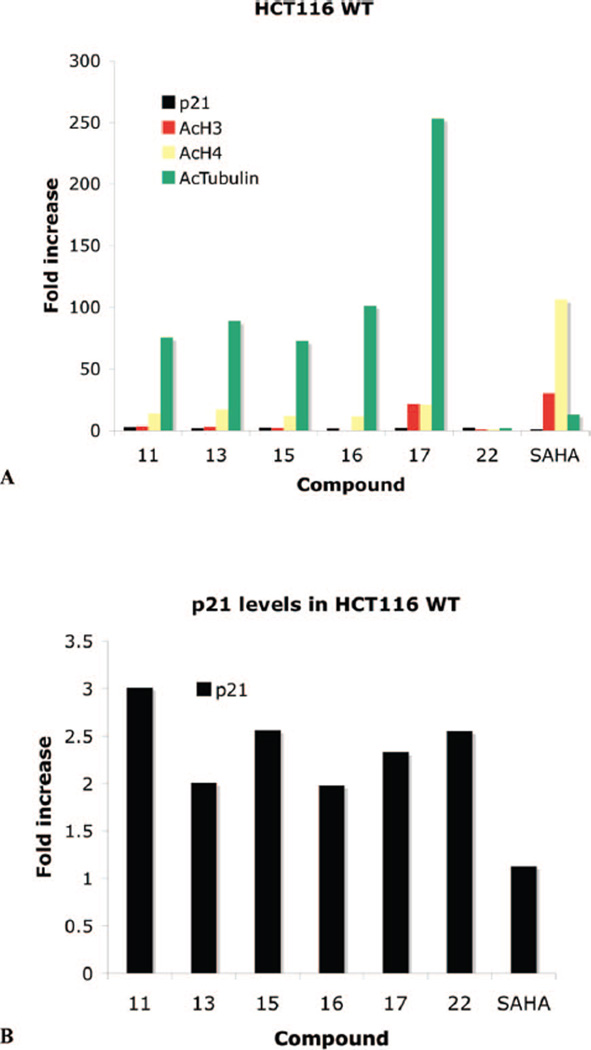
Compounds 9–18, 20, and 22 were next evaluated for their ability to promote in vitro re-expression of the cyclin-dependent kinase inhibitor p21Waf1, in cultured HCT116 human colon carcinoma cells, as shown in Table 2. These analogues were also assayed for their ability to promote increased acetylated histones H3 and H4 (AcH3, AcH4) and acetylated R-tubulin (AcTubulin). Following treatment for 24 h, the effects of 9, 10, 12, 14, 18-20 were unremarkable. However, as summarized in Figure 3, PAHAs 11, 13, 15-17 all promoted a >50-fold increase in acetylated R-tubulin in the HCT116 cell line (Figure 3A). In the case of 17, AcTubulin levels increased more than 253-fold. By contrast, PABA 22 had no significant effect on the acetylation status of this protein. Each compound also produced significant effects on p21waf1 re-expression, and all were more effective than SAHA (3) in this regard (Figure 3B). These analogues also produced slight to moderate increases in the levels of AcH3 and AcH4. Under these conditions, 3 caused no increase in p21waf1, a modest increase (13.2-fold) in AcTubulin, and 30.7- and 106-fold increases in AcH3 and AcH4, respectively.
Table 2.
Fold Increase in the Re-Expression of p21Waf1, Acetylated Histones H3 (AcH3) and H4 (AcH4), and Acetylated α-tubulin (AcTubulin) Proteins in HCT116 Colon Carcinoma Cellsa
| compd | p21waf1 | AcH3 | AcH4 | AcTubulin |
|---|---|---|---|---|
| 9 | 0.41 | 0 | 0.95 | 1.97 |
| 10 | 0.46 | 0 | 1.36 | 6.76 |
| 11 | 3.01 | 3.76 | 14.22 | 75.7 |
| 12 | 0.86 | 0 | 4.74 | 11.98 |
| 13 | 2.01 | 3.34 | 17.4 | 89.01 |
| 14 | 1.64 | 5.19 | 7.82 | 36.3 |
| 15 | 2.56 | 2.38 | 12.06 | 72.9 |
| 16 | 1.98 | 0 | 11.68 | 101.36 |
| 17 | 2.33 | 21.49 | 20.84 | 253.25 |
| 18 | 0.61 | 0 | 3.25 | 11.99 |
| 20 | 0.49 | 0 | 1.95 | 6.07 |
| 22 | 2.55 | 1.19 | 1.12 | 2.27 |
| SAHA | 1.13 | 30.65 | 106.57 | 13.24 |
Cells were treated for 24 h with a 10 µM concentration of compounds 9–18, 20, 22, and SAHA. Data points are single determinations derived from infrared detection and quantification of representative Western blots.
Figure 3.
Fold increase in the re-expression of p21Waf1, acetylated histones H3 (AcH3) and H4 (AcH4), and acetylated R-tubulin (AcTub) proteins in HCT116 colon carcinoma cells. Cells were treated for 24 h with 10 µM compounds 11, 13, 15, 16, 18, 22, and SAHA: (A) p21, AcH3, AcH4, and AcTub values in the HCT166 cell line; (B) amplification of p21 data from part A. Data points are single determinations derived from infrared detection and quantification of representative Western blots.
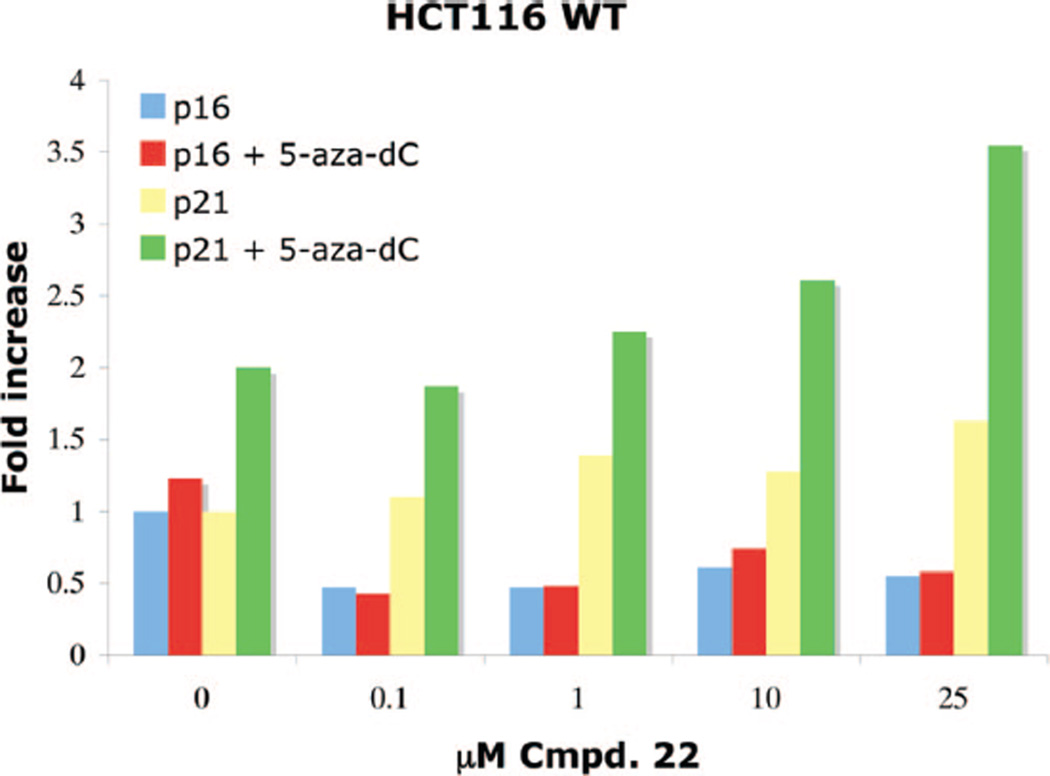
Benzamide 22, which exhibited an IC50 of 4.9 µM against HDAC in the Fluor de Lys assay system, was evaluated in combination with 0.1 µM of the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) for additive or synergistic effects with respect to re-expression of p21waf1, as well as p16, a tumor suppressor gene important for cell cycle regulation (Figure 4). In the HCT116 cell line, no additive effects were observed for the re-expression of p16, whose levels appeared to decrease slightly in the presence of either 22 alone or 22 plus pretreatment with 0.1 µM 5-aza-dC. By contrast, dose-dependent additive effects were observed in the re-expression of p21waf1 when treatment with increasing concentrations of 22 was preceded by treatment with 0.1 µM 5-aza-dC.
Figure 4.
Dose–response data for the re-expression of p16 and p21waf1 proteins in HCT116 human colon carcinoma cells following 24 h of treatment with PABA 22, +/− 5-aza-dC pretreatment. Data points are single determinations derived from infrared detection-based quantification of representative Western blots.
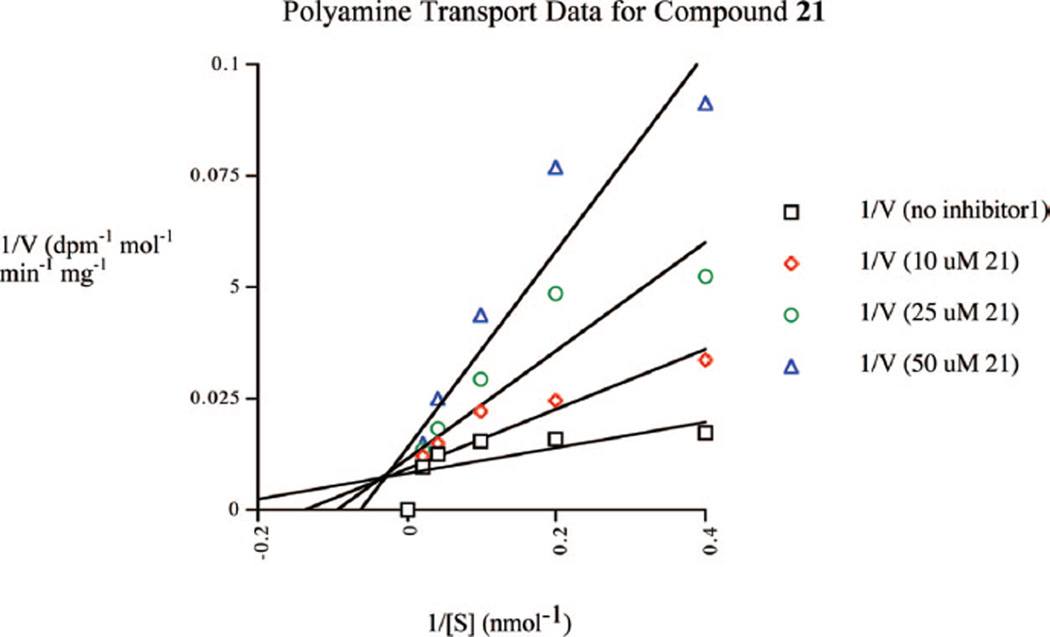
A 14C-spermidine uptake competition assay20,21 was conducted to determine whether PAHAs or PABAs are substrates for the polyamine transport system in the ML-1 acute myelogenous leukemia cell line. In the case of all PAHA analogues tested, no competition for the uptake of 14C-spermidine was observed, suggesting that PAHAs are not taken up into cells by the polyamine transporter. At physiological pH, the hydroxamic acid moiety (pKa ≈ 8.8) is less than 5% ionized. However, in unrelated experiments, we have observed that polyamine analogues with terminal, oxygen-containing substituents that can ionize at physiological pH are poor substrates for the polyamine transport system (unpublished observations). This is consistent with the hypothesis that, rather than being a membrane-bound proteinaceous transporter, the mammalian polyamine transport system may operate by attracting positively charged substrates, followed by endocytosis,20 in which case negatively charged substrates would be less likely to bind to the membrane receptor. However, the lack of transport of these molecules does not formally preclude the two-step pathway proposed by Poulin and colleagues.21 By contrast, benzamide 21 produced a noncompetitive inhibition of 14C-spermidine uptake (Ki = 7.7 µM, Figure 5), suggesting that it is readily accumulated through the polyamine transporter.
Figure 5.
Inhibition of 14C-spermidine uptake by compound 21. The concentration-dependent uptake of 14C-spermidine was measured over a range of concentrations in the presence of 0.0, 10.0, 25.0, and 50.0 µM concentrations of compound 21. Each data point is an average of two determinations that differed by <1% in all cases.
The cytotoxicity of compounds 6– 23 was determined in the HCT116 colon carcinoma cell line using a standard 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reduction assay procedure after 96 h of treatment. The results of these studies are summarized in Table 3. The traditional HDAC inhibitor SAHA (3) was found to have an IC50 value of 1.8 µM in the HCT116 cell line. By contrast, IC50 values for compounds 6–19 and 21–23 were all found to be >100 µM under the same conditions. Compound 20, which inhibited HDAC activity by 58.6% at a concentration of 1.0 µM, was initially found to have IC50 values of 38.6 and 47.2 µM against the HCT116 line. However, in the presence of 1.0 mM aminoguanidine, a diamine oxidase inhibitor, this effect was completely abolished. Compound 20 possesses a terminal primary amine moiety, and thus, the observed cytotoxicity is likely the result of the production of toxic metabolites by serum amine oxidases.
Table 3.
Cytotoxicity Data for PAHA Analogues 6–20 and PABA Analogues 21–23 in HCT116 Human Colon Cancer Cellsa
| compd | % HDAC inhibition at 1.0 µM |
IC50 in wild type HCT 116 cells (µM) |
|---|---|---|
| SAHA | 100 | 1.8 |
| 6 | 55.2 | >100 |
| 7 | 46.3 | >100 |
| 8 | 54.6 | >100 |
| 9 | 73.9 | >100 |
| 10 | 46.7 | >100 |
| 11 | 15.9 | >100 |
| 12 | 41.4 | >100 |
| 13 | 71.3 | >100 |
| 14 | 38.6 | >100 |
| 15 | 6.7 | >100 |
| 16 | 16.5 | >100 |
| 17 | 51.5 | >100 |
| 18 | 88.7 | >100 |
| 19 | 2.0 | ND |
| 20 | 58.6 | 38.6 |
| 20 + 1.0 mM AG | 58.6 | >100 |
| 21 | 28.0 | >100 |
| 22 | 30.8 | >100 |
| 23 | 66.7 (5 µM) | >100 |
Cells were treated for 96 h with a range of concentrations of each test compound, and cell viability was measured using a standard MTS reduction assay. Each data point is the average of two separate experiments that differed by <5% in every case, with each data point within an experiment performed in triplicate. AG = aminoguanidine. ND = not determined.
Discussion
Clinical studies indicate that HDAC inhibitors such as 2 are effective therapies for human cancer,22 but dose-limiting toxicity remains a problem.6,23 HDAC inhibitors partially restore normal cell cycle and apoptotic functions in human tumor cells. In vitro models have demonstrated that synergistic re-expression of genes silenced by promoter hypermethylation can be achieved by the sequential application of deoxynucleotide methyltransferase inhibitors and HDAC inhibitors.24 Phase I trials sequencing 2′-deoxy-5-azacytidine (5-aza-dC) with HDAC inhibitors in patients with acute myelocytic leukemia demonstrate a correlation between chromatin remodeling and clinical efficacy, and robust clinical responses have developed at rates higher than treatment with either demethylating agents or HDAC inhibitors alone.24,25 However, step-down dosing of the HDAC inhibitors in these combinations is often required to minimize toxicity. Consequently, effective HDAC inhibitors having minimal inherent toxicity on their own may provide excellent candidates for combination with agents like 5-aza-dC. Thus, compounds such as those described in this manuscript could be used as part of a combination therapy approach with a lower overall incidence of adverse side effects.
Not surprisingly, in the PAHA series, the length of the linker chain adjacent to the hydroxamic acid metal binding group appears to have a dramatic effect on HDAC inhibitory activity (Table 1). For example, HDAC inhibition increases for compounds 7–9 as the length of the chain increases from five to eight carbons. Compound 18, the most effective inhibitor in this series, has a seven-carbon linker chain. However, the inhibitory activity of 13 cannot be explained on the basis of linker length, suggesting that other factors can contribute to the activity of PAHAs. It is noteworthy that the most active inhibitors, compounds 9, 13, and 18, are significantly more active that compounds 19 and 20, which contain a traditional “cap” group that is thought to bind to a surface region in the HDAC active site.1,2,4 Linker length also appears to play a role in the activity of PABAs 21–23 (Figure 2), although more analogues will need to be examined to make any structure-function correlations.
When the cellular effects of PAHAs 6–20 and PABA 22 are examined, distinct differences from the effects of the traditional HDAC inhibitor SAHA are observed. Since the linker and metal binding moieties of compounds 6–20 are identical to or close homologues of SAHA, these differences in specificity of cellular effects are most likely due to the polyamine portion of the molecules. The compounds shown in Figure 3 produce much greater effects on the levels of AcTubulin than SAHA, 3. These compounds possess either a four-carbon (as in 11, 13, and 15), five-carbon (as in 17), or six-carbon (as in 16) linker chain, which is comparable to the six-carbon linker chain of 3, supporting the contention that the observed effect on AcTubulin is mediated through the polyamine portion of the molecule. Compound 17, with a five-carbon linker, produced the greatest increase in the expression of AcTubulin (>253-fold). It is noteworthy that none of the analogues that increased the levels of AcTubulin have the so-called “cap” group that is present in linker chain of 3 and that is thought to bind to the surface binding area of the HDAC active site. The ability of PAHAs to increase the levels of AcTubulin does not correlate with their ability to inhibit HDAC in the commercial assay containing all of the HDAC isoforms. However, such a correlation may exist with HDAC 6 activity alone, or the increase in AcTubulin may be due to interaction of these PAHAs with another site. Nonetheless, the data suggest that compounds such as 17 selectively inhibit HDAC 6. As was mentioned above, compound 17 produced 51.5% inhibition of HDAC isoforms in a HeLa cell lysate HDAC at 1 µM. In a preliminary study involving recombinant HDAC isoforms 3 and 6, it was determined that 17 produced 99.1% and 99.7% inhibition of HDACs 3 and 6, respectively. This inhibition profile is consistent with the high degree of induction of AcTubulin re-expression by 17. The inhibition of recombinant HDAC 3 and 6 is now being determined for all of the analogues reported in this manuscript, and the results of these experiments are forthcoming.
As shown in Table 2 and Figure 3, 10.0 µM PABA 22 caused a 2.55-fold increase in the expression of p21waf1 in the HCT116 cell line. This compound is structurally similar to 17 except that it has a six-carbon (rather than five-carbon) linker and has a benzamide in place of the hydroxamic acid metal binding moiety. However, unlike 17, PABA 22 did not increase the expression of AcH3 and AcH4 and only increased AcTubulin expression by 2-fold. Compound 22 was also evaluated as an inhibitor of recombinant HDACs 3 and 6, and produced a 1.3% and 11.7% inhibition of these isoforms, respectively. This inhibition profile is consistent with the absence of induction of AcTubulin re-expression by 22. The data shown in Figure 4 demonstrate that 22 reduced the expression of p16 and had only a moderate effect on p21waf1 levels (1.63-fold at 25 µM). When 22 was combined with the known p16/p21waf1 inducer 5-azadC, no re-expression of p16 was observed. However, the combination of 22 and 5-aza-dC produced an additive, dose dependent increase in p21waf1 levels.
The polyamine transport system, which is known to import the natural polyamines and a variety of their analogues, is up-regulated in most tumor cell types.26 As discussed above, none of the PAHA analogues tested were taken up by the polyamine transport system. However, PABA 21 was shown to be an effective noncompetitive inhibitor of the uptake of 14C-spermidine, suggesting that it is a substrate for the polyamine transport system. Because the polyamine transport system is highly active in tumor cells, PABAs related to 21 have the potential to be preferentially imported into tumor cells in high concentrations.
As shown in Table 3, PAHAs 6–20 and PABAs 21–23 exhibited IC50 values greater than 100 µM in the HCT116 colon carcinoma cell line. By contrast, 3 is significantly more cytotoxic, with an IC50 value of 1.8 µM in the HCT116 line. As was pointed out above, there may be advantages to administration of an HDAC inhibitor that effectively promotes the re-expression of aberrantly silenced genes with minimal inherent toxicity. In tumor therapy, such agents could be used in place of HDAC inhibitors with dose limiting side effects, in order to decrease the overall toxicity to normal tissue during combination chemotherapy. In addition, such agents may prove useful in disease states such as diabetes, where HDACs play a role, but cytotoxicity is not an acceptable end point. The synthesis of additional analogues in the PAHA and PABA series is an ongoing concern in our laboratories. In addition, the analogues described herein are now being evaluated as inhibitors of individually expressed HDAC isoforms, which are now available commercially. The results of these studies, as well as in vivo efficacy studies of 17 and 22 in a mouse xenograft model, will be reported in a future manuscript.
Experimental Section
All reagents and dry solvents were purchased from Aldrich Chemical Co. (Milwaukee, WI), Sigma Chemical Co. (St. Louis, MO), or Acros Chemical (Chicago, IL) and were used without further purification except as noted below. Pyridine was dried by passing it through an aluminum oxide column and then stored over KOH. Triethylamine was distilled from potassium hydroxide and stored in a nitrogen atmosphere. Methanol was distilled from magnesium and iodine under a nitrogen atmosphere and stored over molecular sieves. Methylene chloride was distilled from phosphorus pentoxide, and chloroform was distilled from calcium sulfate. Tetrahydrofuran was purified by distillation from sodium and benzophenone. Dimethylformamide was dried by distillation from anhydrous calcium sulfate and was stored under nitrogen. Preparative scale chromatographic procedures were carried out using E. Merck silica gel 60, 230–440 mesh. Thin layer chromatography was conducted on Merck precoated silica gel 60 F-254. Ion exchange chromatography was conducted on Dowex 1X8-200 anion exchange resin. Compounds 26, 30a, and 30b (Schemes 1 and 2) were synthesized as previously described.8
All 1H and 13C NMR spectra were recorded on a Varian Mercury 400 mHz spectrometer, and all chemical shifts are reported as δ values referenced to TMS or DSS. Infrared spectra were recorded on a Nicolet 5DXB FT-IR spectrophotometer and are referenced to polystyrene. In all cases, 1H NMR, 13C NMR, and IR spectra were consistent with assigned structures. Microanalyses were performed by Galbraith Laboratories, Knoxville, TN, and were within 0.4% of calculated values.
Compounds 6–20 were produced using a synthetic scheme that was previously described.7 Spectral data for each of these analogues are given below.
20-{N-[(2,2-Diphenyl)propyl]amino}-8-oxo-9,13,17-triazaeicosanohydroxamic Acid Trihydrobromide (6)
1H NMR (CDCl3) δ 1.37 (m, 4H), 1.40 (m, 4H), 1.71 (m, 2H), 1.88 (m, 4H), 1.97 (t, J = 7.2 Hz, 2H), 2.07 (t, J = 7.2 Hz, 2H), 2.30 (q, J = 7.6 Hz, 2H), 2.85 (m, 12H), 3.10 (t, J = 6.8 Hz, 2H), 3.93 (t, J = 8 Hz, 1H), 7.11 (m, 2H), 7.21 (m, 8H). Anal. (C32H54Br3N5O3) C, H, N.
16-{N-[(2,2-Diphenyl)propyl]amino}-7-oxo-8,12-diazahexade-canohydroxamic Acid Dihydrobromide (7)
1H NMR (CDCl3) δ 1.10 (m, 2H), 1.40 (m, 2H), 1.69 (m, 4H), 1.71 (m, 2H), 1.97 (t, J = 6.8 Hz, 2H), 2.06 (m, 2H), 2.31 (m, 2H), 2.84 (m, 8H), 3.09 (t, J = 6.4 Hz, 2H), 3.94 (t, J = 7.6 Hz, 1H), 7.11 (m, 2H), 7.21 (m, 8H). 13C NMR (400 MHz CDCl3) 13.38, 20.66, 22.79, 22.89, 24.63, 25.02, 25.78, 27.55, 30.88, 35.57, 36.00, 45.19, 46.47, 46.79, 46.96, 48.11, 61.85, 127.15, 127.64, 129.20, 143.68, 174.84, 177.58. IR (cm−1): 3376.6, 2929.7, 2780.5, 1634.5, 1557.5, 1452.4. Anal. (C29H46Br2N4O3) C, H, N.
18-{N-[(2,2-Diphenyl)propyl]amino}-9-oxo-10,14-diazaoctadecanohydroxamic Acid Dihydrobromide (8)
1H NMR (CDCl3) δ 1.11 (m, 6H), 1.38 (m, 4H), 1.54 (m, 4H), 1.70 (m, 2H), 1.96 (t, J = 7.6 Hz, 2H), 2.06 (t, J = 7.2 Hz, 2H), 2.32 (m, 2H), 2.84 (m, 8H), 3.10 (t, J = 6.8 Hz, 2H), 3.98 (t, J = 7.6 Hz, 1H), 7.12 (m, 2H), 7.22 (m, 8H). 13C NMR (CDCl3) δ 13.38, 20.65, 22.78, 22.88, 24.91, 25.33, 25.77, 27.93, 27.99, 30.88, 32.46, 35.77, 35.96, 61.85, 127.15, 127.63, 129.20, 143.68, 174.85, 177.93. IR (cm-1) 3420.8, 2936.6, 2845.4, 2793.5, 1637.5, 1554.5, 1450.8. Anal. (C31H50-Br2N4O3) C, H, N.
19-N-[(2,2-Diphenyl)propyl]amino-10-oxo-11,15-diazanonadecanohydroxamic Acid Dihydrobromide (9)
1H NMR (CDCl3) δ 1.08 (m, 8H), 1.39 (m, 4H), 1.54 (m, 4H), 1.70 (m, 2H), 1.96 (t, J = 7.2 Hz, 2H), 2.06 (t, J = 7.2 Hz, 2H), 2.31 (q, J = 8.0 Hz, 2H), 2.81 (m, 8H), 3.10 (t, J = 6.8 Hz, 1H), 7.11 (m, 2H), 7.14–7.24 (m, 8H). 13C NMR (CDCl3) δ 13.38, 20.65, 22.79, 22.88, 24.96, 25.39, 25.77, 27.99, 28.19, 30.87, 32.42, 35.80, 35.96, 38.75, 45.14, 61.85, 71.23, 109.99, 127.15, 127.63, 129.20, 130.12, 143.67, 166.48, 173.72, 174.86, 178.00. Anal. (C32H52Br2N4O3) C, H, N.
17-N-{4-[(N,N-Dimethylamino)benzyl]amino}-8-oxo-9,13-diazaheptadecanohydroxamic Acid Dihydrobromide (10)
1H NMR (CDCl3) δ 1.12 (m, 4H), 1.40 (m, 4H), 1.60 (m, 4H), 1.72 (m, 2H), 1.98 (t, J = 7.2 Hz, 2H), 2.08 (t, J = 6.8 Hz, 2H), 2.89 (m, 8H), 3.11 (s, 6H), 3.18 (m, 2H), 7.36 (d, J = 8.4 Hz, 2H), 7.43 (d, J = 8.4 Hz, 2H). 13C NMR (CDCl3) δ 13.38, 20.67, 22.83, 22.95, 24.05, 24.88, 25.23, 25.52, 25.77, 27.76, 27.91, 28.09, 31.32, 35.72, 36.01, 61.85, 115.48, 117.49, 120.96, 121.74, 130.02, 131.00, 132.62, 138.98, 141.18, 141.88, 168.77, 177.83. Anal. (C25H47-Br2N5O3) C, H, N.
15-N-[4-(Isopropyl)benzyl]amino-6-oxo-7,11-diazapentadecanohydroxamic Acid Dihydrobromide (11)
1H NMR (CDCl3) δ 1.08 (d, J = 7.2 Hz, 6H), 1.43 (m, 4H), 1.60 (m, 4H), 1.72 (m, 2H), 2.02 (m, 2H), 2.11 (m,2H), 2.89 (m, 6H), 3.12 (m, 2H), 4.05 (s, 2H), 7.26 (s, 4H). IR (cm−1): 3397.7, 2936.2, 2787.0, 1637.0, 1612.9, 1548.1, 1424.8. Anal. (C24H46Br2N4O3) C, H, N.
17-N-[4-(Isopropyl)benzyl]amino-8-oxo-9,13-diazaheptadecanohydroxamic Acid Dihydrobromide (12)
1H NMR (D2O) δ 1.08 (d, J = 7.2 Hz, 6H), 1.14 (m, 4H), 1.24 (m, 4H), 1.61 (m, 4H), 1.75 (m, 2H), 2.01 (t, 2H), 2.12 (t, 2H), 2.89 (m, 6H), 3.12 (t, 2H), 4.12 (s, 2H), 7.27 (s, 4H). IR (cm−1): 3539.4, 3468.1, 3402.4, 2941.3, 2787.0, 1634.5, 1612.9, 1483.2, 1431.3. Anal. (C25H46-Br2N4O3) C, H, N.
15-N-[2-(Phenyl)benzyl]amino-6-oxo-7,11-diazapentadecanohydroxamic Acid Dihydrobromide (13)
1H NMR (CDCl3) δ 1.14 (m, 4H), 1.33 (m, 2H), 1.41 (m, 6H), 1.71 (m, 2H), 1.99 (t, J = 7.2 Hz, 2H), 2.09 (t, J = 7.6 Hz, 2H), 2.65 (t, J = 7.6 Hz, 2H), 2.76 (t, J = 8 Hz, 2H), 3.12 (t, J = 6.8 Hz, 2H), 4.17 (s, 2H), 7.28 (m, 3H), 7.36–7.45 (m, 6H). IR (cm−1): 3539.4, 3461.6, 3414.9, 2953.9, 2851.8, 1636.3, 1612.9, 1561.0, 1457.3, 1431.3. Anal. (C26H40Br2N4O3) C, H, N.
17-N-[2-(Phenyl)benzyl]amino-8-oxo-9,13-diazaheptadecanohydroxamic Acid Dihydrobromide (14)
1H NMR (CDCl3) δ 1.31 (m, 2H), 1.41 (m, 6H), 1.70 (m, 2H), 2.01 (m, 2H), 2.11 (m, 2H), 2.64 (m, 2H), 2.75 (m, 2H), 2.83 (m, 2H), 3.12 (m, 2H), 4.16 (s, 2H), 7.21(m, 1H), 7.42(m, 9H). IR (cm−1): 3552.4, 3474.5, 3414.0, 3228.1, 2955.6, 2806.4, 1637.9, 1612.9, 1554.5, 1450.8, 1431.3. Anal. (C28H44Br2N4O3) C, H, N.
15-N-{2-[(Phenyl)thio]ethyl]amino-6-oxo-7,11-diazapentadecanohydroxamic Acid Dihydrobromide (15)
1H NMR (CDCl3) δ 1.42 (m, 4H), 1.58 (m, 4H), 1.73 (m, 2H), 2.03 (m, 2H), 2.11 (m, 2H), 2.91 (m, 6H), 3.12 (m, 6H), 7.2–7.3 (m, 4H). 13C NMR (CDCl3) δ 22.73, 22.90, 24.52, 24.78, 25.78, 29.39, 35.44, 36.06, 45.22, 47.01, 127.79, 129.73, 130.13, 130.63, 132.78, 177.18. Anal. (C21H38Br2N4O3S) C, H, N.
17-N-{2-[(Phenyl)thio]ethyl]amino-8-oxo-9,13-diazaheptadecanohydroxamic Acid Dihydrobromide (16)
1H NMR (CDCl3) δ 1.15 (m, 4H), 1.41 (m, 4H), 1.57 (m, 4H), 1.72 (m, 2H), 1.99 (t, J = 7.6 Hz, 2H), 2.08 (t, J = 7.2 Hz, 2H), 2.87 (m, 6H), 3.11 (m, 6H), 7.13–7.36 (m, 4H). 13C NMR (CDCl3) δ 24.88, 25.24, 25.78, 27.77, 27.93, 29.39, 35.72, 36.01, 45.20, 46.17, 46.99, 127.80, 129.71, 129.74, 130.60, 130.63. Anal. (C23H42Br2N4O3S) C, H, N.
16-N-[4-(tert-Butyl)benzyl]amino-7-oxo-8,12-diazahexadecanohydroxamic Acid Dihydrobromide (17)
1H NMR (CDCl3) δ 1.14 (s, 9H), 1.42 (m, 4H), 1.59 (m, 4H), 1.70 (m, 2H), 1.99 (m, 2H), 2.08 (t, J = 7.2 Hz, 2H), 2.87 (m, 8H), 3.10 (t, J = 7.2 Hz, 2H), 4.04 (s, 2H), 7.26 (d, J = 8.4 Hz, 2H), 7.42 (d, J = 8.4 Hz, 2H). 13C NMR (CDCl3) δ 30.48, 30.52, 35.51, 126.41, 126.43, 129.88, 129.90, 168.31, and 175.60. Anal. (C25H46Br2N4O3) C, H, N.
16-N-[4-(tert-Butyl)benzyl]amino-9-oxo-10,14-diazaoctadecanohydroxamic Acid Dihydrobromide (18)
1H NMR (CDCl3) δ 1.14 (m, 13H), 1.58 (m, 5H), 1.59 (m, 5H), 1.70 (m, 2H), 1.99 (m, 2H), 2.08 (m, 2H), 2.89 (m, 8H), 3.11 (m, 2H), 4.05 (s, 2H), 7.27 (d, J = 8.4 Hz, 2H), 7.42 (d, J = 8.4 Hz, 2H). 13C NMR (CDCl3) δ 22.80, 24.88, 28.02, 30.50, 35.96, 45.17, 47.05, 126.42, 129.89. Anal. (C27H50Br2N4O3) C, H, N.
15-N-[4-(tert-Butyl)benzyl]amino-7-N-{[4-N,N-(dimethyl)aminobenzyl] amino}-6-oxo-7,11-diazapentadecanohydroxamic Acid Dihydrobromide (19)
1H NMR (CDCl3) δ 1.27 (s, 9H), 1.33 (m, 8H), 1.62 (m, 6H), 2.22 (m, 4H), 2.30 (m, 6H), 2.57 (m, 12H), 3.01 (m, 4H), 3.19 (m, 4H), 4.07 (s, 2H), 4.68 (d, J = 5.6 Hz, 2H), 6.87 (m, 2H), 6.95 (d, J = 5.6 Hz, 4H), 7.25 (m, 4H), 7.40 (d, J = 8.0 Hz, 2H), 7.79 (d, J = 8.0 Hz, 2H), 7.99 (d, J = 7.6 Hz, 4H). Anal. (C33H55Br2N5O3) C, H, N.
17-Amino-9-N-{[4-N,N-(dimethyl)aminobenzyl]amino}-8-oxo-9,13-diazaheptadecanohydroxamic Acid Dihydrobromide (20)
1H NMR (CDCl3) δ 1.11 (m, 6H), 1.26 (m, 4H), 1.52 (m, 4H), 1.73 (m, 1H), 1.82 (m, 1H), 2.19 (m, 2H), 2.81 m, 6H), 3.01 (s, 6H), 3.22 (m, 2H), 7.22 (m, 2H), 7.39 (m, 2H). 13C NMR (CDCl3) δ 22.91, 24.02, 25.12, 27.22, 28.29, 32.4, 32.55, 38.89, 46.51, 115.11, 120.99, 121.00, 128.51, 128.67, 130.12, 139.25, 142.11, 179.23. IR (cm−1): 3407.6, 2940.8, 2860.3, 1712.1, 1615.3, 1514.0. Anal. (C24H45Br2N5O3) C, H, N.
4-{[N-(tert-Butyloxycarbonyl)amino]methyl}benzoic Acid (25)
A mixture of 2.0 g (0.013 mol) of 24 in 100 mL of dioxane/1.0 N sodium hydroxide (2:1) was cooled in an ice bath with stirring. To this mixture a 3.16 g (0.0015 mol) portion of di-tert-butyl dicarbonate was added, and the mixture was allowed to stir for 12 h at room temperature. The dioxane was removed, and the pH of the resulting aqueous solution was adjusted to 3.0 using 1.0 N HCl. The aqueous layer was extracted with three 50 mL portions of ethyl acetate, and the organic layers were combined, washed with brine, and dried over anhydrous magnesium sulfate. Filtration and removal of the solvent under reduced pressure afforded the crude N-Boc protected intermediate, which was purified on a silica gel column using ethyl acetate/hexane (3:4) followed by ethyl acetate/hexane (3:1). The pure compound was obtained as fine white powder (2.12 g) in 63.8% yield. 1H NMR (CDCl3) δ 1.44 (s, 9H), 4.40 (s, 2H) 7.39 (d, J = 8.4 Hz, 2H), 8.08 (d, J = 8.0 Hz, 2H). 13C NMR (CDCl3) 28.59, 44.59, 127.45, 128.56, 130.76, 135.37, 171.41.
A 0.60 g (0.0024 mol) portion of the N-Boc protected intermediate above was dissolved in 10.0 mL of dichloromethane and cooled to 0 °C, and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDCI, 0.503 g, 0.0026 mol) and N-hydroxybenzotriazole (HOBt, 0.35 g, 0.0026 mol) were added with stirring. The mixture was allowed to stir for 15 min at 0 °C, after which time 0.26 mL (0.19 mL, 0.0026 mol) of triethylamine was added. The reaction mixture was stirred for an additional 15 min at 0 °C, and then a 0.360 g portion (0.0026 mol) of o-nitroaniline dissolved in 2 mL of dichloromethane was added, followed by stirring for 8 h at room temperature. The reaction mixture was then concentrated in vacuo to yield a dark-yellow semisolid. The semisolid was dissolved in 50 mL of water and extracted with three 50 mL portions of chloroform, and the combined organic layers were dried over anhydrous magnesium sulfate. Filtration and removal of the solvent then afforded crude 25, which was purified by column chromatography (hexane/ethyl acetate 3:1) to yield pure 25 as yellow crystals (0.68 g, 76.7%). 1H NMR (CDCl3) δ 1.47(s, 9H) 4.43 (d, d, J = 6.0 Hz, 2H), 5.56 (s, 1H), 7.48 (m, 5H), 8.06 (d, J = 8.4 Hz, 1H), 8.19 (d, J = 8.0 Hz, 2H).
17-N-[4-(tert-Butyl)benzyl]amino-1-N-[4-(N-(2-aminophenyl) benzamido)methyl]amino-1,5-dioxo-6,10-diazatetradecanamide Dihydrobromide (21)
Compound 21 was synthesized from 25 and 26 using methodology described by our laboratory7 and a previously published benzamide synthesis.14,15 In our hands, these syntheses provided 21 but in poor overall yield. Thus, the complete experimental details are not described in this manuscript. An improved synthesis used to produce 22 and 23 is described below. 1H NMR (D2O) δ (ppm) 1.14 (s, 9H), 1.59–1.90 (comlex m, 10H), 2.99 (m, 10H), 3.53 (s, 1H), 3.61 (s, 1H), 4.04 (s, 2H), 6.81 (m, 2H), 7.23 (m, 4H), 7.41 (m, 2H), 7.66 (m, 4H). Anal. (C37H54Br2N6O3) C, H, N.
2-[N-(tert-Butyloxycarbonyl)amino]aniline (28)
A 1.0 g portion of benzene-1,2-diamine 27 (0.009 mol) was dissolved in 30 mL of chloroform, and the reaction mixture was cooled to 0 °C. To this mixture was added an aqueous solution of sodium bicarbonate (0.78 g, 0.0054 mol) and sodium chloride (0.54 g, 0.000 92mol), and the mixture was allowed to stir at 0 °C for 30 min. Di-tert-butyl dicarbonate (2.018, 0.0092 mol) was dissolved in 20 mL of chloroform, and the solution was slowly added to the mixture. The mixture was allowed to stir at 0 °C for an additional 10 min, warmed to room temperature, and then refluxed for 12 h. The mixture was then cooled to room temperature and extracted with three 30 mL portions of chloroform. The combined organic layers were washed with 50 mL of saturated sodium bicarbonate and 50 mL of saturated sodium chloride solution and then dried over anhydrous magnesium sulfate. The combined organic layers were then filtered, and the solvent was removed in vacuo to yield crude 28. The crude compound was purified on a silica gel column eluted with hexane/ethyl acetate (3:1, then 2:1) to yield compound 28 as an off-white solid (1.55 g, 80.1%). 1H NMR (CDCl3) δ 1.51 (s, 9H), 3.73 (br, 2H), 6.21 (br, 1H), 6.78 (m, 2H), 7.01 (m, 1H), 7.28 (d, J = 9.2 Hz, 1H). 13C NMR (CDCl3) δ 28.0, 29.9 116, 124, 127, 129.4, 129.9, 158. IR (cm−1) 3435, 1735.
17-{N-[4-(tert-Butyl)benzyl]-N-[2-(mesitylene)sulfonyl]}amino-13-N-[2-(mesitylene)sulfonyl]-1-N-[4-(carboxy)benzyl]amino-1,8-dioxo-9,13-diazaheptadecanamide Methyl Ester (31a)
A 0.341 g (0.000 42 mol) of 30a was dissolved in 3 mL of dichloromethane, and to this mixture was added 2 mL of dichloromethane containing 29 (0.08 g, 0.0005 mol), HOBT (0.067 g, 0.0005 mol), and triethylamine (0.050 g, 0.0007 mol). The reaction mixture was cooled to 0 °C, and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) (0.163 g, 0.000 85 mol) was added in 5 mL of dichloromethane. Following the addition of EDCI, the resulting mixture was stirred at 0 °C for 10 additional min, and the mixture was allowed to warm to room temperature and stirred for 12 h. The resulting dichloromethane solution was washed with 20 mL of 0.5 M HCl, 20 mL of water, and 20 mL of saturated NaHCO3. The organic layer was dried over anhydrous magnesium sulfate and filtered, and the dichloromethane was removed under reduced pressure. The crude product was purified by column chromatography (hexane/ethyl acetate 2:1, then 1:2) to yield 31a as a white solid (0.383 g, 95.3%) 1H NMR (CDCl3) δ 1.27 (m, 4H), 1.32 (s, 9H), 1.37 (m, 4H), 1.53 (m, 4H), 1.70 (m, 2H), 2.19–2.25 (m, 4H), 2.37 (s, 3H), 2.61 (s, 6H), 3.13–3.24 (m, 6H), 3.44 (t, 2H), 3.76 (s, 3H), 4.29 (s, 2H), 4.83 (s, 2H), 7.14–7.37 (m, 8H), 7.74–8.0 (m, 3H), 8.39 (m, 1H). 13C NMR (CDCl3) δ 21.9, 22.8, 25.2, 28.2, 31.9, 37.1, 43.9, 47.7, 48.2, 48.7, 52.5, 53.7, 128.8, 129.7, 133.4, 137.6, 142.9, 144.6, 172.0, 172.7.
17-{N-[3,3-(Diphenyl)propyl]-N-[2-(mesitylene)sulfonyl]}amino-13-N-[2-(mesitylene)sulfonyl]-1-N-[4-(carboxy)benzyl]amino-1,8-dioxo-9,13-diazaheptadecanamide Methyl Ester (31b)
Compound 31b was synthesized from 30b and 29 exactly as described for the synthesis of 31a in 92.4% yield. 1H NMR (CDCl3) δ 1.25–1.37 (m, 10H), 1.55–1.67 (m, 6H), 1.98–2.08 (m, 4H), 2.20 (s, 3H), 2.31 (s, 3H), 2.43 (s, 6H), 2.56 (s, 6H), 2.90 (q, J = 6 Hz, 2H), 3.05, (m, 2H), 3.15–3.22 (m, 6H), 3.66 (m, 1H), 3.906 (s, 3H), 4.14–4.46 (m, 2H), 5.96 (t, 1H), 6.32 (t, 1H), 6.90 (m, 4H), 7.00 (m, 4H), 7.12–7.21 (m, 6H), 7.29–7.36 (m, 2H), 7.96–8.00 (m, 2H). 13C NMR (CDCl3) δ 21.16, 22.91, 23.08, 24.46, 24.72, 25.67, 28.82, 28.91, 29.91, 32.74, 36.42, 36.59, 36.71, 43.26, 44.09, 45.29, 45.79, 48.86, 52.32, 126.65, 127.63, 127.70, 128.79, 130.10, 130.16, 132.25, 132.31, 133.35, 140.22, 142.64, 142.83, 143.83, 144.13, 167.08, 173.46. IR (cm−1) 3386, 3295, 2969, 2843, 1721, 1652, 1543, 1313, 1147, 913, 730.
17-{N-[4-(tert-Butyl)benzyl]-N-[2-(mesitylene)sulfonyl]}amino-13-N-[2-(mesitylene)sulfonyl]-1-N-[4-(carboxy)benzyl]amino-1,8-dioxo-9,13-diazaheptadecanamide (32a)
A 0.383 g (0.0004 mol) portion of 31a was dissolved in 12 mL of tetrahydrofuran:water (4:2) and cooled to 0 °C, and 6 mL of 1.0 N LiOH was added to the mixture dropwise. The solution was warmed to room temperature and allowed to stir for 16 h, during which time the reaction was monitored by TLC. The mixture was again cooled to 0 °C, neutralized by the dropwise addition of 2.0 N HCl, and extracted with three 20 mL portions of ethyl acetate. The ethyl acetate layers were combined, washed with brine, and dried over anhydrous magnesium sulfate. Removal of the solvent in vacuo yielded compound 32a as a cloudy yellow oil (0.372 g, 98.4%) that was of sufficient purity to be used in the next reaction without further purification. 1H NMR (CDCl3) δ 1.22 (m, 4H), 1.33 (s, 9H), 1.41 (m, 4 h), 1.49 (m, 4H), 1.72 (q, 2H), 2.20 (m, 4H), 2.34 (s, 6H), 2.66 (s, 12H), 3.08 (m, 6H), 3.39 (t, 2H), 4.29 (s, 2H), 4.37 (s, 2H), 7.09 (m, 2H), 7.19 (m, 4H), 7.37 (m, 2H), 7.54 (m, 2H), 7.68 (m, 1H), 7.99 (m, 2H), 8.27 (m, 1H).
17-{N-[3,3-(Diphenyl)propyl]-N-[2-(mesitylene)sulfonyl]}amino-13-N-[2-(mesitylene)sulfonyl]-1-N-[4-(carboxy)benzyl]amino-1,8-dioxo-9,13-diazaheptadecanamide (32b)
Compound 32b was made from 31b exactly as described for the synthesis of 32a in 98.1% yield. 1H NMR (CDCl3) δ 1.28–1.33 (m, 10H), 1.56–1.64 (m, 6H), 2.01 (q, J = 7.6 Hz, 2H), 2.10 (t, J = 7.6 Hz, 2H), 2.25 (s, 3H), 2.32 (s, 3H), 2.43 (s, 6H), 2.53 (s, 6H), 2.91 (t, J = 8 Hz, 2H), 3.08 (m, 4H), 3.06–3.16 (m, 4H), 3.72 (t, J = 7.6 Hz,1H), 4.40 (s, 2H), 6.0 (br, 1H), 6.97 (d, J = 8.0 Hz, 4H), 7.04 (d, J = 8.0 Hz, 4H), 7.14 (m, 2H), 7.18 (m, 4H), 7.33 (m, 2H), 7.68 (t, 1H), 7.94 (m, 2H). 13C NMR (CDCl3) δ 21.00, 21.16, 22.83, 23.05, 25.43, 25.58, 28.59, 29.91, 32.80, 36.47, 43.37, 43.60, 48.68, 102.29, 126.65, 127.60, 127.76, 128.79, 130.61, 132.29, 132.36, 133.07, 140.25, 142.94, 143.73, 174.07, 174.29, 176.96.
17-{N-[4-(tert-Butyl)benzyl]-N-[2-(mesitylene)sulfonyl]}amino-13-N-[2-(mesitylene)sulfonyl]-1-N-[4-(N-(2-aminophenyl)benzamido)methyl]amino-1,8-dioxo-9,13-diazaheptadecanamide (33a)
Compound 33a was synthesized from 32a and 28 using the procedure described for the synthesis of 31a in 80.7% yield. 1H NMR (CDCl3) δ 1.27 (s, 9H), 1.33 (m, 8H), 1.62 (m, 6H), 2.22 (m, 4H), 2.30 (m, 6H) 2.57 (m, 12H), 3.01 (m, 4H), 3.19 (m, 4H), 4.07 (s, 2H), 4.18 (d, J = 5.6 Hz, 2H), 6.87 (m, 2H), 6.95 (d, J ) 5.6 Hz, 4H), 7.25 (m, 4H), 7.40 (J = 7.8 Hz, 2H), 7.99 (d, J = 7.6 Hz, 4H).
17-{N-[3,3-(Diphenyl)propyl]-N-[2-(mesitylene)sulfonyl]}amino-13-N-[2-(mesitylene)sulfonyl]-1-N-[4-(N-(2-aminophenyl)benzamido)methyl]amino-1,8-dioxo-9,13-diazaheptadecanamide (33b)
Compound 33b was synthesized from 32b and 28 using the procedure described for the synthesis of 31a in 84.2% yield. 1H NMR (CDCl3) δ 1.37 (m, 8H), 1.49–1.53 (m, 11H), 1.64 (m, 4H), 2.04 (m, 6H), 2.21 (s, 3H), 2.31 (s, 3H), 2.43 (s, 6H), 2.54 (s, 6H), 2.90 (t, J = 8 Hz, 2H), 2.92 (t, J = 8 Hz, 2H), 3.13–3.20 (m, 6H), 3.64 (t, J = 7.6 Hz,1H), 4.45 (d, J = 6 Hz, 2H), 6.05 (broad s, 1H), 2.65 (broad s, 1H), 6.90 (d, J = 5.6 Hz,2H), 6.98 (d, J = 5.6 Hz, 4H), 7.14–7.31 (m, 12H), 7.31 (m1H), 7.65 (m, 1H), 7.89 (d, J = 8 Hz, 2H), 9.4 (br, 1H). 13C NMR (400 MHz CDCl3) δ 14.14, 21.19, 22.91, 23.08, 24.42, 25.72, 28.52, 28.86, 36.54, 43.19, 44.07, 45.17, 48.84, 125.70, 126.65, 127.63, 127.89, 128.03, 128.79, 132.27, 132.32, 140.21, 140.19, 142.66, 143.12, 143.78, 173.69, 183.70. IR (cm−1) 3439, 3021, 2921, 2843, 1652, 1526, 1313, 1213, 1095, 850.
17-N-[4-(tert-Butyl)benzyl]amino-1-N-[4-(N-(2-aminophenyl) benzamido)methyl]amino-1,8-dioxo-9,13-diazaheptadecanamide Dihydrobromide (22)
A 4.2 g portion of phenol (0.44 mol) was dissolved in 20 mL of 30% HBr in acetic acid in a stoppered flask. Compound 33a (0.200 g, 0.0002 mol) in 15 mL of ethyl acetate was then added in three portions over a period of 3 h. After the addition was complete, the reaction mixture was stirred for an additional 15 h at room temperature, then cooled to 0 °C and diluted with 30 mL of water. The aqueous phase was washed with two 30 mL portions of ethyl acetate before being lyophylized to give the crude product as yellow solid. This crude product was washed with methanol and filtered to yield the tetrahydrobromide salt of 22 (0.099 g, 74.2%) as an off-white solid. An analytical sample of 22 was prepared by recrystallization from aqueous ethanol. 1H NMR (D2O) δ 1.15 (s, 9H), 1.21 (m, 4H), 1.34–1.71 (m, 10H), 1.97 (t, 1H), 2.11 (m, 3H), 2.85 (m, 6H), 3.07 (m, 2H), 3.97 (s, 1H), 4.05 (s, 1H), 4.23 (m, 2H), 7.19 (d, J = 8 Hz, 2H), 7.24–7.34 (m, 4H), 7.44 (m, 2H), 7.61 (d, J = 8 Hz, 2H), 7.84 (d, J = 7.6 Hz, 2H). Anal. (C40H60Br2N6O3) C, H, N.
17-N-[3,3-(Diphenyl)propyl]amino-1-N-[4-(N-(2-aminophenyl)benzamido)methyl]amino-1,8-dioxo-9,13-diazaheptadecanamide Dihydrobromide (23)
Compound 23 was prepared from 33b exactly as described for the synthesis of 22 in 76.7% yield. An analytical sample of 23 was prepared by recrystallization from aqueous ethanol. 1H NMR (D2O) δ 1.12 (m, 4H), 1.20 (m, 4H), 1.53 (m, 4H), 1.69 (m, 2H), 2.01 (t, J = 7.2 Hz, 2H), 2.14 (m, 2H), 2.32 (m, 2H), 2.70–2.84 (m, 8H), 3.09 (t, J = 7.2 Hz, 2H), 3.95 (t, J = 7.2 Hz, 1H), 4.29 (s, 2H), 6.64 (d, J = 8.8 Hz,1H), 6.83 (d, J = 8.8 Hz, 1H), 6.80 (m, 1H), 6.98 (m, 1H), 7.13 (m, 2H), 7.26 (m, 6H), 7.34 (m, 4H), 7.77 (d, J = 8 Hz, 2H). 13C NMR (D2O) δ 16.92, 20.52, 22.73, 22.83, 25.35, 25.75, 27.95, 28.07, 30.86, 35.71, 35.79, 35.89, 42.79, 45.06, 46.37, 46.72, 46.90, 48.06, 57.58, 113.83, 115.49, 117.48, 121.42, 126.64, 127.12, 127.58, 128.19, 128.65, 129.16, 129.22, 130.13, 130.31, 132.61, 143.58, 144.54, 176.85, 177.44, 177.82. IR (cm−1) 3408, 2934, 2847, 1469, 1378. Anal. (C44H60Br2N6O3) C, H, N.
Histone Deacetylase Activity Assay
Compounds 6–23 were evaluated for their ability to inhibit isolated HDAC in a commercially available assay (Fluor de Lys assay system, Biomol International LP, Plymouth Meeting, PA), employing 1 and 2 as positive controls.8 The reaction mixture contains a HeLa cell nuclear extract and a commercial substrate containing acetylated lysine side chains. The substrate and extract are incubated in the presence of the appropriate concentration of the inhibitor. Deacetylation of the substrate followed by mixing with the provided developer generates a fluorophore, and comparison of inhibited vs control relative fluorescence using a standard plate reader was employed to determine percent HDAC activity remaining. For studies involving individually expressed HDACs 3 and 6, the appropriate HDAC protein was purchased from BPS Bioscience (San Diego, CA), and a comparable solution of the pure isoform was substituted for the HeLa cell assay in the Fluor de Lys kit. All determinations were carried out in triplicate, and reported values are the average of these determinations, which in no case varied by more than 3%.
Cell Lines and Drug Treatment
HCT116 colon carcinoma cells were provided by Dr. Bert Vogelstein.27 Cells were maintained in McCoy’s 5A medium supplemented with 10% fetal calf serum, at 37 °C, 5% CO2. Seeded cells were allowed to adhere overnight, then treated with 1 (Wako Pure Chemicals, Richmond, VA), 2 (Mitsui Pharmaceuticals, Chiba, Japan), SAHA (BioVision, Mountain View, CA), or the desired test compound for the concentration and time indicated in the figure captions. For combination studies, attached cells were incubated with 0.1 µM 5-aza-dC (Sigma) for 24 h, after which medium was removed and replaced with fresh medium containing the appropriate concentration of PABA 22 for an additional 24 h.
Histone Preparation
Histones were prepared by a modification of a previously described method.8,28 Cells were washed in 2 mL of HBSS and disrupted by 1 mL of ice-cold lysis buffer A (10 mM Tris, pH 7.6, 5 mM butyric acid, 1% Triton X-100, 1 mM MgCl2, and 1 mM PMSF). Nuclei were collected by centrifugation at 14 000 rpm for 15 min. The pellet was resuspended once with 250 µL of ice-cold lysis buffer B (10 mM Tris, pH 7.6, 0.25 M sucrose, 3 mM CaCl2, and 5 mM butyric acid). Sulfuric acid was added to a concentration of 0.4 N, and the tubes were incubated at 4 °C for overnight. Debris was pelleted by centrifugation, and the supernatant was collected. Histones were precipitated by addition of 10 volumes of acetone and incubated at −20 °C overnight. Pellets were collected by centrifugation, briefly dried under vacuum, and resuspended in doubly distilled H2O.
Protein Expression Analysis
Following treatment, HCT116 cells were lysed in buffer M (25 mM HEPES, pH 7.9, 150 mM NaCl, 0.5 mM EDTA, 0.1% Triton-X, 10% glycerol, 0.1 mg/ml BSA, 1 mM DTT) containing an EDTA-free protease inhibitor cocktail, at 4 °C for 20 min. Lysate was clarified by centrifugation at 14000g for 15 min at 4 °C, and the resulting supernatant was used for analysis. Total protein concentration was determined using the BioRad DC assay (Hercules, CA), with absorbance measured using a spectrophotometer at a wavelength of 750 nm. Absorbance was converted to protein content using a bovine serum albumin standard curve. Total cellular proteins (30 µg per lane) were separated on 10% Bis-Tris NOVEX gels (Invitrogen, Carlsbad, CA), transferred to Immunoblot PVDF membrane (Bio-Rad), and proteins of interest were visualized by Western blot analysis using the following primary antibodies: acetylhistone H3 (06-599) (diluted 1:1000) and acetylhistone H4 (06-866) (diluted 1:1000), from Upstate Biotechnologies (Billerica, MA); p21Waf1 (556431) (diluted 1:500) from BD Pharmingen (San Jose, CA); p16 (36-5600) (1:1000) from Zymed (San Francisco, CA); acetylated R-tubulin (Sigma No. T-6793) (1:2000); and _-actin (sc-1615) (diluted 1:1500) from Santa Cruz (Santa Cruz, CA). Following washes, blots were incubated with species-specific, fluorophore-conjugated secondary antibodies to allow visualization and quantification of immunoreactive proteins using the Odyssey infrared detection system and software (LI-COR, Lincoln, NE).
PAHA and PABA uptake by the polyamine transporter was determined by measuring the ability of each compound to compete with the uptake of 14C-spermidine, as previously described.29
Cytoproliferative Responses to PAHA and PABA Exposure
Cell proliferation was quantified using the CellTiter 96 One Solution MTS assay (Promega, Madison, WI). Cells were seeded in triplicate in 96-well plates at a density of 3000 cells per well and allowed to attach overnight. Medium was aspirated and replaced with 100 µL of fresh medium containing the appropriate concentration of HDAC inhibitor, PAHA, or PABA. Following incubation at 37 °C, 5% CO2, 20 µL of CellTiter 96 One Solution was added per well and incubated 1.5 h at 37 °C and absorbance measured at 490 nm.
Footnotes
This manuscript is dedicated to Professor James K. Coward on the occasion of his retirement.
Abbreviations: HAT, histone acetyltransferase; HDAC, histone deacetylase, SAHA, suberoylanilide hydroxamic acid; PAHA, polyaminohydroxamic acid; PABA, polyaminobenzamide; 5-aza-dC, 5-aza-2′-deoxycytidine; AcTubulin, acetylated R-tubulin; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt.
References
- 1.Johnstone RW. Histone Deacetylase Inhibitors: Novel Drugs for the Treatment of Cancer. Nat. Rev. Drug. Discovery. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 2.Marks PA, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone Deacetylases and Cancer: Causes and Therapies. Nat. Rev. Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T, Ando T, Tsuchiya K, Fukazawa N. Synthesis and Histone Deacetylase Inhibitory Activity of New Benzamide Derivatives. J. Med. Chem. 1999;42:3001–3003. doi: 10.1021/jm980565u. [DOI] [PubMed] [Google Scholar]
- 4.Grozinger CM, Schrieber SL. Deacetylase Enzymes: Biological Functions and the Use of Small Molecule Inhibitors. Chem. Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 5.Weinmann H, Ottow E. Recent Advances in Medicinal Chemistry of Histone Deacetylase Inhibitors. Annu. Rep. Med. Chem. 2004;39:185–196. [Google Scholar]
- 6.(a) For a toxicological profile of 1 and recent clinical studies involving 2 and 3, see the following: Vanhaecke T, Papeleu P, Elaut G, Rogiers V. Trichostatin A-like Hydroxamate Histone Deacetylase Inhibitors as Therapeutic Agents: Toxicological Point of View. Curr. Med. Chem. 2004;11:1629–1643. doi: 10.2174/0929867043365099. Fouladi M. Histone Deacetylase Inhibitors in Cancer Therapy. Cancer Invest. 2006;24:521–527. doi: 10.1080/07357900600814979.
- 7.Bieliauskas AV, Weerasinghe SVW, Pflum MKH. Structural Requirements of HDAC Inhibitors: SAHA Analogues Functionalized Adjacent to the Hydroxamic Acid. Bioorg. Med. Chem. Lett. 2007;17(8):2216–2219. doi: 10.1016/j.bmcl.2007.01.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varghese S, Gupta D, Baran T, Jiemjit A, Gore SD, Casero RA, Jr, Woster PM. Alkyl Substituted Polyaminohydroxamic Acids: A Novel Class of Targeted Histone Deacetylase Inhibitors. J. Med. Chem. 2005;48:6350–6365. doi: 10.1021/jm0505009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casero RA, Jr, Woster PM. Terminally Alkylated Polyamine Analogues as Chemotherapeutic Agents. J. Med. Chem. 2001;44:1–29. doi: 10.1021/jm000084m. [DOI] [PubMed] [Google Scholar]
- 10.Valasinas A, Reddy VK, Blohkin AV, Basu HS, Bhattacharya S, Sarkar A, Marton LJ, Frydman B. Long-Chain Polyamines (Oligoamines) Exhibit Strong Cytotoxicities against Human Prostate Cancer Cells. Bioorg. Med. Chem. 2003;11:4121–4131. doi: 10.1016/s0968-0896(03)00453-x. [DOI] [PubMed] [Google Scholar]
- 11.Frydman B, Porter CW, Maxuitenko Y, Sarkar A, Bhattacharya S, Valasinas A, Reddy VK, Kisiel N, Marton LJ, Basu HS. A Novel Polyamine Analog (SL-11093) Inhibits Growth of Human Prostate Tumor Xenografts in Nude Mice. Cancer Chemother. Pharmacol. 2003;51:488–492. doi: 10.1007/s00280-003-0598-8. [DOI] [PubMed] [Google Scholar]
- 12.Ha HC, Yager JD, Woster PM, Casero RA. Structural Specificity of Polyamines and Polyamine Analogues in the Protection of DNA from Strand Breaks Induced by Reactive Oxygen Species. Biochem. Biophys. Res. Commun. 1998;244:298–303. doi: 10.1006/bbrc.1998.8258. [DOI] [PubMed] [Google Scholar]
- 13.Cullis PM, Green RE, Merson-Davies L, Travis NG. Chemical Highlights of Polyamine Transport. Biochem. Soc. Trans. 1998;26:595–601. doi: 10.1042/bst0260595. [DOI] [PubMed] [Google Scholar]
- 14.Wunsch E, Graf W, Keller O, Keller W, Wersin G. On the Synthesis of Benzyloxycarbonyl Amino Acids. Synthesis. 1986:958–960. [Google Scholar]
- 15.Nagaoka Y, Maeda a T., Kawai Y, Nakashima D, Oikawa T, Shimoke T, Ikeuchi I, Kuwajima H, Uesato S. Synthesis and Cancer Antiproliferative Activity of New Histone Deacetylase Inhibitors: Hydrophilic Hydroxamates and 2-Aminobenzamide-Containing Derivatives. Eur. J. Med. Chem. 2006;41:697–708. doi: 10.1016/j.ejmech.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Bamford MJ, Alberti MJ, Bailey N, Davies S, Dean DK, Gaiba A, Garland S, Harling JD, Jung DK, Panchal TA, Parr CA, Steadman JG, Takle AK, Townsend JT, Wilson DM, Witherington J. (1H-Imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol- 3-ylamine Derivatives: A Novel Class of Potent MSK-1 Inhibitors. Bioorg. Med. Chem. Lett. 2005;15:3402–3406. doi: 10.1016/j.bmcl.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q, Yang YT, Chen CS, Davis M, Byrd JC, Etherton MR, Umar A, Chen CS. Zn2+-Chelating Motif-Tethered Short- Chain Fatty Acids as a Novel Class of Histone Deacetylase Inhibitors. J. Med. Chem. 2004;47:467–474. doi: 10.1021/jm0303655. [DOI] [PubMed] [Google Scholar]
- 18.Yajima H, Takeyama M, Kanaki J, Nishimura O, Fujino M. Studies on Peptides. LXXX. NG-Mesitylene-2-sulfonylarginine. Chem. Pharm. Bull. 1978;26:3752–3757. [Google Scholar]
- 19.Roemmele RC, Rapoport H. Removal of N-Arylsulfonyl Groups from Hydroxy R-Amino Acids. J. Org. Chem. 1988;53:2367–2371. [Google Scholar]
- 20.Belting M, Mani K, Jonsson M, Cheng F, Sandgren S, Jonsson S, Ding K, Delcros JG, Fransson LA. Glypican-1 Is a Vehicle for Polyamine Uptake in Mammalian Cells: A Pivotal Role for Nitrosothiol-Derived Nitric Oxide. J. Biol. Chem. 2003;278:47181–47189. doi: 10.1074/jbc.M308325200. [DOI] [PubMed] [Google Scholar]
- 21.Soulet D, Gagnon B, Rivest S, Audette M, Poulin R. A Fluorescent Probe of Polyamine Transport Accumulates into Intracellular Acidic Vesicles via a Two-Step Mechanism. J. Biol. Chem. 2004;279:49355–49366. doi: 10.1074/jbc.M401287200. [DOI] [PubMed] [Google Scholar]
- 22.Kouraklis G, Theocharis S. Histone Deacetylase Inhibitors: A Novel Target of Anticancer Therapy. Oncol. Rep. 2006;15:489–494. [PubMed] [Google Scholar]
- 23.Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Joseph Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, Melillo G, Elsayed Y, Monga M, Kalnitskiy M, Zwiebel J, Sausville EA. Phase I and Pharmacokinetic Study of MS-275, a Histone Deacetylase Inhibitor, in Patients with Advanced and Refractory Solid Tumors or Lymphoma. J. Clin. Oncol. 2005;23:3912–3922. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 24.Cameron EE, Baylin SB, Herman JG. p15(INK4B) CpG Island Methylation in Primary Acute Leukemia Is Heterogeneous and Suggests Density as a Critical Factor for Transcriptional Silencing. Blood. 1999;94:2445–2451. [PubMed] [Google Scholar]
- 25.Corn PG, Smith BD, Ruckdeschel ES, Douglas D, Baylin SB, Herman JG. E-Cadherin Expression Is Silenced by 5′ CpG Island Methylation in Acute Leukemia. Clin. Cancer Res. 2000;6:4243–4248. [PubMed] [Google Scholar]
- 26.Seiler N, Delcros JG, Moulinoux JP. Polyamine Transport in Mammalian Cells. An Update. Int. J. Biochem. Cell Biol. 1996;28:843–861. doi: 10.1016/1357-2725(96)00021-0. [DOI] [PubMed] [Google Scholar]
- 27.Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, Kinzler KW, Vogelstein B, Lengauer C. Targeted Inactivation of p53 in Human Cells Does Not Result in Aneuploidy. Cancer Res. 2002;62:1129–1133. [PubMed] [Google Scholar]
- 28.Cousens LS, Gallwitz D, Alberts BM. Different Accessibilities in Chromatin to Histone Acetylase. J. Biol. Chem. 1979;254:1716. [PubMed] [Google Scholar]
- 29.Aziz SW, Yatin M, Worthen DR, Lipke DW, Crooks PA. A Novel Technique for Visualizing the Intracellular Localization and Distribution of Transported Polyamines in Cultured Pulmonary Artery Smooth Muscle Cells. J. Pharm. Biol. Anal. 1998;17:307–320. doi: 10.1016/s0731-7085(98)00016-8. [DOI] [PubMed] [Google Scholar]