Abstract
Background: Clarithromycin resistance in Helicbacter pylori has been found to be associated with point mutations in 23s rRNA gene leads to reduced affinity of the antibiotic to its ribosomal target or changing the site of methylation. The aim of this study was to determine the most important point mutations in 23s rRNA gene in H. pylori that are closely related to clarithromycin resistance among such isolates.
Methods: Sixty three H. pylori isolates, obtained from gastric biopsy speciemens in Kerman, Iran, were used to evaluate their susceptibility to clarithromycin by disk diffusion test, and to detect the most common point mutations in 23s rRNA gene associated with clarithromycin resistance by Polymerase chain reaction-amplification and restriction fragment length polymorphism (PCR-RFLP) and 3'-mismatch PCR.
Results: 31.7% of the H. pylori isolates were resistant to clarithromycin, and each of the resistant isolate had at least one of the most common point mutations in 23s rRNA gene associated with calrithromycin resistance.
Conclusion: According to our results three common point mutation in 23s rRNA gene in H. pylori are closely related to clarithromycin resistance. There was an absolute relation between 23s rRNA gene point mutations and clarithromycin resistance in this study. Helicbacter pylori resistance to clarithromycin can cause failure in the eradications of the bacteria. The resistance of the bacteria is expanding in most parts of the world including Iran.
Key Words: Clarithromycin, point mutations, Helicobacter pylori
Introduction
Helicobacter pylori is a microaerophilic gram-negative organism involved in many digestive system diseases, such as peptic ulcer, gastritis, or mucosa-associated lymphoid tissue (MALT) lymphoma, or acting as a risk factor in the development of gastric cancer.1 The prevalence of H. pylori infection varies greatly among different countries, as in many developing countries it is over 70%, while in most industrialized nations it is 20% to 50%.2
Eradication of H. pylori is an important component of treatments for peptic ulcer disease and other gastrointestinal disorders.3Triple or quadruple therapy regimen containing a proton-pump inhibitor (PPI) and antibiotics, mainly clarithromycin and metronidazole, are currently in use.4 The inhibition of protein synthesis is the functional mechanisms of the macrolides, causing the separation of peptidyl-tRNA from the ribosome during the elongation reaction.5
One of the most common components of the H. pylori infections therapy regimens is clarithromycin. The resistance to macrolides such as clarithromycin in H. pylori has been demonstrated to occur at different rates (1 to 10%) in different countries, and is an important cause of H. pylori therapeutics regimens failure. Furthermore, macrolide-resistant H. pylori mutants are simply obtained by in vitro selection.5
Macrolide resistance is caused by several mechanisms such as the lack of macrolide binding to the ribosomal target, inactivation of the macrolides by enzymes, reduced or lack of bacterial membrane permeability, and macrolides active efflux.5 The widespread use of clarithromycin for the treatment of H. pylori infection has resulted in the development of resistance.6 Clarithromycin resistance (ClaR) of H. pylori is mainly caused by point mutations of the genomic 23s rRNA, the main component of the 50S subunit, mostly at position 2142/43 (A2142 to G/C/T; A2143 to G/C) in the peptidyl-transferase region of the V domain, thereby preventing drug binding. ClaR is increasing due to widespread use of macrolides for other diseases in the western world.7
There are some methods to detect the point mutations in genes such as sequencing, and amplification and restriction fragment length polymorphism (RFLP). In this study we used the RFLP method to detect the point mutations in 23s rRNA gene in our local H. pylori isolates.8 Clarithromycin is recognized as the key antibiotic for the treatment of H. pylori infections, as has a powerful bactericidal effect in vitro compared with the other available macrolides.9 Therefore, the present study aimed at evaluating the ClaR rates in local H. pylori isolates and the probable molecular mechanisms of such a resistance. Specifically, the study aimed at determining the most important point mutations in 23s rRNA gene that are closely related to clarithromycin resistance among H. pylori isolates in Kerman, Iran.
Materials and Methods
Bacteria
Sixty three H. pylori isolates were obtained from 191 patients' biopsy samples referred to the Endoscopy Division Unit of Afzalipour Hospital in Kerman, Iran. The biopsy samples were cultivated in Brucella Agar medium (Merck, Germany) supplemented with 10% defibrinated sheep blood (Darvash, Iran) and three antibiotics including Vancomycin (10 mg/l), Amphotricin B (10 mg/l) and Trimetoprim (5 mg/l) (Sigma, USA). The inoculated plates were incubated at 37°C under microaerophilic atmosphere provided by anerocult C (Merck, Germany) for 3-5 days. The isolates were recognized as H. pylori by urease, catalase, oxidase positive and gram negative staining tests.10
Antibiotic Susceptibility Tests
The susceptibility of the isolates to clarithromycin was evaluated by disc diffusion method. There is no an standard method to evaluate the susceptibility of H. pylori to antibiotics. We used the clinical and laboratory standards institute (CLSI) -recommended method called Modified Disc Diffusion method. In this method a microbial suspension with turbidity equals to four McFarland (12 x108 CFU/ml) and cultivated in Muller-Hinton agar (Merck, Germany) supplemented with 10% defibrinated sheep blood (Darvash, Iran). The 2 μg clarithromycin disc (Mast, England) were placed in the plates and incubated in 37°C under microaerophilic atmosphere for three days. Any inhibition zone was considered susceptible.10,11
DNA Extraction
DNA was extracted from all 63 H. pylori isolates using Bioneer genomics kit for DNA extraction (Bioneer, South Korea) according to the manufacturer’s instruction.
Amplification and Restriction Fragment Length Polymorphism (RFLP)
Two sets of primers were used in this study (table 1).
Table 1.
Primers used for amplifications. Primers CLA 18 and CLA 21 were used in polymerase chain reaction-amplification and restriction fragment length polymorphism (PCR-RFLP) to obtain a 1.4 kbp amplified fragment. Primers CLA 18 and CLA 3 were used in 3′-mismatched PCR to obtain a 700 bp amplified fragments.
| Set 1 | |||
|---|---|---|---|
| Cla18 | AGTCGGGACCTAAGGCGAG | 1400 bp | 7 |
| Cla21 | TTCCCGCTTAGATGCTTTCAG | ||
| (set2) | |||
| Cla18 | AGTCGGGACCTAAGGCGAG | 700 bp | 11 |
| Cla3 | AGGTCCACCACGGGGTCTTG |
The first set (cla18, cla21) was used to amplify a 1400 bp fragment from an internal region of 23s rRNA gene followed by digestion with BsaI & MboII (Fermentas, Lithuania). The 1400 bp fragment normally has one restriction site for BsaI enzyme. If the gene is wild type, the enzyme produces a 1000 bp and a 400 bp fragments.
If the A2143G point mutation occurs in 1400 bp fragment, the enzyme find two restriction sites and produces three fragments: a 700 bp, a 400 bp, and a 300 bp one. The 1400 bp fragment normally has no restriction site for MboII enzyme, therefore, if the gene is wild type, the 1400 bp remains undigested. But, if the A2142G point mutation exist, the enzyme find one restriction site in the 1400 bp fragment and digest it to two 700 bp fragments that look as one overlapping band in electrophorsis gel.7
The second set (cla18, cla3) was used to do 3'-mismatch PCR to detect A2142C point mutation. In this case, if the gene was of wild type there was no fragment, and if the A2142C point mutation took place, a 700 bp fragment was produced.12
Polymerase chain reaction condition was as follows for the amplification of the 1400 bp fragment: reactions were carried out in Primus thermo cycler (MWG-Biotech, Germany) in 50 μl mixtures containing 25 μl PCR master mix (CinnaGen Inc, Iran), 19 μl sterile deionized water, two μl template DNA and two μl of each oligonucleotide primer (4 μl totally). Initial denaturation at 94°C for five min followed by 30 cycles of denaturation at 94°C for one min, annealing for one min at 58°C, extension at 72°C for one min. The final extension step was extended to five min at 72°C.
The RFLP protocol was as follows: 10 μl of the 1400 bp fragment was added to two PCR microtubes, and five units of each enzyme was added to the micritubes and incubated at 37°C for 16 hours.
3'-mismatch PCR condition was as follow: reactions were carried out in Primus thermo cycler (MWG-Biotech, Germany) in 25 μl mixtures containing 12 μl PCR master mix (CinnaGen Inc, Iran), 10 μl sterile deionized water, one μl template DNA and one μl of each oligonucleotide primer. Initial denaturation at 94°C for five min followed by 30 cycles of denaturation at 94°C for one min, annealing for one min at 55°C, extension at 72°C for one min. The final extension step was extended to five min at 72°C.
Electrophorsis
The PCR products were separated on 1.5% and the PCR-RFLP products were separated on 2% agarose gels (Cinna gen, Iran) after being stained with ethidium bromide (Merck, Germany) in TBE 1X (Tris/borate/EDTA) buffer under 100 volts electricity flow. Bands were visualized under UV gel documentation and photographed.
Results
Twenty out of 63 (31.7%) of the H. pylori isolates were resistant to clarithromycin. There was no significant relation between gender, age or the history of antibiotic consumption by the patients and resistance to calrithromycin. All of the 20 ClaR isolates had at least one of the three common point mutation in 23s rRNA gene, while none of the ClaS isolates had such a point mutation (table 2).
Table2.
The frequency and (rate) of clarithromycin susceptibility test for H. pylori isolates in both resistant and sensitive isolates in Kerman, Iran.
| CLA susceptibility | Number (%) |
23 s
rRNA
point
mutation |
Number (%) |
|---|---|---|---|
| R | 20 (31.7%) | + | 20 (100%) |
| - | 0 (0%) | ||
| S | 43 (69.3%) | + | 0 (0%) |
| - | 43 (100%) |
CLA: Clarithromycin, R: Resistant, S: sensitive
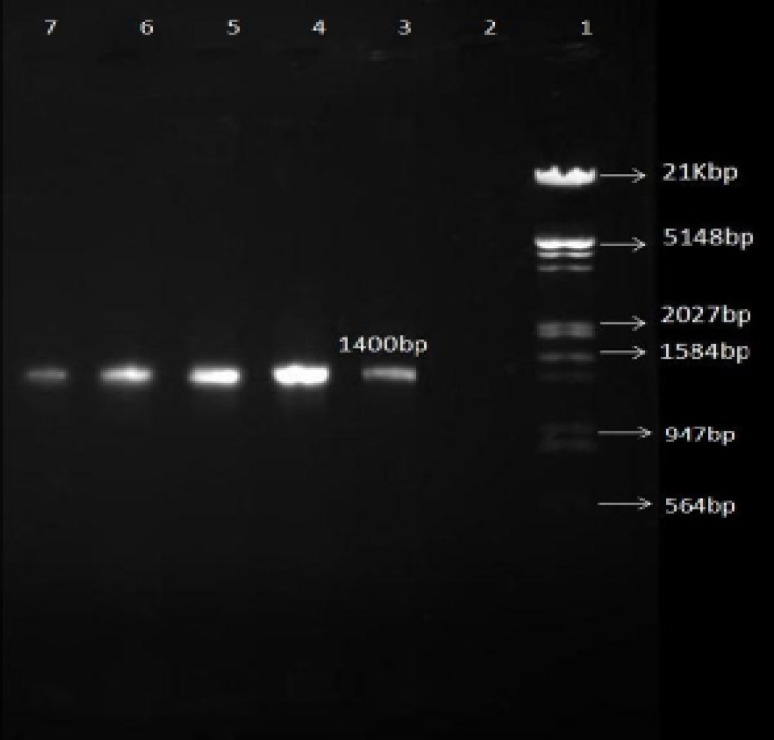
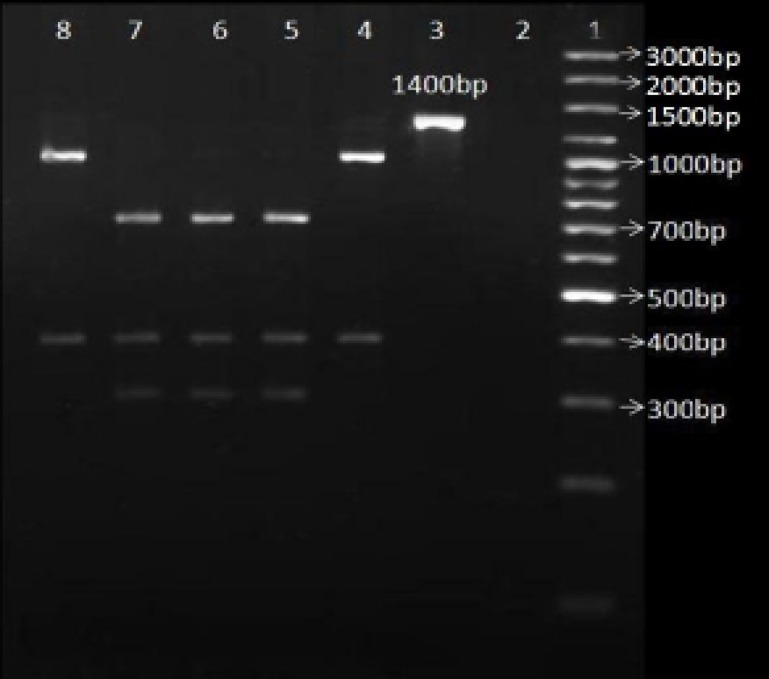
All of the 63 H. pylori isolates were positive for the 1400 bp fragment (figure1). Fifteen percent of the ClaR isolates (three out of 20 isolates) had the A2143G point mutation (figure 2). There was a significant relation between the gender of the patients and the A2143G point mutation. Three out of 38 (7.9%) of the strains isolated from the female population had this point mutation, whereas no such a mutation was found in the strains isolated from the male population. There was no significant relation between age or the history of antibiotics consumption and the A2143G point mutation.
Figure 1.
Gel electerophorsis of 1400 bp fragment PCR products from 23s rRNA gene for RFLP. All 63 H. pylori isolates were positive.
Line 1:HindΙΙΙ/EcoRΙ 21Kbp DNA marker (fermentas); Line 2: Negative control; Line 3-7: Different samples
Figure 2.
PCR-RFLP patterns of 1400 bp fragments after digestion with BsaΙ enzyme in order to detect A2143G point mutation in 23s rRNA gene.
Line 1: 3000 bp DNA marker (fermentas); Line 2: Negative control (only enzyme); Line 3: only PCR product (1400 bp fragment); Line 4, 5, 6, 7, 8: 1400 bp fragment+BsaΙ; Line 4, 8: clarithromycin-sensitive H. pylori isolates; Line 5, 6, 7: clarithromycin-resistant H. pylori isolates.
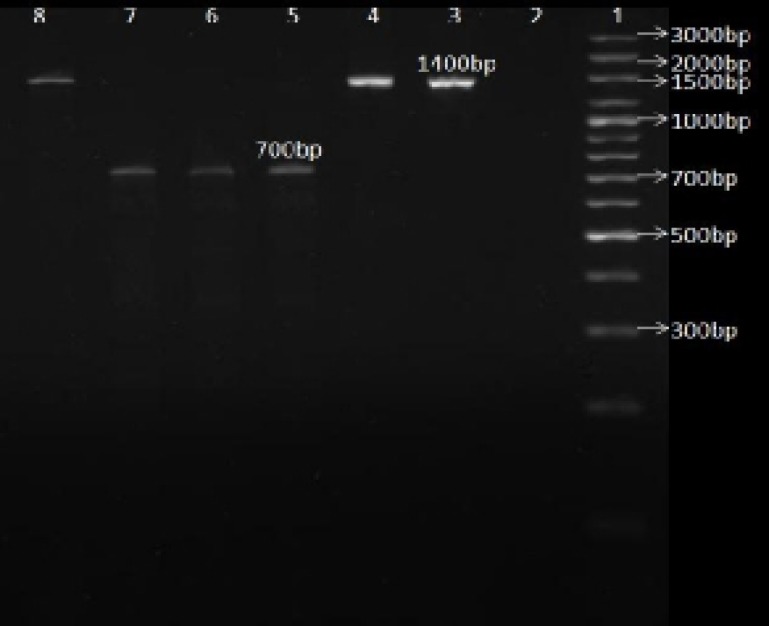
Fifty five percent of the ClaR isolates (11 out of 20 isolates) had the A2142G point mutation (figure 3). There was no significant relation between gender, age or the history of antibiotics consumption of the patients and this mutation.
Figure 3.
PCR-RFLP patterns of the 1400 bp fragments digested with MboΙΙ enzyme in order to detect A2142G point mutation in 23s rRNA gene.
Line1: 3000 bp DNA marker (fermentas); Line 2: Negative control (only enzyme); Line 3: only PCR product (1400 bp); Line 4, 5, 6, 7, 8: 1400 bp fragment+MboΙΙ; Line 4, 8: clarithromycin-sensitive H. pylori isolates; Line 5, 6, 7: clarithromycin-resistant isolates
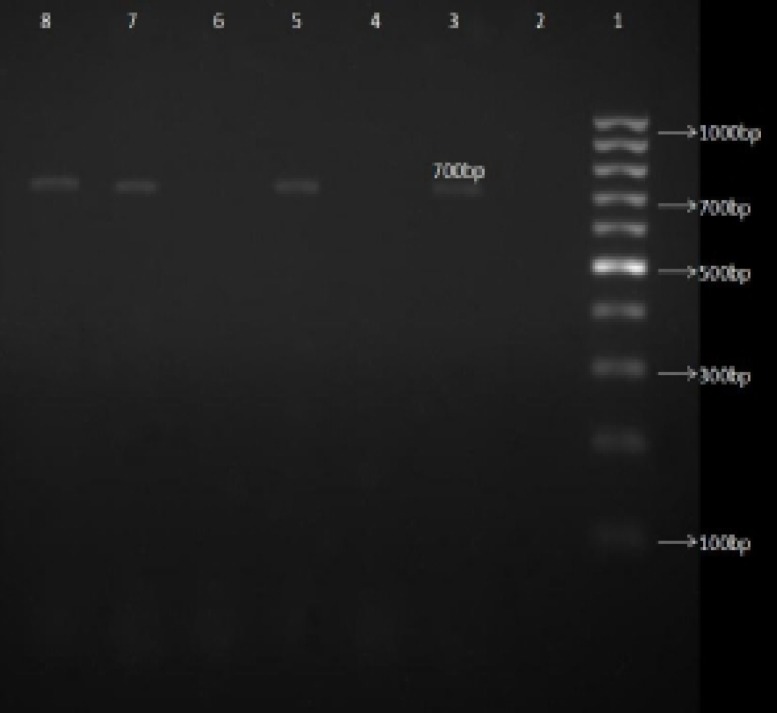
Thirty percent of the ClaR isolates (six out of 20 isolates) were positive for the A2142C point mutation (figure 4). There was no significant relation between age, gender or the history of antibiotics consumption of the patients and this mutation.
Figure 4.
Gel electerophorsis of 3'-mismatch PCR products in order to detect A2142C point mutation in 23s rRNA gene.
Line1: 1000 bp DNA marker (fermentas); Line 2: Negative control; Line 3, 5, 7, 8: clarithromycin-resistant H. pylori isolates without A2142G or A2143G; Line 4: clarithromycin-sensitive H. pylori isolates; Line 6: ClaR H. pylori isolates with A2143G mutation
The A2142C point mutations occurred only in ClaR isolates without A2142G or A2143G (table 3).
Table3.
Results obtained with the PCR-RFLP and the 3′-mismatched PCR methods for the clinical isolates tested according to the clarithromycin resistance.
| mutation | Number (%) | Digestion with: BsaΙ/MboΙΙ | 3'mismatch PCR | Cla susceptibility |
|---|---|---|---|---|
| A2143G | 3 (15%) | +/- | - | ClaR |
| A2142G | 11 (55%) | -/+ | - | ClaR |
| A2142C | 6 (30%) | -/- | + | ClaR |
CLa: Clarithromycin, R: Resistant
Discussions
Resistance of H. pylori to antibiotics has been increasing in most parts of the world including Iran.11,13-15 Clarithromycin resistances is a serious concern for doctors who are using the drug as one of the most important therapeutic components for H. pylori-induced gastric ulcer. There are ever-increasing requests from physicians for a reliable standard antimicrobial susceptibility test for H. pylori against clarithromycin, but that would be hard to do because of its fastidious properties and its time-consuming culture. Furthermore, success in H. pylori culture is dependent on the microbiology laboratory technicians' skills.16 Clarithromycin resistance rates are varied across the world. For example Elviss et al in London reported 11% resistance to clarithromycin,17 or Bagalan et al announced 27.6% resistance.18 Also, the rate of clarithromycin resistance varies in different cities in Iran. For example Kohanteb et al reported 9.4% resistance in Shiraz (2007),19 while Mohammadi et al showed 20% resistance to clarithromycin in Tehran (2005), and in more recent studies Siyavoshi et al (2010) in Tehran reported 7.3% resistance.11,15
Clarithromycin is a macrolide, that due to its high prices, was not used commonly in Iran in the past years. However, after its production in the country in recent years, it has been used routinely in the treatment of H. pylori infections. So, the emergence of ClaR isolates is inevitable. It is also has been shown that countries with a high consumption of other macrolides have a higher rates of clarithromycin resistance.20 Macrolides such as erythromycin and clarithromycin inhibit nascent peptide chain elongation by interacting with the 50S ribosomal subunit and stimulating the release of peptidyl-tRNA from the A site.21 Biochemical studies have demonstrated a direct interaction of clarithromycin and its chief metabolite, 14-hydroxyclarithromycin, with 50S ribosomal subunits isolated from H. pylori.22,23 The antibacterial activity of clarithromycin is better than that of erythromycin. One reason for such a difference is the synergistic phenomenon between clarithromycin and one its metabolites 14-hydroxyclarithromycin, which leads to a considerable post antibiotic effect. The second reason is higher hydrophobicity of clarithromycin, which leads to a better penetration through the cell membranes than that of erythromycin. The third reason is clarithromycin activity, which is less influenced by acidity than that of erythromycin.23
Versalovic and colleagues were the first to announce that the clarithromycin resistance of H. pylori was associated with a point mutation in the V domain of 23S rRNA. They discovered A to G point mutations at positions identical to E.coli 23S rRNA positions 2058 and 2059, and then called these positions 2143 and 2144 according to the entire H. pylori 23S rRNA sequence.24
The present study focused on the three common point mutations, namely A2143G, A2142G and A2142C, which according to a sizable number of previous reports are the most common mutations associated with clarithromycin resistance. All of 20 ClaR isolates had at least one of these three mutations. Therefore, there was an absolute association between these three point mutations in 23s rRNA gene and Clarithromycin resistance in the isolates.
In agreement with the findings by Alarcon et al,16 the present study showed that the A2142C point mutation in 23s rRNA existed only on ClaR isolates without A2142G or A2143G point mutations in 23s rRNA (table 3).
A number of other investigator reported other point mutations in 23s rRNA gene that were associated with clarithromycin resistance as well. For example Hao et al. in China reported three novel point mutations including C2245T, G2244A and T2289C that were associated with clarithromycin resistance in their local isolates.25 Also, Khan et al. showed that T2182C point mutation in 23s rRNA was associated with clarithromycin resistance in Bangladesh.26 Therefore, it is important to realize that the three common point mutations that the present study focused on are not the only reason of clarithromycin resistance, and there could be some other point mutations in 23s rRNA gene associated with such a resistance.
Some other mechanisms have been suggested for clarithromycin resistance, of which one is efflux pumps. Hirata et al. suggested a contribution of efflux pumps to the clarithromycin resistance in Japan.27 Since there was no significant relation between gender, age, or the history of antibiotics consumption of the patients and resistance to clarithromycin, it seems that spontaneous mutations are responsible for such a resistance among the microbial population. The importance of such a resistance was revealed when a number of studies reported that resistance to clarithromycin was equal to the whole therapeutic regime failure.28
Conclusion
The high rate of clarithromycin resistance in the isolates in the present study is a serious alarm, and in agreement with clinical colleagues' views that many of their patients do not respond to clarithromycin anymore. Point mutations in 23s rRNA are closely related to such a resistance. With daily increase in the use of clarithromycin in therapeutic regime for H. pylori in Iran, the rate of H. pylori resistance rate to the drug is increasing. Therefore, it seems necessary to do antibiotic susceptibility tests for H. pylori before therapy begins.
Acknowledgment
The authors would like to thank the Research Council of Kerman University of Medical Sciences for their financial supports.
Conflict of Interest: None declared
References
- 1.Cameron EA, Powell KU, Baldwin L, et al. Helicobacter pylori: antibiotic resistance and eradication rates in Suffolk, UK, 1991–2001. J Med Microbiol. 2004;53:535–8. doi: 10.1099/jmm.0.05499-0. [DOI] [PubMed] [Google Scholar]
- 2.Liu Z, Shen J, Zhang L, et al. Prevalence of A2143G mutation of H. pylori-23S rRNA in Chinese subjects with and without clarithromycin use history. BMC Microbiol. 2008;8:81. doi: 10.1186/1471-2180-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korn A, Garber A, Goodman S. Pharmachogenomics-based treatment of Helicobacter pylori infection. Asessment program. 2008;23:1–21. [Google Scholar]
- 4.Soltermann A, Perrena A, Schmida S, et al. Assessment of Helicobacter pylori clarithromycin resistance mutations in archival gastric biopsy samples. Swiss Med Wkly. 2005;135:327–32. doi: 10.4414/smw.2005.10866. [DOI] [PubMed] [Google Scholar]
- 5.Occhialini A, Urdaci M, Doucet-populaire F, et al. Macrolide resistance in Helicobacter pylori: rapid detection of point Mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–8. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hultén K, Gibreel A, Sköld O, Engstrand L. Macrolide Resistance in Helicobacter pylori: Mechanism and Stability in Strains from Clarithromycin-Treated Patients. Antimicrob Agents Chemother. 1997;41:2550–3. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versalovic J, Shortridge D, Kibler K, et al. Mutations in 23S rRNA Are Associated with Clarithromycin Resistance in Helicobacter pylori. Antimicrob Agents and Chemother. 1996;40:477–80. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374–84. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De FrancescoV, Margiotta M, Zullo A, et al. Prevalence of primary clarithromycin resistance in Helicobacter pylori strains over a 15 year period in Italy. J Antimicrob Chemother. 2007;59:783–5. doi: 10.1093/jac/dkm005. [DOI] [PubMed] [Google Scholar]
- 10.Mishra KK, Srivastava S, Garg A, Ayyagari A. Antibiotic susceptibility of Helicobacter Pylori clinical isolats: Comparative evalution of disk-diffusion and E-test methods. Curr Microbiol. 2006;53:329–34. doi: 10.1007/s00284-006-0143-1. [DOI] [PubMed] [Google Scholar]
- 11.Mohammadi M, Doroud D, Mohajerani N, Massarrat S. Helicobacter pylori antibiotic resistance in Iran. World J Gastroenterol. 2005;11:6009–13. doi: 10.3748/wjg.v11.i38.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alarcón T, Domingo D, Prieto N, López-Brea M. PCR using 3-mismatched primers to detect A2142C mutation in 23S rRNA conferring resistance to clarithromycin in Helicobacter pylori clinical isolates. J Clin Microbiol. 2000;38:923–5. doi: 10.1128/jcm.38.2.923-925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savari M, Abdollahi H, Zahedi MJ, Darvish-Moghadam S, Hayat-Bakhah M. Antibiotic-resistance Patterns of Helicobacter pylori isolates Obtained from Patients in Kerman- 2009. Journal of Kerman University of Medical Sciences. 2011;18(1):73–82. [Google Scholar]
- 14.Abdollahi H, Savari M, Zahedi MJ, Darvish-Moghadam S, Hayat-Bakhah MA. Study of rdxA gene deletion in metronidazole resistant and sensitive Helicobacter pylori isolates in Kerman, Iran. Jundishapur J Microbiol. 2011;4(2):99–104. [Google Scholar]
- 15.Siavoshi F, Saniee P, Latifi-Navid S, et al. Increase in resistance rates of H.pylori isolates to metronidazole and tetracycline-comparison of three 3-year studies. Arch Iranian med. 2010;13:177–87. [PubMed] [Google Scholar]
- 16.Kim KS, Kang JO, Eun CS, et al. Mutations in the 23S rRNA Gene of Helicobacter pylori Associated with Clarithromycin Resistance. J Korean Med Sci. 2002;17:599–603. doi: 10.3346/jkms.2002.17.5.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elviss NC, Owen RJ, Breathnach A, et al. Helicobacter pylori antibiotic-resistance patterns and risk factors in adult dyspeptic patients from ethnically diverse populations in central and south London during 2000. J Med Microbiol. 2005;54:567–74. doi: 10.1099/jmm.0.45896-0. [DOI] [PubMed] [Google Scholar]
- 18.Baglan PH, Bozdayi G, Ozkan M, et al. Clarithromycin Resistance Prevalence and Icea Gene Status in Helicobacter Pylori Clinical Isolates in Turkish Patients with Duodenal Ulcer and Functional Dyspepsia. J Microbiol. 2006;44:409–16. [PubMed] [Google Scholar]
- 19.Kohanteb J, Bazargana I, Saberi-Firoozi M, Mobasser A. Antimimicrobial susceptibility testing of Helicobacter pylori to selected agents by agar dillution method in Shiraz, Iran. Indian J Med Microbiol. 2007;25:374–7. doi: 10.4103/0255-0857.37342. [DOI] [PubMed] [Google Scholar]
- 20.Grove D, Koutsouridis G. Increasing resistance of Helicobacter pylori to clarithromycin: is the horse bolting? Pathology. 2002;34:71–73. doi: 10.1080/00313020120105624. [DOI] [PubMed] [Google Scholar]
- 21.Menninger R. Functional consequences of binding macrolides to ribosomes. J Antimicrob Chemother. 1985;16:23–34. doi: 10.1093/jac/16.suppl_a.23. [DOI] [PubMed] [Google Scholar]
- 22.Goldman RC, Zakula D, Flamm R, et al. Tight binding of clarithromycin, its 14-R-hydroxy metabolite, and erythromycin to Helicobacter pylori ribosomes. Antimicrob Agents Chemother. 1994;38:1496–500. doi: 10.1128/aac.38.7.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia HX, Buckley M, Keane CT, O'Morain CA. Clarithromycin resistance in Helicobacter pylori: prevalence in untreated dyspeptic patients and stability in vitro. J antimicrob chemother. 1996;37:473–81. doi: 10.1093/jac/37.3.473. [DOI] [PubMed] [Google Scholar]
- 24.Taylor D, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–8. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao Q, Li Y, Zhang ZJ, et al. New mutation points in 23S rRNA gene associated with Helicobacter pylori resistance to clarithromycin in northeast China. World J Gastroenterol. 2004;10:1075–7. doi: 10.3748/wjg.v10.i7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan R, Nahar S, Sultana J, et al. T2182C Mutation in 23S rRNA Is Associated with Clarithromycin Resistance in Helicobacter pylori Isolates Obtained in Bangladesh. Antimicrob Agents Chemother. 2004;48:3567–9. doi: 10.1128/AAC.48.9.3567-3569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirata K, Suzuki H, Nishizawa T, et al. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol. 2010;25:S75–9. doi: 10.1111/j.1440-1746.2009.06220.x. [DOI] [PubMed] [Google Scholar]
- 28.Tankovic J, Lamarque D, Lascols C, et al. Clarithromycin resistance of Helicobacter pylori has a major impact on the efficacy of the omeprazole-amoxicillin-clarithromycin therapy. Pathol Biol (Paris) 2001;49:528–33. doi: 10.1016/s0369-8114(01)00209-7. [DOI] [PubMed] [Google Scholar]