Abstract
Staphylococcus epidermidis is the most important member of coagulase negative staphylococci responsible for community and hospital acquired infections. Most clinical isolates of S. epidermidis are resistant to methicillin making these infections difficult to treat. In this study, correlation of methicillin resistance phenotype was compared with methicillin resistance (mecA) gene carriage in 55 clinical isolates of S. epidermidis. Susceptibility was measured by disc diffusion using methicillin discs, and minimum inhibitory concentrations (MIC) were measured using broth microdilution. Methicillin resistance gene (MecA gene) carriage was detected by specific primers and PCR. Disc susceptibility results showed 90.9% resistance to methicillin. Considering a MIC of 4 µg/ml, 78.1% of the isolates were methicillin resistant, 76.36% of which carried the mecA gene. On the other hand, when a breakpoint of 0.5 µg/ml was used, 89.09% were methicillin resistant, of which 93.75% were mecA positive. There was a better correlation between MIC of 0.5 µg/ml with disc diffusion results and mecA gene carriage. The findings suggest that despite the usefulness of molecular methods for rapid diagnosis of virulence genes, gene carriage does not necessarily account for virulence phenotype. Ultimately, gene expression, which is controlled by the environment, would determine the outcome.
Key Words: Staphylococcus epidermidis, methicillin, minimum inhibitory concentration, mecA
Introduction
Coagulase-negative staphylococci (CoNS) are the major causes of hospital acquired infections, and are often isolated from neonates, immunocompromised individuals and patients with indwelling prosthetic devices.1,2 Among the CoNS, Staphylococcus epidermidis is frequently associated with bacteremia, urinary tract infections and infections associated with indwelling medical devices.3 The major concern with regard to the treatment of staphylococcal infections is antibiotic resistance among the clinical isolates. Indeed, over 90% of all nosocomial isolates are resistant to penicillin and an increasing number are becoming resistant to the semisynthetic, β-lactamase resistant derivatives represented by oxacillin.4
It has been reported that presence of the methicillin resistance gene (mecA) gene, which encodes penicillin binding protein (PBP) 2a, correlates with oxacillin (methicillin) resistance in CoNS.5 Moreover, methicillin-resistant strains are often resistant to other drugs, and therapeutic options in such cases are often limited to glycopeptid antibiotics such as vancomycin.4 Hence, it is important for clinical laboratories to distinguish between methicillin-susceptible and methicillin-resistant CoNS. Methicillin minimum inhibitory concentration (MIC) breakpoint (4 µg/ml), which was first recommended by the National Committee for Clinical Laboratory Standards (NCCLS), lacked sensitivity, and was unable to classify many mecA-positive CoNS as methicillin resistant.6,7 Consequently, it was suggested that lowering methicillin MIC breakpoint may significantly improve the accuracy of the susceptibility tests.8,9 Accordingly, the NCCLS redefined methicillin susceptibility breakpoints for CoNS, so that organisms for which methicillin MIC is >0.5 µg/ml are considered resistant and those for which the MIC is 0.25 µg/ml or lower are considered susceptible.10
A number of factors including hyperproduction of β-lactamase and alteration of PBPs,11 and auxiliary genes such as femA, mecR and other β-lactamase genes,12 may affect methicillin-resistance gene expression. In addition, methicillin-resistance is influenced by culture conditions such as temperature, medium, pH and NaCl content in the medium. These factors complicate the detection of methicillin-resistance, especially for strains with low level resistance.11-13 In this report, we compared mecA gene carriage with different MIC breakpoints for methicillin resistance in 55 local clinical isolates of S. epidermidis.
Materials and Methods
Sixty nine clinical isolates of coagulase negative staphylococci were collected from three hospitals (Talghani, Imam Hossein and Boo Ali) in Tehran during January to October 2007. The majority of the isolates was from blood (46.7%), followed by wound and wound exudates (22.2%), catheters (8.9%) and the rest were from unknown clinical sources. Identification of S. epidermidis was carried out using the standard biochemical tests including catalase, DNase and coagulase production, growth and fermentation of mannitol on mannitol salt agar, and susceptibility to bacitracin and novobiocin. Bacteria were maintained in Lauria Bertani broth containing 8% dimethylsulfoxide (DMSO) at –80°C. Staphylococcus aureus (ATCC 25923) was used as control for antibiotic susceptibility assays. Susceptibility of the isolates to methicillin (5 µg) and vancomycin (30 µg) was determined by disc diffusion using the NCCLS guidelines.14 Antibiotic discs were obtained from Padtan Teb, Iran. Minimum inhibitory concentrations for methicillin were determined by broth microdilution within the range of 64-0.125 µg/ml.
For PCR experiments, DNA was extracted by boiling. Briefly, a loopful of colonies from an overnight growth on nutrient agar was transferred into 250 µl of distilled water and boiled at 100°C for 20 min. The lysate was then centrifuged at 12000 g for 10 min, and 10 µl of the supernatant was used directly as DNA template in PCR reaction mixtures. The presence of mecA was detected by specific primers (forward: GTA GAA ATG ACT GAA CGT CCG ATA A and reverse: CCA ATT CCA CAT TGT TTC GGT CTA A) resulting in amplification of a 310 bp PCR product.15 Reaction mixtures (25 µl) contained 10 µl genomic DNA, 20 pM of each oligonucleotide primer, 1u Taq polymerase (Cinnagen, Iran), 200 µM of dNTP mix and 1.5 mM MgCl2 in the reaction buffer provided by the manufacturer. Amplifications were performed using a Thermal Cycler (Techne TC-312, England) with the following program: an initial denaturation at 94°C for 2 min followed by 30 cycles of amplification (1 min denaturation at 94°C, 1 min annealing at 55°C, 2 min extension at 72°C) and a final extension period of 5 min at 72°C. The PCR products were electrophoresed on a 1% agarose gel in a 0.5 X tris-borate-EDTA buffer and stained with ethidium bromide. Gene Ruler 100 bp DNA ladder (Fermentas) was used as DNA size marker.
Results
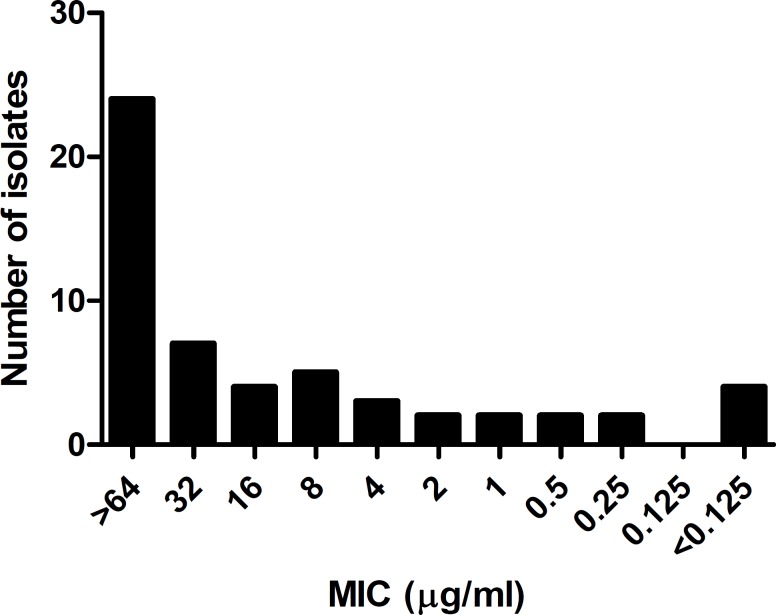
Of the 69 CoNS clinical isolates, 55 were identified as S. epidermidis. Disc diffusion results showed that 50 isolates (90.9%) were resistant to methicillin, and all of them were sensitive to vancomycin. There was no relation between methicillin resistance and the type of infection. The MIC values obtained for methicillin were interpreted with two sensitivity breakpoints; 4 µg/ml (test group A) and 0.5 µg/ml (test group B) (figure 1). Among the methicillin resistant isolates in group A, 43 (78.1%) had MIC values of >4 µg/ml of which, 42 (97.67%) carried the mecA gene. Three of the seven isolates, which were methicillin resistant by disc diffusion but had MIC values lower than 4 µg/ml, were mecA positive. Of the five isolates, which were sensitive by both phenotypic methods, 4 were mecA positive. On the other hand, when breakpoint of 0.5 µg/ml was chosen as the cut off point, 49/55 (89.09%) were resistant to methicillin of which, 46 (93.88%) carried the mecA gene. Of the 6 remaining methicillin susceptible isolates in group B, three carried the mecA gene and three were mecA negative. Overall, comparison of the MIC values in the two groups with the disc susceptibility results showed a better agreement with the 0.5 µg /ml breakpoint. Comparison of the PCR results with the disc susceptibility assay also showed a closer agreement for group B where 46/55 (86.64%) organisms were methicillin resistant and carried the mecA gene. On the other hand, in group A, 42/55 isolates (76.36%) were methicillin resistant/mecA positive (figure 2). These results indicate that the 0.5 µg/ml breakpoint is a more realistic value for determining methicillin resistance in clinical isolates of S. epidermidis as suggested before.
Figure 1.
Distribution of minimum inhibitory concentrations (MICs) for methicillin in 55 clinical isolates of Staphylococcus epidermidis.
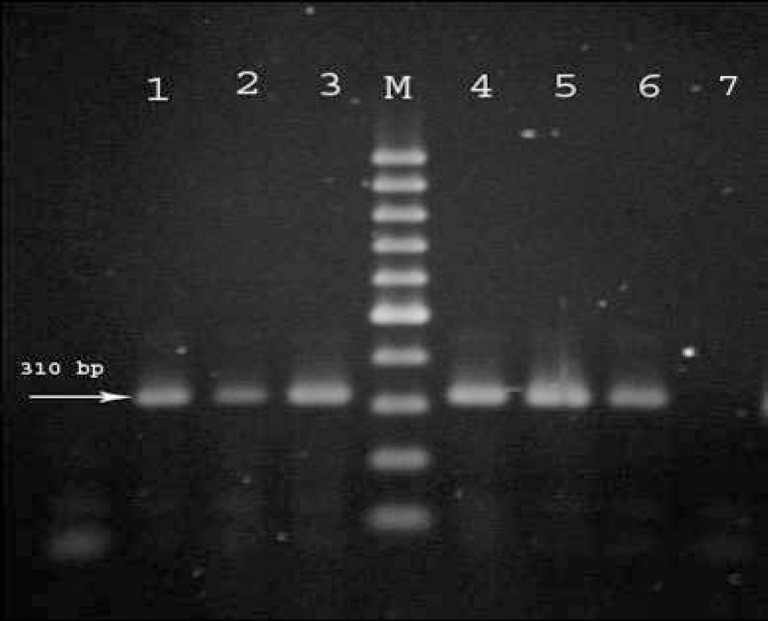
Figure 2.
Amplification of the methicillin resistance (mecA) gene in a number of Staphylococcus epidermidis isolates. Lanes 1-6; mecA positive isolates, lane 7; mecA negative control, M; 100 bp DNA ladder marker
Discussion
Detection of methicillin resistance in staphylococci is complex, mainly because it is often heterogeneous, and only 1 in 104 to 108 cells in a bacterial population expresses the trait. 11 The previously used NCCLS breakpoints for methicillin resistance (4 and 2 µg/ml) were shown to significantly underestimate the degree of true methicillin resistance among CoNS. Hence, the NCCLS redefined the breakpoints for methicillin susceptibility to MIC values of ≥0.5 µg/ml and organisms with MICs ≤0.25 µg/ml were considered susceptible.7,9,10 Another phenotypic method for successful prediction of methicillin resistance in CoNS is the simultaneous use of cefoxitin and oxacillin discs. However, interpretation of inhibition zones are often in dispute, and prediction of methicillin susceptibility is not 100% accurate.5,11
Although culture-based methods are generally reliable for detecting methicillin-resistant staphylococci, detection of the mecA gene by PCR has been considered as the gold standard, and a number of investigators have found a complete agreement between methicillin resistance phenotype and mecA gene presence.8,9,12,16 Concordance between mecA gene carriage and resistance phenotype in CoNS using 2 µg/ml MIC breakpoint showed12-16% false susceptibility, and lowering the MIC breakpoint to 0.25 µg/ml greatly improved the accuracy of the MIC test performance.7-9 In our experiments with clinical isolates of S. epidermidis, using mecA gene carriage as standard, MIC value of 4 (or 2) µg/ml resulted in 11% false susceptibility, and MIC values of 0.5 (or 0.25) µg/ml showed a more accurate profile for methicillin susceptibility. However, unlike other investigations, we did not find complete agreement between mecA gene carriage and MIC phenotype even at lower MIC values. In the case of methicillin resistant mecA negative isolates, it has been suggested that mechanisms such as β-lactamase hyperproduction and alteration of PBPs other than PBP 2a may be responsible for the resistance phenotype.11,13 In methicillin sensitive mecA positive isolates, mecA gene is not consistently expressed and auxiliary genes such as femA, mecR and other β-lactamase genes may participate in the control of gene expression.12
Conclusion
The findings of this study indicate that the choice of correct MIC breakpoints is important for the detection of methicillin resistance in clinical isolates of S. epidermidis, and can lower the number of false sensitive isolates. There was a better agreement between MIC of <0.5 µg/ml and presence of the mecA gene compared to higher MIC values (2 and 4 µg/ml). They also show that gene carriage does not necessarily account for resistance phenotype, and environment would ultimately control gene expression.
Acknowledgment
The authors wish to thank Shahid Beheshti University Research Council for providing a special grant to finance this research.
Conflict of Interest: None declared
References
- 1.Heilmann C, Schweitzer O, Gerke C, et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–91. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 2.Cerca N, Pier GB, Vilanova M, et al. Influence of batch or fed-batch growth on Staphylococcus epidermidis biofilm formation. Lett Appl Microbiol. 2004;39:420–4. doi: 10.1111/j.1472-765X.2004.01601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wieser M, Busse HJ. Rapid identification of Staphylococcus epidermidis. Int J Syst Evol Microbiol. 2000;3:1087–93. doi: 10.1099/00207713-50-3-1087. [DOI] [PubMed] [Google Scholar]
- 4.Mason WJ, Blevins JS, Beenken K, et al. Multiplex PCR protocol for the diagnosis of staphylococcal infections. J Clin Microbiol. 2001;39:3332–38. doi: 10.1128/JCM.39.9.3332-3338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrazi B, Fermepin MR, Malimovka A, et al. Accuracy of cefoxitin disc testing for characterization of oxacillin resistance mediated by penicillin-binding protein 2a in coagulase-negative staphylococci. J Clin Microbiol. 2006;44:3634–9. doi: 10.1128/JCM.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Committee for Clinical Laboratory Standards (NCCLS) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 2nd ed: approved standard M100-S8. PA: Villanova; 1999. [Google Scholar]
- 7.Tenover FC, Jones RN, Swenson JM, et al. Methods for improved detection of oxacillin resistance in coagulase-negative staphylococci: results of a multicenter study. J Clin Microbiol. 1999;37:4051–58. doi: 10.1128/jcm.37.12.4051-4058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall SA, Wilke WW, Pfaller MA, Jones RN. Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE program. Diagn Microbiol Infect Dis. 1998;30:205–14. doi: 10.1016/s0732-8893(97)00212-5. [DOI] [PubMed] [Google Scholar]
- 9.Hussain Z, Stoakes L, Massey V, et al. Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J Clin Microbiol. 2000;38:752–4. doi: 10.1128/jcm.38.2.752-754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards (NCCLS) Performance standards for antimicrobial susceptibility testing: ninth information supplement document M100-S10. PA: Wayne; [Google Scholar]
- 11.Chambers HF. Methicillin resistant staphylococci. Clin Microbiol Rev. 1988;1:173–86. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–91. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabath LD. Mechanisms of resistance to to β-lactam antibiotics in strains of Staphylococcus aureus. Ann Intern Med. 1982;97:339–44. doi: 10.7326/0003-4819-97-3-339. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 15th ed. Approved standard M7-A6. Wayne, PA: Clinical and Laboratory Standards Institute; 2005. [Google Scholar]
- 15.Eftekhar F, Mirmohammadi Z. Evaluation of biofilm production by Staphylococcus epidermidis isolates from nosocomial infections and skin of healthy individuals. Internatl J Med & Med Sci. 2009;1:438–41. [Google Scholar]
- 16.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assay and the antibiotic susceptibility patterns of and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–8. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]