Abstract
Objective
The goal of this preliminary study was to use metagenomic approaches to investigate the taxonomic diversity of microorganisms in patients with bisphosphonate-related osteonecrosis of the jaw (BRONJ).
Study Design
Samples of saliva for planktonic microbial analysis and biofilm cultivation were collected from 10 patients (5 with BRONJ and 5 non-BRONJ control subjects) who met all ascertainment criteria. Prophage induction experiments—16S rRNA polymerase chain reaction and 454 pyrosequencing—and epifluorescent microscopy were performed for characterization and enumeration of microbes and viruses.
Results
Three phyla of microbes—Proteobacteria (70%), Firmicutes (26.9%), and Actinobacteria (1.95%)—dominated all BRONJ samples and accounted for almost 99% of the total data. Viral abundance was ~1 order of magnitude greater than microbial cell abundance and comprised mainly phage viruses.
Conclusions
Individuals with jaw osteonecrosis harbored different microbial assemblages than nonaffected patients, and in general viral abundance and prophage induction increased with biofilm formation, suggesting that biofilm formation encouraged lysogenic interactions between viruses and microbial hosts and may contribute to pathogenicity.
Millions of Americans are currently receiving antiresorptive drugs to treat common bone disorders such as osteoporosis and skeletal complications associated with osseous metastasis and multiple myeloma.1 Of the antiresorptive drugs, nitrogen-containing bisphosphonates (BP), with their high affinity for bone and long safety record, constitute the largest class; they can be administered orally or intravenously and are the most widely used because they are relatively inexpensive and used across a broad spectrum of osteoporosis types, including postmenopausal, male, and steroid-induced osteoporosis.2 However, a serious adverse effect of BP therapy is bisphosphonate-related osteonecrosis of the jaw (BRONJ), which clinically is often characterized by exposed nonvital jaw bone (sequestrum) in the oral cavity.3 Severe BRONJ cases can interfere with rheumatologic or oncologic care of patients. Importantly, there are no universally accepted protocols for treatment of BRONJ and no known cure, so some patients never experience disease resolution, and the toll on affected individuals and the health care system is significant.
The pathogenesis of BRONJ has not been clearly understood and is thought to be multifactorial.4 Mechanisms for BRONJ pathogenesis that have been proposed in the literature include direct toxicity of BP to oral tissues, antiangiogenic effects of BP, inflammation associated with bone microcracks, suppression of bone turnover and wound healing, secondary infection, and an altered immune response.5 Investigators have questioned why the jaw bones are predominantly affected by osteonecrosis when BP drugs disseminate to all bones of the body. There is now mounting evidence that the reason this complication primarily affects jaw bones, and usually after invasive dental procedures or oral trauma, is because microbial biofilms in the mouth and saliva gain access to the exposed jaw bone and play a significant role in the necrosis of the bone and thus the etiopathogenesis of this condition.6–8 Recent investigations have shown that BP drugs inhibit oral wound healing and facilitate bacterial colonization on bony surfaces where both BP and biofilm bacteria colocalize.9,10 Therefore, the clinical problem of BRONJ is essentially a nonhealing biofilm-mediated osteomyelitis of the jaw.11–13
A microbial biofilm is a community of microorganisms attached to a surface and surrounded by a matrix of extracellular polymeric substance.14 Biofilm organisms differ considerably from their plank-tonic counterparts because they are characterized by a community of cells that are attached to a surface, are embedded in a matrix of extracellular polymeric substances that they have produced to connect to and communicate with each other, and exhibit an altered phenotype in terms of growth rate, gene transcription and antimicrobial resistance.15,16 A major limitation in the field of BRONJ biofilm research and one that has hindered targeted therapeutics is our current knowledge gap of the oral microbial ecology in affected patients. Microbial studies of BRONJ have been predominantly cross-sectional descriptive and histomorphologic studies, and little is known about the oral microbial community composition, taxonomic diversity, or functional potential of associated microorganisms in affected patients. This has restricted targeted therapeutics for this disease.
Therefore, the goal of the present study was to use metagenomic approaches to investigate the taxonomic diversity of microorganisms in patients with BRONJ. Furthermore, we sought to evaluate the role of viruses, which has never been described in this condition, and to assess the occurrence of lysogeny to determine if prophage potentially contributes to the pathogenicity of community microbes. This line of investigation could provide rationale in the future for BRONJ therapeutics and targeted antimicrobial therapy. Our working hypotheses were that individuals with BRONJ harbor different microbial assemblages than nonaffected patients, that changes in microbial assemblages occur when host cells shift from a primarily planktonic (e.g., saliva) to surface-attached (e.g., biofilms) lifestyle, and that BRONJ patients have a higher proportion of lysogenic bacteria and their integrated prophage could influence host pathogenicity.
MATERIALS AND METHODS
Clinical design and extraction of microbes and viruses from saliva and saliva-induced biofilms
Institutional Review Board approval was obtained for this study (approval no. UP-09-00026). Samples of saliva for microbial analysis and biofilm cultivation were collected from 10 individuals recruited for this study at the Herman Ostrow School of Dentistry, University of Southern California. Five patients affected by BRONJ and 5 non-BRONJ control patients met all of the inclusion and exclusion criteria for the study, and informed consent was obtained from each patient. No patients with cognitive, language, and hearing problems were recruited, to ensure that all patients could provide their own consent. The individuals in the BRONJ and control groups were matched by age, sex, ethnicity, disease, and BP use, including type, dose, and duration. For inclusion in this study, BRONJ diagnosis had to be established by standard clinical and radiographic protocol as per the American Association of Oral and Maxillofacial Surgeons diagnostic criteria.3 All patients in the study were diagnosed by one of the study investigators (P.P.S.). Male and female adult patients with a history of intravenous or oral BP therapy for ≥1 year were recruited for this study, and stage I-III lesions of BRONJ were used for study inclusion to ensure that all patients had bone exposed in the oral cavity for >8 weeks as per the definition of BRONJ. To minimize confounding variables or effect modifiers, patients were excluded from the study if they: 1) had active cancer or recent cancer (<3 year history); 2) had salivary gland hypofunction or xerostomia as determined by clinical criteria17 regardless of the underlying pathology or medication associated with these conditions; 3) had any oral or gastrointestinal pathology, such as self-reported active reflux or reflux history, or mucositis determined by clinical examination, which could potentially alter oral microbial community composition; 4) had an active oral or systemic infection; 5) had received any local or systemic antimicrobial therapy for any condition within 6 months; or 6) were at ≥1 year of BP therapy discontinuation.
Standardized documentation was also obtained for all patients for history of present illness, medication, medical and dental history, and review of systems. In addition, lesion parameters, such as stage, size, location, morphology, duration, and visual analog scale pain (0–10) for BRONJ patients were also measured to ensure thorough and consistent clinical data collection. A head and neck exam and panoramic radiographic imaging were performed and documented for all patients as part of routine clinical protocol. After collection, all saliva samples were assigned a coded number to be used for the specimen from that point on to blind future investigation and remove any unique identifiers associated with each patient.
Salivary flow rates vary significantly among individuals and in the same individual under different conditions.18 Therefore, patients were instructed not to eat, clean, or rinse their mouth 1 hour before saliva collection, because these activities can affect the microbial environment. Saliva for all study subjects was collected at the same time by a standardized draining method as described previously,19 with patients sitting and in the upright head position. Saliva was collected for 5 minutes in a Proflow sialometer (Amityville, NY) or until ≥0.5 mL of saliva was collected (0.1 mL/min average). Samples were immediately transfered to sterile chilled plastic tubes with 10% glycerol added and then frozen with liquid nitrogen and stored in an −80°C freezer until being transported to the J. Craig Venter Institute in San Diego for further analysis. Before analysis, the saliva samples were slowly thawed, combined with a nutrient-depleted Todd-Hewitt (TH) buffer, and mechanically homogenized. Density-gradient centrifugation through Nycodenz (Axis-Shield, Oslo, Norway) was used to separate microbial cells (collected from the interface) and viruses (collected from the supernatant). The virus fraction was passed through a series of filters of decreasing pore size (1.2, 0.8, and 0.2 µm) to remove residual cellular contamination, and the flow-through was collected for final purification and microscopic enumeration. A fraction of the microbial cells was reserved for microscopic enumeration and the remainder was passed through the same series of filters that were used for virus purification. The 0.22-µm filters were reserved for microbial community DNA extraction. Saliva samples were used to create anaerobic biofilm starter cultures with TH media, ascorbic acid, and cysteine HCl. Starter cultures were incubated at 37°C overnight under anaerobic conditions to mimic in vivo conditions because the organisms associated with BRONJ identified to date are predominantly anaerobes or facultative anaerobes in areas of sequestrum with little blood supply or oxygen. Cultures were mixed, transfered to separate 12-well plates and incubated under anaerobic conditions. Media was refreshed until mature biofilms formed (~5 days). Microbial cells and viruses were extracted from biofilms as described for the saliva samples.
Prophage induction experiments
Prophage induction experiments were conducted to determine the occurrence of lysogeny in saliva and biofilms, for both normal individuals and those with BRONJ. For the BRONJ samples, the mutagen mitomycin C was added to saliva diluted in nutrient-depleted TH buffer to a final concentration of 0.5 µg/mL. Control samples had no mitomycin C treatment. Control and BRONJ samples were incubated overnight in the dark at 37°C. Induction of biofilms was performed in the same manner with the following exceptions: 1) mitomycin C was added to TH media and was applied evenly to the top of the biofilms; and 2) incubations were carried out under anaerobic conditions. Viruses were extracted from BRONJ and control samples as described above.
16S rRNA PCR and 454 pyrosequencing
One control sample and 1 BRONJ sample were selected for sequence-based analysis. Microbial DNA was extracted from the 0.2-µm filters as described previously20 from 1 control sample and 1 BRONJ sample for the following subsamples: ambient saliva, BRONJ saliva, control saliva, ambient biofilm, BRONJ biofilm, and control biofilm. Ambient refers to populations of naturally occurring microbes or viruses in unaltered samples of saliva and biofilms regardless of disease status. The 16S V3V5 variable region was simultaneously amplified and barcoded according to the National Institutes of Health Human Microbiome Project protocol. A total of 12 16S amplicons of both normal and BRONJ subsamples were pooled with barcoded viral metagenomes and sequenced with 454 GS FLX titanium pyrosequencing.
Purification of viral particles
Viral fractions which flowed through 0.2-µm filters were then passed through a 37% sucrose cushion to separate viral particles from residual cellular material. We have found that pelleting viruses through sucrose, rather than purification via CsCl-gradient centrifugation, dramatically increases the recovery of intact viruses and therefore total DNA yield.21 Pelleted viruses were resuspended in Tris-EDTA buffer.
Enumeration of microbes and viruses
Microbial cells and viruses were filtered onto 0.02-µm aluminum oxide filters, stained with Sybr Green and enumerated with the use of epifluorescence microscopy (Zeiss LSM710 microscope; Carl Zeiss Microscopy, Thornwood, NY) as previously described.22
Analysis of 16S rRNA sequence data
The Chimera Slayer program23 was used to identify chimeric 16S sequences, which were subsequently removed. Taxonomic assignments (genus level) were made to the remaining sequences using the Ribosomal Database Project (RDP) classifier.24
Statistical analysis
Statistical analysis was performed with SPSS statistical software version 20.0 (IBM, Armonk, NY). Frequency tables were generated where appropriate and distribution assessed. Mean values, standard deviations, and standard errors of the means were calculated for applicable data. Paired t test was used for assessment of statistical significance between groups when data being compared had a parametric distribution. Statistical significance was accepted at P values of <.01.
RESULTS
Clinical design and extraction and enumeration of microbes from saliva and saliva-induced biofilms
Table I summarizes the key demographic and clinico-pathologic features of the study patients. The mean age of patients in the study was 77 years old, and there was a 3:2 female-to-male ratio. All patients were white, and BP use among patients included oral alendronate for osteoporosis in the women or intravenous zoledronic acid for prostate cancer among the men. For clarification, the men with prostate cancer in our study were all >3 years cancer free, so they met the first exclusion criteria, and no patients had active periodontal or odontogenic infection as per the fourth exclusion criteria. All stages of BRONJ were represented, and all cases of BRONJ affected the posterior mandible, which is consistent with the literature as the most common anatomic site of involvement. In the BRONJ group, 2 patients were edentulous, and the average number of teeth in each patient in this group was 14 (range 0–25), compared with the control group where there were no edentulous patients and the average number of teeth in each patient was 22 (range 8–26). Epifluorescence counts were obtained for viruses and microbial cells for saliva samples and for viruses only for biofilm samples (Table II). A greater quantity of microbes and viruses were present in the normal saliva samples than in BRONJ samples, and in general viral abundance increased with biofilm formation. Viral abundance in the normal and BRONJ saliva samples were ~1 order of magnitude greater than microbial cell abundance (P < .01).
Table I.
Demographic and clinicopathologic features of study patients
| Patient | Age/sex/race | Disease, site | BP route, dose, duration | Reason for BP use |
|---|---|---|---|---|
| 1 | 80/F/W | ONJ stage I, left posterior mandible | Alendronate orally, 70 mg/wk, 12 mo | Osteoporosis |
| 2 | 75/M/W | ONJ stage II, left posterior mandible | Zoledronic acid IV, 4 mg/mo, 18 mo | Prostate cancer |
| 3 | 80/F/W | ONJ stage I, right posterior mandible | Alendronate orally, 70 mg/wk, 12 mo | Osteoporosis |
| 4 | 74/M/W | ONJ stage II, left posterior mandible | Zoledronic acid IV, 4 mg/mo, 36 mo | Prostate cancer |
| 5 | 75/F/W | ONJ stage III, right posterior mandible | Alendronate orally, 70 mg/wk, 36 mo | Osteoporosis |
| 6 | 81/F/W | Non-ONJ control | Alendronate orally, 70 mg/wk, 12 mo | Osteoporosis |
| 7 | 77/M/W | Non-ONJ control | Zoledronic acid IV, 4 mg/mo, 16 mo | Prostate cancer |
| 8 | 82/F/W | Non-ONJ control | Alendronate orally, 70 mg/wk, 13 mo | Osteoporosis |
| 9 | 71/M/W | Non-ONJ control | Zoledronic acid IV, 4 mg/mo, 35 mo | Prostate cancer |
| 10 | 76/F/W | Non-ONJ control | Alendronate orally, 70 mg/wk, 38 mo | Osteoporosis |
Table II.
Abundance of microbes and viruses in saliva and biofilm samples
| Sample type | Avg. microbes | Avg. viruses | Avg. PI BRONJ (V) | Avg. PI Control* (V) |
|---|---|---|---|---|
| Normal saliva | 1.4 × 107 | 1.5 × 108 | 1.2 × 108 | 1.6 × 108 |
| BRONJ saliva | 8.2 × 106 | 7.2 × 107 | 7.6 × 108 | 1.1 × 1010 |
| Normal biofilm | NA | 7.1 × 108 | 7.1 × 1010 | 1.6 × 1010 |
| BRONJ biofilm | NA | 8.1 × 108 | 5.3 × 107 | 1.6 × 107 |
PI, Prophage induction; V, viruses.
Non–mitomycin C.
Prophage induction experiments
Stronger prophage induction responses were observed in the biofilm samples than in the planktonic saliva samples, as evidenced by larger increases in viral abundance (Table II). Viral abundance decreased in the mitomycin C–treated BRONJ saliva samples compared with control samples (P < .01), and the reverse trend was observed for BRONJ biofilm samples, though not statistically significant.
Microbial diversity
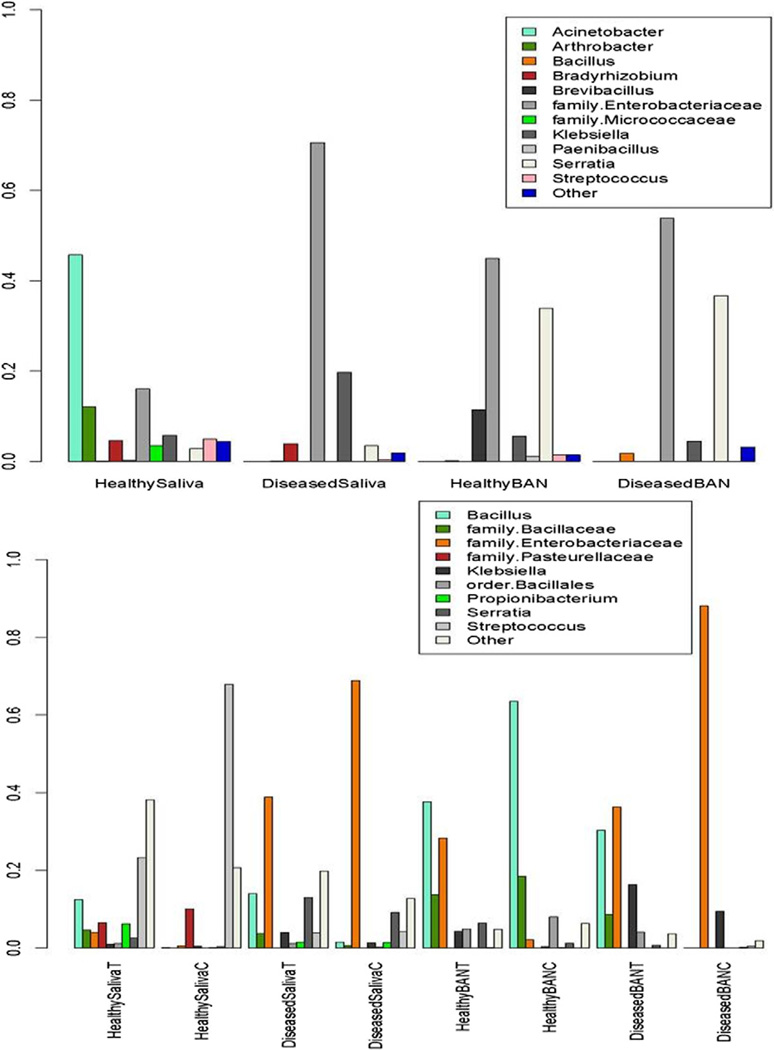
A total of 31,722 chimera-free 16S reads were produced, and taxonomic classifications were assigned to the reads using the RDP classifier (Figure 1). Not all of the 16S sequences were assigned taxonomy to the genus level (with high confidence) by RDP, and such sequences were assigned to their most resolved taxonomic level (e.g., family Bacillaceae). Three phyla—Proteobacteria (70%), Firmicutes (26.9%), and Actinobacteria (1.95%)—dominated all samples and accounted for almost 99% of the total data. In the Proteobacteria phylum, the major genera representations were from Klebsiella and Serratia, both of them members of the family Enterobacteriaceae and facultative anaerobes. In the Firmicutes phylum, the major representations were from Streptococcus (family Streptococcaceae) and Bacillus (family Bacillaceae). The number and proportion of the various taxonomic groups across the samples also indicate a higher microbial diversity in saliva compared with biofilms. Many of the dominant phyla and genera seen in these data have been observed previously in microbiome studies of the oral cavity and saliva,25,26 though at different abundance levels. Although Serratia has not been previously observed as a dominant component of oral microflora, it is an opportunistic pathogen and members of this group have been isolated from subgingival biofilms.27
Fig. 1.
Proportions of various taxonomic groups across the ambient samples (top graph) and mitomycin C control/BRONJ samples (bottom graph) for control and BRONJ saliva and biofilms. BAN, Biofilms; BANT, BRONJ biofilms; BANC, control biofilms. Only groups that accounted for ≥1% of the data in ambient samples and ≥5% of the data in control/BRONJ samples are shown. The category “other” denotes the remainder of the data in each case.
DISCUSSION
To date, the paucity of investigation into the role of oral microbes in the pathogenesis of BRONJ has hindered accurate disease characterization, treatment, and prognostication. Furthermore, research on the infectious aspects of jaw diseases in general has focused on the internal development and the pathogenicity of dental biofilms, and relatively little attention has been given to the source of the biofilm microorganisms, such as saliva.28 Oral bacteria exist in a planktonic state in saliva before colonizing oral and dental surfaces as biofilms, such as exposed bone and soft tissue surfaces in patients with BRONJ. Therefore, a better understanding of salivary pathogens and saliva-derived biofilms could lead to new approaches for diagnosing and managing oral infectious diseases.28 Human viruses are also frequent inhabitants of the mouth, yet until now no information existed on the presence or the role that viruses may play in BRONJ despite the fact that viruses are known to heavily influence gene transfer among microbes, virulence, and the population pathobiology of microbial biofilms.
In this preliminary study we found that individuals with BRONJ harbored different microbial assemblages than nonaffected patients and that ambient viral abundance increased substantially with biofilm formation. This trend is often observed in other natural environments.29 The finding of Proteobacteria, Firmicutes, and Actinobacteria being the dominant phylotypes found in patients is consistent with recent in vivo molecular findings of affected bone and soft tissue specimens in BRONJ patients.30,31 This suggests that saliva may be one source for bone colonization and infection in patients with BRONJ in addition to other possible sources such as odontogenic or periradicular infections.32 Further, planktonic salivary diversity may be influenced by the microbial niche of the dentition. We found that BRONJ patients had fewer teeth on average and some were edentate compared with the control group. Because patients with teeth have a different microbial flora than edentulous individuals, this could influence microbial findings. However, the small size of the patient population, the nature, and the scope of the present study does not allow for any statistical analysis or meaningful conclusions in this respect.
Our results indicate that the majority of microbes in affected patients are facultative anaerobes, making them ideal organisms for surviving in oxygen-depleted areas of necrotic bone that lack adequate blood supply, which is a hallmark of BRONJ. These findings provide insight into targeted antimicrobial therapy and indicate that qualitative rather than quantitative changes in community composition are most related to disease or disease-free status, and that fundamental changes in microbial composition, community subsets, and viral assemblages occur when host cells shift from a primarily planktonic (saliva) to a surface (biofilm) lifestyle.
A possible limitation of our study is the number of 16S reads that were obtained, averaging ~3,000 per sample. As a reference, the National Institutes of Health’s Human Genome Project has been generating similar reads per samples from various human micro-biome samples, including the oral cavity, and investigators have shown that 1,000 sequences per sample is sufficient to address the taxonomy question.33 Additionally, metagenomic identification of bacteria in general determines the presence of specific DNA but not viability; because bacterial DNA can remain PCR-detectable for up to 1 year after cell death, metagenomics may overestimate current bacterial load.34 Further, a single microbiologic examination can not determine if specific bacteria are permanent or transient residents or are primary pathogens or merely bystanders to disease. Laboratory contamination or contamination by nonhuman bacteria is also a potential problem in studies such as this one, which we sought to minimize by our methodology. Our results of the types of bacteria present suggest low contamination because they are consistent with recent reports of identified oral species in a similar context.30,31 However, the lack of detection of any obligate anaerobes in our study, in contrast to those earlier studies, is a limitation and is likely due to the nature of our study, which involved analysis of salivary pathogens and not BRONJ lesions directly. Finally, our results showing differences in microbial and viral abundance or morphotypes between samples or groups may be due to inherent differences in adherence or biofilm formation potential of organisms. Such differences should not be considered to be solely due to other covariates such as disease state.
Metagenomic approaches applied in this study allowed us to identify clinically relevant microbes in patients with BRONJ. Microbial cultures in patients with BRONJ traditionally have not been helpful in directing therapy, because specific pathogens have not been identified and most of the associated biofilm organisms are noncultivable35—thus the need for metagenomic approaches. Importantly, viruses had not been identified or described in this condition before the present study. Their presence may account for some of the virulence and pathobiology of biofilm microbes. The presence of viruses existing as prophage could also account for some of the antibiotic resistance that has been reported in BRONJ-related microbes.31 Collectively, this translational research has overcome critical barriers to progress in this field and informed our understanding of the pathogenesis of BRONJ. Future investigations with larger populations conducted prospectively to correlate metagenomic findings with disease progression and treatment response over time may allow for more accurate diagnostics and prognostication. Continuing in this line of investigation will be critical in the future for informing clinical approaches to disease intervention and the realization of targeted antimicrobial therapeutics for jaw osteonecrosis.
Statement of Clinical Relevance.
There is an urgent need to better understand the pathogenesis of jaw osteonecrosis associated with bisphosphonates (BP) therapy to inform clinical decision making. We apply metagenomic approaches to the study of jaw osteonecrosis, allowing us to identify clinically relevant microbes and viruses.
Acknowledgments
Supported by a Zumberge Award from the University of Southern California and an Loan Repayment Program grant from the National Institute on Minority Health and Health Disparities, National Institutes of Health.
REFERENCES
- 1.Licata AA. Discovery, clinical development, and therapeutic uses of bisphosphonates. Ann Pharmacother. 2005;39:668–677. doi: 10.1345/aph.1E357. [DOI] [PubMed] [Google Scholar]
- 2.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruggiero SL, Fantasia J, Carlson E. Bisphosphonate-related osteonecrosis of the jaw: background and guidelines for diagnosis, staging and management. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:433–441. doi: 10.1016/j.tripleo.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Assouline-Dayan Y, Chang C, Greenspan A, Shoenfeld Y, Gershwin ME. Pathogenesis and natural history of osteonecrosis. Semin Arthritis Rheum. 2002;32:94–124. [PubMed] [Google Scholar]
- 5.Migliorati CA, Epstein JB, Abt E, Berenson JR. Osteonecrosis of the jaw and bisphosphonates in cancer: a narrative review. Nat Rev Endocrinol. 2011;7:34–42. doi: 10.1038/nrendo.2010.195. [DOI] [PubMed] [Google Scholar]
- 6.Sedghizadeh PP, Kumar SK, Gorur A, Schaudinn C, Shuler CF, Costerton JW. Identification of microbial biofilms in osteonecrosis of the jaws secondary to bisphosphonate therapy. J Oral Maxillofac Surg. 2008;66:767–775. doi: 10.1016/j.joms.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 7.Hansen T, Kunkel M, Springer E, Walter C, Weber A, Siegel E. Osteonecrosis of the jaws in patients treated with bisphosphonates—histomorphologic analysis in comparison with infected osteoradionecrosis. J Oral Pathol Med. 2006;35:155–160. doi: 10.1111/j.1600-0714.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 8.Hansen T, Kunkel M, Springer E, Walter C, Weber A, Siegel E, Kirkpatrick CJ. Actinomycosis of the jaws—histopathological study of 45 patients shows significant involvement in bisphosphonate-associated osteonecrosis and infected osteoradionecrosis. Virchows Arch. 2007;451:1009–1017. doi: 10.1007/s00428-007-0516-2. [DOI] [PubMed] [Google Scholar]
- 9.Kos M. Bisphosphonates promote jaw osteonecrosis through facilitating bacterial colonisation. Med Hypotheses. 2011;77:214–215. doi: 10.1016/j.mehy.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Ganguli A, Steward C, Butler SL, Philips GJ, Meikle ST, Lloyd AW, Grant MH. Bacterial adhesion to bisphosphonate coated hydroxyapatite. J Mater Sci Mater Med. 2005;16:283–287. doi: 10.1007/s10856-005-0625-x. [DOI] [PubMed] [Google Scholar]
- 11.Sedghizadeh PP, Kumar SK, Gorur A, Schaudinn C, Shuler CF, Costerton JW. Microbial biofilms in osteomyelitis of the jaw and osteonecrosis of the jaws secondary to bisphosphonate therapy. J Am Dent Assoc. 2009;140:1259–1265. doi: 10.14219/jada.archive.2009.0049. [DOI] [PubMed] [Google Scholar]
- 12.Kumar SK, Gorur A, Schaudinn C, Shuler CF, Costerton JW, Sedghizadeh PP. The role of microbial biofilms in osteonecrosis of the jaw associated with bisphosphonate therapy. Curr Osteoporos Rep. 2010;8:40–48. doi: 10.1007/s11914-010-0008-1. [DOI] [PubMed] [Google Scholar]
- 13.Reid IR. Osteonecrosis of the jaw: who gets it, and why? Bone. 2009;44:4–10. doi: 10.1016/j.bone.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 15.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005;437:41–47. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- 17.Navazesh M. ADA Council on Scientific Affairs and Division of Science. How can oral health care providers determine if patients have dry mouth? J Am Dent Assoc. 2003;134:613–620. doi: 10.14219/jada.archive.2003.0229. [DOI] [PubMed] [Google Scholar]
- 18.Dawes C. Physiological factors affecting salivary flow rate, oral sugar clearance, and the sensation of dry mouth in man. J Dent Res. 1987;66:648–653. doi: 10.1177/00220345870660S107. [DOI] [PubMed] [Google Scholar]
- 19.Navazesh M, Kumar SMS. Measuring salivary flow: challenges and opportunities. J Am Dent Assoc. 2008;139:35S–40S. doi: 10.14219/jada.archive.2008.0353. [DOI] [PubMed] [Google Scholar]
- 20.Allen LZ, Ishoey T, Novotny MA, McLean JS, Lasken RS, Williamson SJ. Single virus genomics: a new tool for virus discovery. PLoS ONE. 2011;23:e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 22.Noble RT, Fuhrman JA. Use of Sybr Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 23.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Human Microbiome Consortium. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbosa FC, Irino K, Carbonell GV, Mayer MP. Characterization of Serratia marcescens isolates from subgingival biofilm, extraoral infections and environment by prodigiosin production, serotyping, and genotyping. Oral Microbiol Immunol. 2006;21:53–60. doi: 10.1111/j.1399-302X.2005.00254.x. [DOI] [PubMed] [Google Scholar]
- 28.Slots J, Slots H. Bacterial and viral pathogens in saliva: disease relationship and infectious risk. Periodontol 2000. 2011;55:48–69. doi: 10.1111/j.1600-0757.2010.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009;11:2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 30.Wei X, Pushalkar S, Estilo C, Wong C, Farooki A, Fornier M, et al. Molecular profiling of oral microbiota in jawbone samples of bisphosphonate-related osteonecrosis of the jaw. Oral Dis. 2012;18:602–612. doi: 10.1111/j.1601-0825.2012.01916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji X, Pushalkar S, Li Y, Glickman R, Fleisher K, Saxena D. Antibiotic effects on bacterial profile in osteonecrosis of the jaw. Oral Dis. 2011;18:85–95. doi: 10.1111/j.1601-0825.2011.01848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saber MH, Alonaizan FA, Schwarzberg K, Kelley ST, Sedghizadeh PP, Levy TA, et al. Bacterial flora of dental periradicular lesions analyzed by the 454-pyrosequencing technology. J Endod. doi: 10.1016/j.joen.2012.06.037. In press. [DOI] [PubMed] [Google Scholar]
- 33.Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young G, Turner S, Davies JK, Sundqvist G, Figdor D. Bacterial DNA persists for extended periods after cell death. J Endod. 2007;33:1417–1420. doi: 10.1016/j.joen.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Fantasia JE. Bisphosphonates-what the dentist needs to know: practical considerations. J Oral Maxillofac Surg. 2009;67:53–60. doi: 10.1016/j.joms.2009.01.011. [DOI] [PubMed] [Google Scholar]