Abstract
Homologous to the E6-associated protein carboxyl terminus domain containing 3 (HECTD3) is an E3 ubiquitin ligase with unknown functions. Here, we show that HECTD3 confers cancer cell resistance to cisplatin. To understand the molecular mechanisms, we performed a yeast two-hybrid analysis and identified mucosa-associated lymphoid tissue 1 (MALT1) as an HECTD3-interacting protein. HECTD3 promotes MALT1 ubiquitination with nondegradative polyubiquitin chains by direct interacting with the MALT1 through its N-terminal destruction of cyclin domain. HECTD3 does not target MALT1 for degradation but stabilize it. HECTD3 depletion dramatically decreases the levels of MALT1 in MCF7 and HeLa cells treated with cisplatin, which is correlated to an increase in apoptosis. Knockdown of MALT1 likewise increases cisplatin-induced apoptosis in these cancer cells. However, HECTD3 over-expression leads to a decreased cisplatin-induced apoptosis, whereas overexpression of MALT1 partially rescues HECTD3 depletion-induced apoptosis. These findings suggest that HECTD3 promotes cell survival through stabilizing MALT1. Our data have important implications in cancer therapy by providing novel molecular targets.
Introduction
Ubiquitination is a posttranslational protein modification involved in regulating a variety of cellular processes, including cell cycle and apoptosis that play key roles in cancer development. Ubiquitin (Ub) chains are assembled in a three-step enzymatic reaction carried out by Ub-activating enzymes (E1), Ub-conjugating enzymes (E2), and Ub-protein ligases (E3). E3 ligases play important roles in cancer development because they control substrate specificity and many have been shown to act as oncoproteins or tumor suppressors because of their genetic and expression alterations in cancer [1]. Ub is conjugated either as a single moiety (mono-Ub) or as poly-Ub chains that are generally linked through K48, K63, or other lysine residues. Different types of poly-Ub chains have different functions: K48-linked poly-Ub chains target substrates for proteasomal degradation, whereas K63-linked poly-Ub chains lead to non-degradative signaling processes [2].
The homologous to the E6-associated protein carboxyl terminus domain containing 3 (HECTD3) E3 ligase contains an HECT domain at the C terminus and a destruction of cyclin (DOC) domain at the N terminus that is responsible for substrate recognition in several E3 ligases such as the anaphase-promoting complex subunit 10 (APC10/DOC1) [3], PARC, CUL7, and HERC2 [4]. Likewise, an N-terminal truncated HECTD3 has been shown to target Trio-associated repeat on actin (Tara) for Ub-mediated degradation [5], and HECTD3 was reported to interact with and ubiquitinate Syntaxin 8 [6]. However, to date, the functions of HECTD3 have not been clearly illustrated.
Mucosa-associated lymphoid tissue 1 (MALT1) is well known to mediate the T cell antigen receptor- and B cell antigen receptor-induced signaling to the transcription factor nuclear factor-kappa B (NF-κB). MALT1 is ubiquitinated by TRAF6 with K63-linked poly-Ub chains, which activates the NF-κB pathway [7]. Additionally, MALT1 can function as a paracaspase to cleave multiple NF-κB inhibitors, including A20 [8], RelB [9], and CYLD [10] in response to T cell antigen receptor signaling. Finally, MALT1 interacts with Caspase-8 and promotes Caspase-8-mediated FLIPL cleavage [11]. These studies suggest that MALT1 may regulate apoptosis.
Here, we demonstrate that HECTD3 promotes cell survival from cisplatin via a direct interaction with MALT1. HECTD3 modifies MALT1 with non-degradative poly-Ub chains and increases protein stability of MALT1. These results suggest that HECTD3 is a pro-survival E3 ligase and provides novel potential therapeutic targets for cancer.
Materials and Methods
Antibodies
The anti-HECTD3 rabbit polyclonal antibody (Ab) was generated using a synthesized peptide from the C terminus of HECTD3 (753CRKLTRFEDFEPSDSR768; Invitrogen, Grand Island, NY). The anti-β-actin mouse monoclonal Ab AC-15 (#A5441), the anti-Flag rabbit polyclonal Ab (#F7425), the anti-PARP (rabbit, #9915), anti-cl-Caspase-7 (rabbit, #9491), anti-Caspase-9 (rabbit, #9502), anti-Caspase-8 (mouse, #9746), and anti-Caspase-3 (rabbit, #9915) Abs are from Cell Signaling (Danvers, MA). Anti-MALT1 (mouse, #sc46677), anti-ERα (rabbit, #7207), and anti-human influenza hemagglutinin (HA) (rabbit, #sc-805) Abs are from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-glutathione S -transferase (GST) (rabbit, #G7781) and anti-Flag M2 monoclonal Ab (mouse, #F3165) were from Sigma (St Louis, MO).
Plasmids
The full-length HECTD3 gene was cloned into the BamHI and SacII sites of the pLenti6/V5-D vector (Invitrogen). To facilitate protein immunoprecipitation (IP), a Flag tag was added to the N terminus of HECTD3 as necessary. The deletion and truncation mutants of Flag-HECTD3 (Δ512–861 and Δ109–393) were generated by a polymerase chain reaction-based approach. The pEBG vector was used to express GST-fused truncated HECTD3 proteins and DD of MALT1. Dr Daniel Krappmann generously provided pEF 3xFlag-MALT1 as a gift (German National Research Center for Environment and Health). HA-Ub and mutants (K0, K48-only, K63-only, K48R, and K63R) were gifts from Dr James Zhijian Chen (University of Texas Southwestern Medical Center).
Cell Culture and Transfection
MCF7 was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% FBS, 0.1 mM NEAA, 1.5 g/l sodium bicarbonate, and 1 mM sodium pyruvate. MDA-MB-231 was cultured in DMEM supplemented with 2 mM l-glutamine, 0.1 mM NEAA, 1.5 g/l sodium bicarbonate, 1 mM sodium pyruvate, and 10% FBS. HeLa and HEK293T cells were cultured in DMEM supplemented with 5% FBS.
Plasmid and siRNA transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. All chemically synthesized siRNA were purchased from Ambion (Austin, TX) and transfected at 25 nM final concentration. The siRNA target sequence for human HECTD3 gene are 5′-GCGGGAACTAGGGTTGAAT-3′ (Hsi#1) and 5′-GGTATTTCACCTCTTAAGA-3′ (Hsi#2). The siRNA target sequences for the human MALT1 gene are 5′-GATCGAGACAGTCAAGATA-3′ (#1) and 5′-GCATTGCCTCTATACCAGA-3′ (#2). The siRNA target sequence for human CASP8 gene is 5′-GATAATCAACGACTATGAA-3′.
Yeast Two-Hybrid Screening
We used the Gal4-based yeast Matchmaker Two-Hybrid Systems to screen the substrates for HECTD3. The N terminus of HECTD3 without the HECT domain (HΔ512–861) was used as bait. The DNA fragment encoding HΔ512–861 was cloned into EcoRI and BamHI sites of pGBK7 vector, and the bait protein was expressed as a fusion to the Gal4 DNA-binding domain. The library of prey proteins was expressed as fusions to the Gal4 activation domain. Screening and validation were performed according to the manufacturer's protocols.
IP and GST Pull-down
Our previous studies described IP using anti-Flag M2-Agarose (A2220; Sigma) and GST pull-down experiments [12]. The antiHECTD3 rabbit polyclonal Ab was used to immunoprecipitate the endogenous HECTD3 protein from HeLa. Rabbit IgG was used as the negative control. The beads were washed five times with 500 µl of 1x cell lysis buffer. Proteins were resuspended with 30 µl of sodium dodecyl sulfate (SDS) sample loading buffer and analyzed by Western blot.
Purification of the HECTD3 DOC Domain and the MALT1 DD
DNA fragments representing the DOC domain of HECTD3 (240–378) was polymerase chain reaction-amplified from the full-length HECTD3 plasmid and inserted into the pGEX-6P-1 between the BamHI and EcoRI sites. The construct was verified by DNA sequencing. For protein expression, Escherichia coli strain Rosetta 2 (DE3) (Novagen, Philadelphia, PA) bearing the expression plasmid were grown at 37°C to 0.8 absorbance of OD600, then induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside at 15°C for 12 hours, and harvested by centrifugation. After sonication of the bacteria, soluble DOC domain was affinity purified through a glutathione Sepharose 4B column (GE HealthCare, Pittsburgh, PA). The GST-DOC fusion protein was digested with the PreScission protease (GE HealthCare) and reloaded to the glutathione Sepharose column to remove the free GST. The flow-through fractions containing the DOC domain was concentrated and further purified through a S200 Superdex column (GE HealthCare). The MALT1 DD was expressed and purified as a His-tag fusion protein as described [13].
BIAcore Analysis
The affinity and kinetic analyses of the interactions between the HECTD3 DOC domain and the DD of MALT1 were determined at 25°C using the surface plasmon resonance (SPR) detections with a BIAcore 3000 System (GE HealthCare). Briefly, MALT1 DD was immobilized (∼460 RU) onto a CM4 sensor chip (GE HealthCare), as described for other systems [14]. The HECTD3 DOC domain (8–0.5 µM) was injected as the analyte sample with two-fold dilutions. Association and dissociation rates (Kon/Koff), as well as the dissociation constant (Kd), were obtained by global fitting of the SPR data from multiple concentrations to a simple 1:1 Langmuir binding model, using the BIA evaluation software 4.1 (GE HealthCare).
MALT1 Ubiquitination
HEK293T cells were transiently transfected with HA-Ub and other plasmids as necessary in six-well plates. Two days after transfection, cells were harvested in 150 µl of SDS lysis buffer (50 mM Tris-Cl, pH 6.8, 1.5% SDS). The samples were boiled for 15 minutes. We diluted 100 µl of protein lysate with 1.2 ml of EBC/BSA buffer (50 mM Tris-Cl, pH6.8, 180 mM NaCl, 0.5% CA 630, 0.5% BSA) and incubated with anti-Flag M2-Agarose overnight at 4°C with rotation. The beads were collected by centrifugation at 10,000g for 30 seconds at 4°C and washed three times with 1 ml of ice-cold EBC/BSA buffer. Proteins were resuspended with 30 µl of SDS sample loading buffer and analyzed by Western blot using the anti-HA Ab.
Apoptosis Measurement
Cells were transfected with the HECTD3 siRNA and control siRNA for 2 days. Different concentrations of cisplatin (Sigma) were added for 24 hours, if applied. The cell viability was measured by the sulforhodamine B (SRB) assay as described in our previous report [15]. Cleaved PARP and/or cleaved Caspase-3/7 protein levels were detected by Western blot.
Statistical Analysis
All experiments were repeated two to three times. Apoptosis experiments were conducted in triplicate. When appropriate, the data were pooled to generate means ± SD and analyzed by t test.
Results
HECTD3 Promotes Cancer Cell Survival from Cisplatin
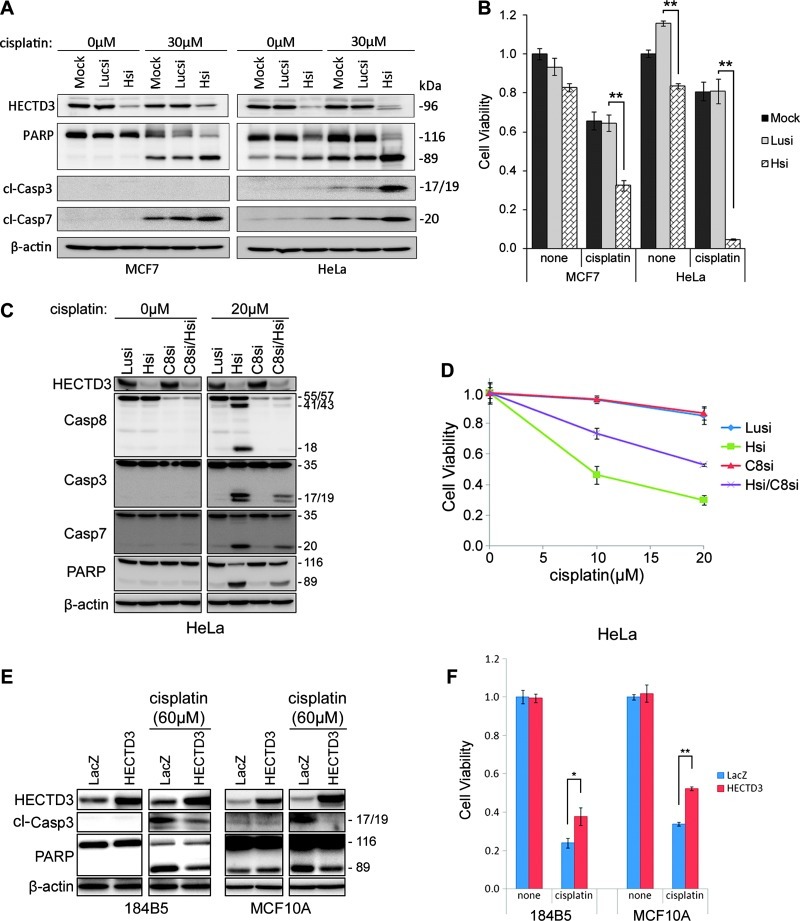
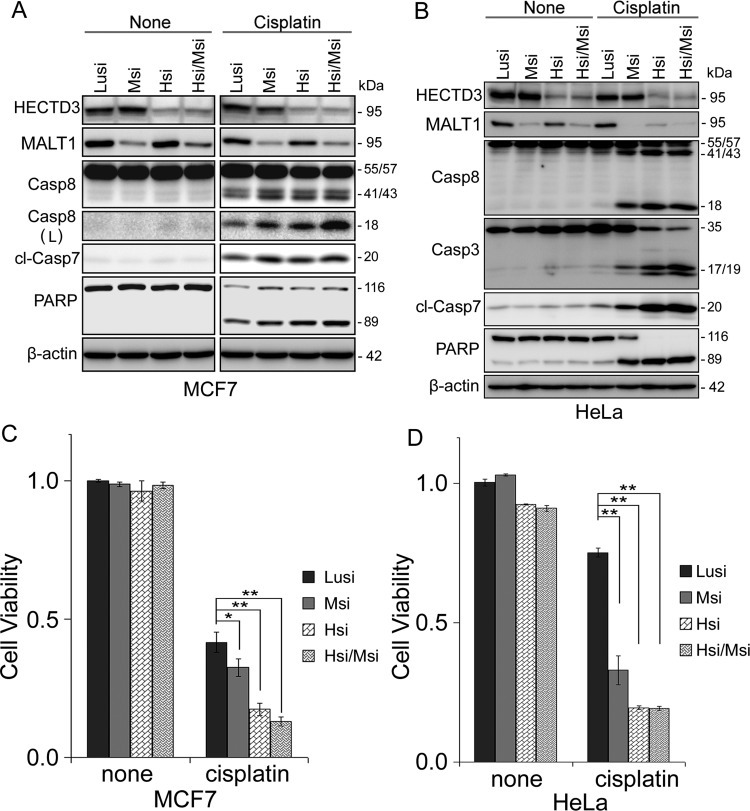
To determine HECTD3′s role in cancer development and therapy, we knocked down endogenous HECTD3 in the MCF7 breast cancer cell line and HeLa and then treated the cells with cisplatin, a common DNA-damaging chemotherapeutic drug. In these cancer cells, HECTD3 depletion dramatically increased the efficacy of cisplatin as determined by PARP/Caspase-3/7 cleavage and cell viability assays (Figure 1, A and B). Similar results were observed in the prostate cancer cell line PC-3 using two different HECTD3 siRNAs (Figure W1, A and B) and the breast cancer cell line MDA-MB-231 using HECTD3 shRNA (Figure W1, C and D). HECTD3 stable depletion also sensitized MDA-MB-231 to cisplatin in severe combined immunodeficiency (SCID) mice (Figure W1E).
Figure 1.
HECTD3 promotes cell survival from cisplatin. (A) HECTD3 knockdown by siRNA sensitized MCF7 and HeLa to cisplatin-induced apoptosis, as detected by the PARP and Caspase-3/7 cleavage. MCF7 is a Caspase-3-deficient cell line [17]. Cisplatin was added for 1 day after cells were transfected with siRNA for 2 days. (B) HECTD3 knockdown by siRNA significantly sensitized MCF7 and HeLa to cisplatin-induced apoptosis, as determined by the loss of cell viability. Cell viability was detected by the SRB assay. **P < .01 (t test). (C) HECTD3 knockdown sensitized HeLa to cisplatin-induced apoptosis in a Caspase-8-dependent manner. Caspase-8 was knocked down by siRNA. Apoptosis was measured by the cleavage of Caspase-8, Caspase-3, Caspase-7, and PARP. (D) HECTD3 knockdown sensitized HeLa to cisplatin-induced apoptosis and was rescued by the depletion of Caspase-8. Cell viability was detected by the SRB assay. (E) HECTD3 overexpression decreased cisplatin-induced apoptosis in 184B5 and MCF10A. HECTD3 was stably overexpressed in 184B5 and MCF10A by lentivirus. Apoptosis was measured by the cleavage of Caspase-3 and PARP. (F) HECTD3 overexpression decreased cisplatin-induced apoptosis in 184B5 and MCF10A. Cell viability was detected by the SRB assay. *P < .05, **P < .01 (t test).
HECTD3 depletion itself also slightly decreased cell viability in HeLa (Figure 1B) and MDA-MB-231 (Figure W1D) and also sensitized HeLa cells to other chemotherapeutic drugs, such as docetaxel (Figure W2, A and B), and γ radiation (Figure W2C). However, HECTD3 depletion did not affect γ radiation-induced foci formation of γ-H2Ax and BRCA1/Rad51 (Figure W2D). These results indicated that radiosensitization activity resulting from HECTD3 depletion is not caused by an impaired homologous recombination, a major mechanism required for repairing DNA damage caused by ionizing radiation.
We found that depletion of Caspase-8 largely blocked cisplatin-induced apoptosis in HECTD3-depleted HeLa cells (Figure 1, C and D). Similar results were observed in MCF7 (Figure W3, A and B). A Caspase-8 inhibitor also rescued apoptosis of both MCF7 and HeLa cells induced by cisplatin and HECTD3 depletion (Figure W3, C and D). We concluded that cisplatin-induced cell apoptosis is executed through activation of Caspase-8.
When HECTD3 was overexpressed in immortalized breast epithelial cell lines 184B5 and MCF10A, cell growth was not affected (Figure 1, E and F). However, after the cells were challenged with cisplatin, the cells with HECTD3 overexpression displayed a significant decrease in cisplatin-induced apoptosis, as detected by the cleavage of Caspase-3 and PARP and the loss of cell viability (Figure 1, E and F).
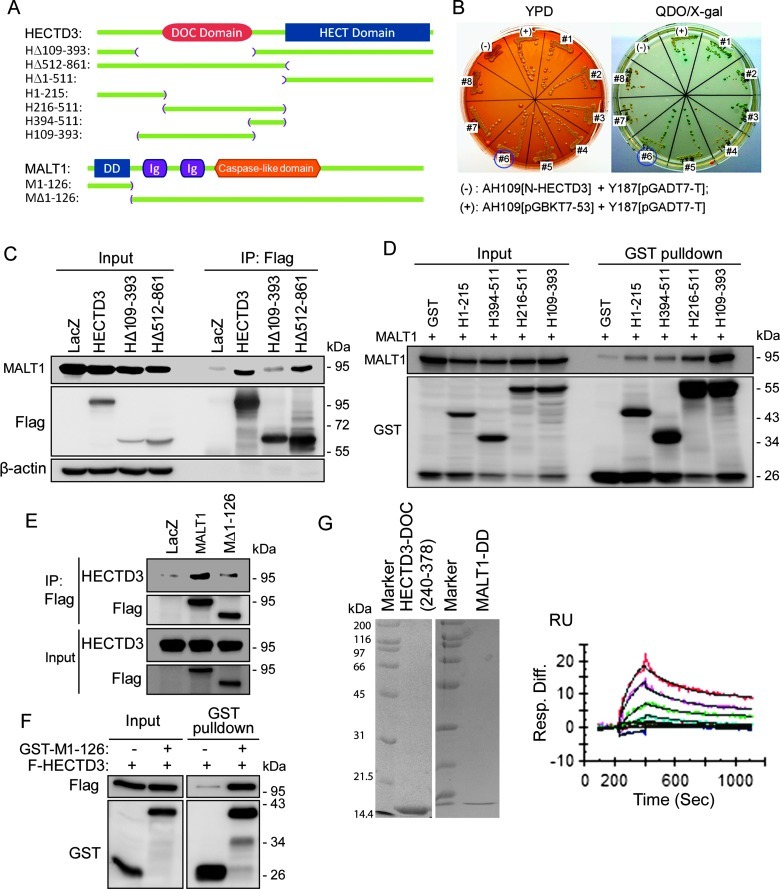
HECTD3 Interacts with MALT1 through DOC/DD
HECTD3 possesses an HECT domain at the C terminus and a DOC domain at the N terminus (Figure 2A), which is the protein interaction domain that may be responsible for recruiting substrates. To identify substrates for HECTD3, we used N-terminal HECTD3 (HΔ512–861) as a bait to screen HECTD3-interacting proteins using the matchmarker yeast two-hybrid (Y2H) system. We identified and validated MALT1 (#6), PON2, SF3B1, and FBP2 as candidate HECTD3-interacting proteins under the most stringent condition (-Ade/-His/-Trp/-Leu/X-α-Gal) (Figure 2B).
Figure 2.
HECTD3 interacts with MALT1 through DOC/DD. (A) Schematic representation of HECTD3 and MALT1 proteins and their truncation mutants. (B) HECTD3 interacts with MALT1 by Y2H. HECTD3Δ512–861 was used as the bait for screening. Eight candidates were confirmed on the plate. The interaction between p53 and the simian virus 40 large T antigen was used as the positive control. The HECTD3Δ512–861 and the simian virus 40 large T antigen were used as the negative control. Yeast was cultured on plates with complete media (YPD) and on plates with QDO (-Ade/-His/-Trp/-Leu) media plus X-α-Gal. All yeast clones grew well in YPD media, but only positive clones grew well in QDO media and are in blue. (C) HECTD3 interacts with MALT1 through the 109–393 region, which contains the DOC domain. Flag-HECTD3 and its mutants were co-transfected with MALT1 into HEK293T as indicated for 2 days, and IP was performed using the anti-Flag M2 beads. MALT1 was probed by the anti-MALT1 Ab. HECTD3Δ110–393 significantly decreased the binding with MALT1 compared to HECTD3. Deletion of the HECT domain in HECTD3 did not affect the protein interaction. LacZ was used as the negative control. (D) The 109–393 region of HECTD3 is sufficient for MALT1 binding as determined by the GST pull-down assay. Different GST-fused HECTD3 fragments were co-expressed with MALT1 in HEK293T and precipitated by glutathione beads. GST alone was used as the negative control. (E) MALT1 interacts with HECTD3 through DD. Flag-MALT1 and Flag-MALT1ΔDD (Δ1–126) were co-transfected with HECTD3 into HEK293T, and IP was performed with anti-Flag M2 beads. Deletion of DD from MALT1 decreased the binding to HECTD3. (F) DD of MALT1 is sufficient for HECTD3 binding. GST-fused MALT1 DD and the GST negative control were co-transfected with Flag-HECTD3 into HEK293T for 2 days. GST pull-down assay was used to detect protein-protein interaction. (G) Binding affinity and kinetic analysis of the interactions between the HECTD3 DOC domain and the MALT1 DD. The concentrations for the injected HECTD3 DOC samples were from 8 to 0.5 µM, with two-fold dilutions. Global fitting of data to a 1:1 binding model is shown in black. The purified recombinant proteins for HECTD3 DOC and MALT1 DD are shown on the left side, as analyzed by SDS-polyacrylamide gel electrophoresis.
The protein-protein interaction between HECTD3 and MALT1 was further confirmed by IP experiments. Flag-HECTD3 and MALT1 were co-transfected into HEK293T cells. When Flag-HECTD3 was immunoprecipitated by anti-Flag Ab, MALT1 was co-immunoprecipitated (Figure 2C). In consistency with the Y2H results, the N-terminal HECTD3 (HΔ512–861) is sufficient for binding to MALT1. When the DOC domain is deleted from HECTD3 (HΔ109–393), the protein interaction was largely abolished (Figure 2C). Additionally, we performed GST pull-down experiments through generation of a series of truncated HECTD3 GST-fusion protein and demonstrated that HECTD3 (109–393) is sufficient for MALT1 interaction (Figure 2D). Together, these results suggest that HECTD3 interacts with MALT1 through the DOC domain.
We further performed reciprocal IP experiments by transfecting Flag-MALT1 and HECTD3 into HEK293T cells. When Flag-MALT1 was immunoprecipitated by anti-Flag Ab, HECTD3 was co-immunoprecipitated (Figure 2E). MALT1 contains a death domain (DD) at its N terminus (Figure 2A), and the MALT1ΔDD (1–126) mutant did not interact with HECTD3 (Figure 2E), suggesting that the DD of MALT1 is essential for interaction with HECTD3. Moreover, DD is also sufficient to bind to HECTD3 as the GST-DD protein pulled down HECTD3, whereas GST alone did not (Figure 2F).
Finally, we expressed and purified recombinant HECTD3-DOC and MALT1-DD from bacteria and tested the direct protein-protein interaction by SPR detection with a BIAcore 3000 System (GE HealthCare). A Kd of 2.1 x 10-6 indicated a strong binding between HECTD3-DOC (240–378) and MALT1-DD (Figure 2G). Taken together, our experiments suggest that MALT1 directly interacts with HECTD3 DOC through DD.
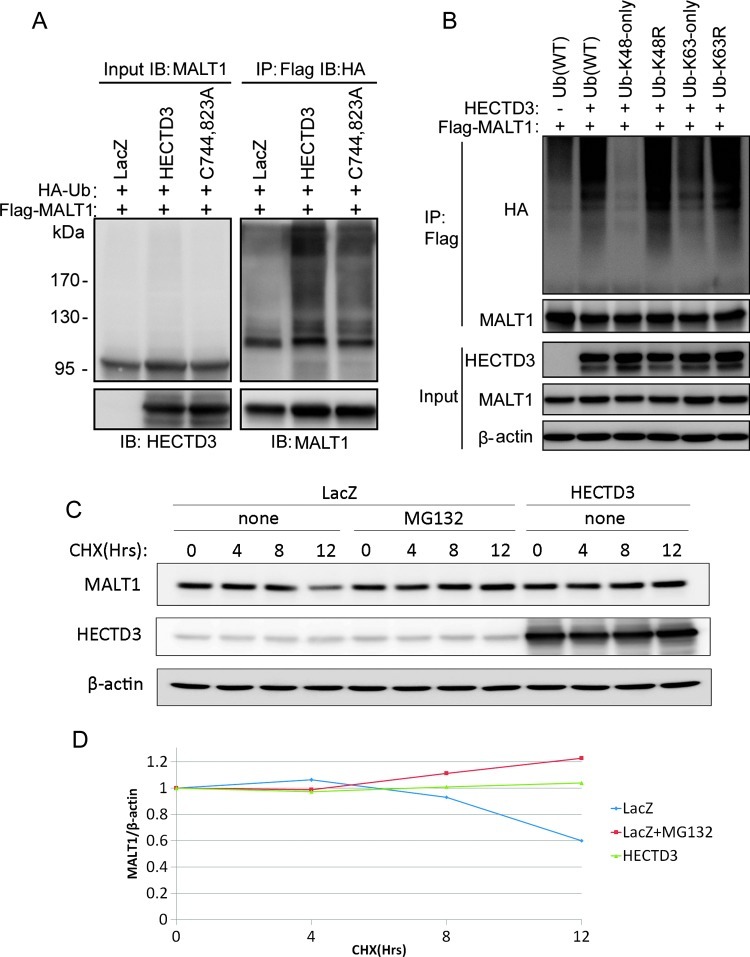
HECTD3 Promotes Non-K48-Linked MALT1 Polyubiquitination
Because the HECTD3 E3 ligase interacts with MALT1, we next determined whether HECTD3 ubiquitinates MALT1. The expression constructs for HECTD3, Flag-MALT1, and HA-Ub were cotransfected into HEK293T cells. Flag-MALT1 was immunoprecipitated with anti-Flag Ab-conjugated M2 beads under a denatured condition. The ubiquitinated MALT1 proteins were detected by Western blot with the anti-HA Ab. As shown in Figure 3A, HECTD3 significantly increased the ubiquitination of MALT1 compared to the vector control. Interestingly, the catalytic inactive HECTD3 mutant (C744, 823A) also increased the MALT1 ubiquitination, although less significant than that by the wild type (WT) HECTD3. These results suggested that HECTD3 may directly ubiquitinate MALT1 and may also facilitate MALT1 ubiquitination by other E3 ligases.
Figure 3.
HECTD3 promotes MALT1 non-K48-linked polyubiquitination and increases the MALT1 protein stability. (A) HECTD3 increased MALT1 ubiquitination in E3 ligase-independent manner in HEK293T cells. HEK293T cells were co-transfected with expressing plasmids for HA-Ub, HECTD3-C744, 823A (a catalytic inactive HECTD3 mutant), and Flag-MALT1 as indicated for 2 days. IP was performed with the anti-Flag M2 beads under a denaturing condition. Western blot was performed with the indicated Abs. (B) HECTD3 promotes MALT1 non-K48-linked polyubiquitination. HA-tagged Ub and mutants (K48 only, K63 only, K48R, and K63R), HECTD3, and Flag-MALT1 were co-transfected into HEK293T. K48-only Ub hardly supported HECTD3-mediated MALT1 ubiquitination. K63-only Ub partially supported HECTD3-mediated MALT1 ubiquitination. Both K48R and K63R Ub did not decrease HECTD3-mediated MALT1 ubiquitination. (C) HECTD3 overexpression decreased the endogenous MALT1 protein degradation in HEK293FT cells, as detected by the CHX chase assay. MG132 blocked the degradation of MALT1. The cells were incubated with 50 µg/ml CHX for different times (4, 8, and 12 hours) and were collected for Western blot. β-actin was used as the control. (D) Quantitative data of C. The protein half-life of MALT1 is more than 12 hours in HEK293T cells. HECTD3 overexpression or MG132 delayed the degradation of MALT1.
To determine the HECTD3-mediated MALT1 poly-Ub chain linkage, we used several Ub mutants in which K48 and K63 residues were substituted with arginine (K48R and K63R), respectively, or in which all K residues were substituted with arginine except K48 and K63 (K48 only and K63 only). As shown in Figure 3B, HECTD3-mediated MALT1 poly-Ub chains are not through K48 because K48-only Ub does not, but K48R Ub supports HECTD3-mediated MALT1 ubiquitination. In addition, HECTD3-mediated MALT1 poly-Ub chains are partially through K63 because both K63-only Ub and K63R Ub partially support MALT1 ubiquitination (Figure 3B). Our results indicated that HECTD3 may also ubiquitinate MALT1 through a yet identified Ub linkage other than K48 and K63, because both K48R and K63R support MALT1 ubiquitination by HECTD3.
HECTD3 Increases the MALT1 Protein Stability
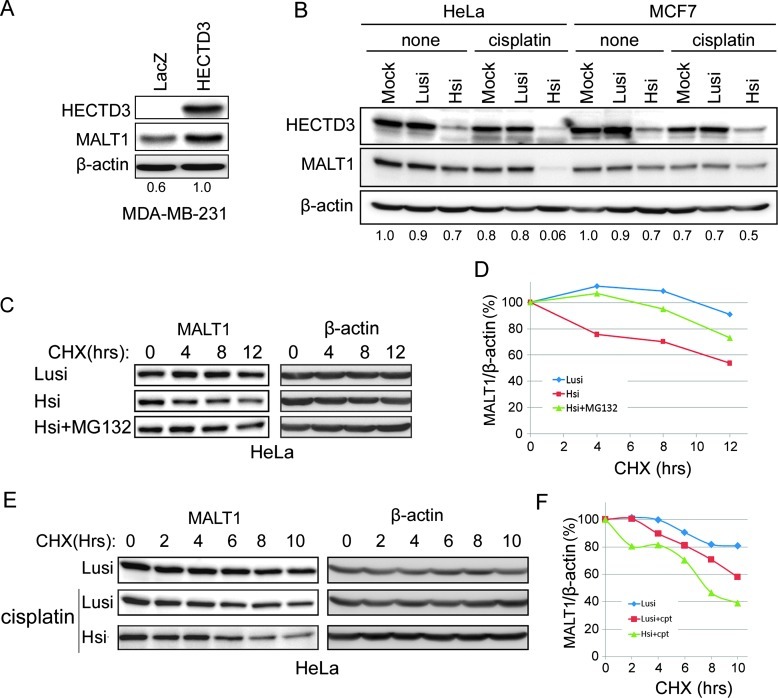
K48-linked polyubiquitination is well known to target substrate proteins for proteasomal degradation. Because HECTD3-mediated MALT1 ubiquitination is not through K48, HECTD3 may not promote MALT1 degradation. Indeed, both endogenous and exogenous MALT1 proteins showed a long protein half-life (>12 hours) in HEK293T cells, as determined by the cycloheximide (CHX) chase assays (Figures 3, C and D, and W4A). MG132, a proteasome inhibitor, can protect MALT1 from degradation. Surprisingly, HECTD3 increased the MALT1 protein stability in HEK293T cells (Figures 3, C and D, and W4A) and HECTD3 overexpression also increased the steady level of MALT1 in MDA-MB-231 cells (Figure 4A).
Figure 4.
HECTD3 increases the MALT1 protein stability. (A) Overexpression of HECTD3 increased the endogenous MALT1 protein level in MDA-MB-231. The ratios between MALT1 and β-actin are shown below the panel. (B) Knockdown of HECTD3 in both HeLa and MCF7 decreased the endogenous protein levels of MALT1, especially when the cells were treated with cisplatin. The ratios between MALT1 and β-actin are shown below the panel. (C) Knockdown of HECTD3 decreased the protein half-life of MALT1, as measured by CHX chase assays. HeLa cells were transfected with anti-HECTD3 and luciferase control siRNAs for 2 days. MG132 partially rescued the HECTD3 depletion-induced MALT1 degradation. (D) Quantitative results of C by the ImageJ software. The β-actin-normalized MALT1 protein levels at 0 hour were defined as 100% for each panel. (E) Cisplatin decreased the protein half-life of MALT1. HeLa cells were transfected with HECTD3 and control siRNAs for 2 days. The cells were incubated with 30 µM cisplatin and 50 µg/ml CHX for different times (2–10 hours) and were harvested for Western blot. (F) Quantitative results of E by the ImageJ software. The β-actin-normalized MALT1 protein levels at 0 hour were defined as 100% for each panel.
To investigate whether endogenous HECTD3 regulates the protein stability of MALT1, we knocked down HECTD3 in both HeLa and MCF7 and treated the cells with cisplatin. Knockdown of HECTD3 modestly decreased the protein levels of MALT1 in both cell lines, and the decrease became very dramatic in the presence of cisplatin (Figure 4B). CHX chase assays were again performed to measure the MALT1 protein half-lives. Depletion of HECTD3 in HeLa cells increased the MALT1 protein proteasomal degradation that was blocked by MG132 (Figure 4, C and D). Interestingly, cisplatin treatment also decreased the MALT1 protein half-life in HeLa (Figure 4, E and F). Depletion of HECTD3 in combination with cisplatin further shortened the MALT1 protein half-life in HeLa (Figure 4, E and F). Taken together, our results indicated that HECTD3-MALT1 interactions stabilize MALT1 in cells.
MALT1 Promotes Cancer Cell Survival from Cisplatin
The function of MALT1 in cancer chemotherapeutic drug resistance has not been previously tested. We knocked down MALT1 in MCF7 by two different siRNAs, treated the cells with cisplatin, and examined apoptosis by Caspase-8 and PARP cleavage and cell viability. As a result, knockdown of MALT1 sensitized MCF7 to cisplatin (Figure W4B). As shown in Figure 5, A and B, MALT1 depletion partially mimicked the results of HECTD3 depletion in both MCF7 and HeLa. Knockdown of either HECTD3 or MALT1 increased the cleavage of Caspase-8, Caspase-3/7, and PARP in the presence of cisplatin. The cell viability assays further confirmed these results. Depletion of MALT1 decreased cell viability in the presence of cisplatin in both MCF7 and HeLa (Figure 5, C and D). However, MALT1 depletion showed weaker proapoptotic effects compared to HECTD3 depletion in both cell lines (Figure 5).
Figure 5.
Knockdown of MALT1 sensitizes MCF7 and HeLa to cisplatin. (A) Knockdown of MALT1 by siRNA in MCF7 increased cisplatin-induced apoptosis as detected by Caspase-8/7 and PARP cleavage. HECTD3 siRNA was used as the positive control. (B) Knockdown of MALT1 by siRNA in HeLa increased cisplatin-induced apoptosis as detected by Caspase-8/3/7 and PARP cleavage. (C) Knockdown of MALT1 by siRNA in MCF7 significantly increased cisplatin-induced loss of cell viability as measured by SRB assays. MALT1 is a weaker pro-survival protein compared to HECTD3. (D) Knockdown of MALT1 by siRNA in HeLa significantly increased cisplatin-induced loss of cell viability as measured by SRB assays.
HECTD3 Promotes the Cancer Cell Survival through MALT1
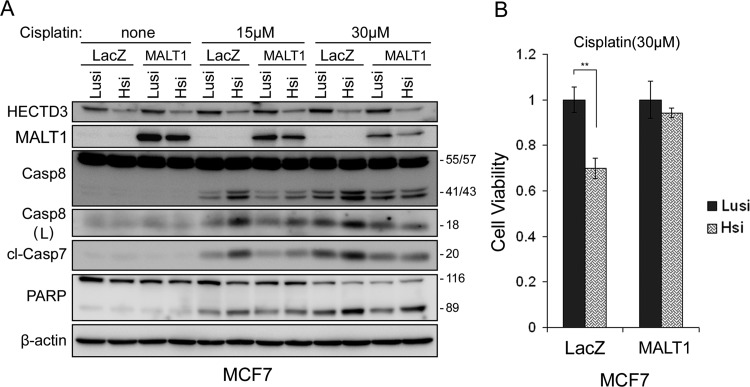
Given that both HECTD3 and MALT1 have pro-survival function and that HECTD3 protects MALT1 from degradation, it seems plausible that HECTD3 functions through MALT1. To test this hypothesis, MALT1 was stably overexpressed in MCF7. Knockdown of HECTD3 decreased the exogenous MALT1 protein levels (Figure 6A). Importantly, overexpression of MALT1 obviously reduced apoptosis (Caspase-8/7 and PARP cleavage) induced by HECTD3 siRNA in combination with cisplatin (Figure 6A). In consistency with this result, the loss of cell viability induced by HECTD3 siRNA and cisplatin was largely rescued by MALT1 overexpression in MCF7 (Figure 6B).
Figure 6.
MALT1 overexpression rescued HECTD3 depletion-induced increase of cisplatin sensitivity in MCF7. (A) Stable MALT1 over-expression decreased Caspase-8/7 and PARP cleavage induced by ablation of HECTD3 and cisplatin. MALT1 was transduced into MCF7 by the lentiviral system. LacZ was used as a negative control. Blasticidin (10 µg/ml)-resistant cell populations were used for siRNA transfection for 2 days and cisplatin treatment for overnight. Western blot was performed with the indicated Abs. (B) Stable MALT1 overexpression decreased loss of cell viability induced by ablation of HECTD3 and cisplatin. Cell viability was measured by SRB assays.
Discussion
HECTD3 is a novel HECT-type E3 ligase that, as we have demonstrated in this study, protects cancer cells from cisplatin-mediated apoptosis. Several lines of evidence support the notion that HECTD3 functions partially through MALT1: 1) HECTD3 interacts with MALT1 as determined by Y2H, CO-IP, GST pull-down, and SPR experiments; 2) HECTD3 promoted MALT1 non-degradative polyubiquitination and increased the protein stability of MALT1, especially in the presence of cisplatin; 3) the knockdown of either HECTD3 or MALT1 sensitized cancer cells to cisplatin; and 4) overexpression of MALT1 partially rescued the apoptosis induced by HECTD3 ablation in the presence of cisplatin.
HECTD3 has been shown to ubiquitinate Tara [5] and Syntaxin-8 [6]. However, neither Tara nor Syntaxin-8 has been implicated in drug resistance in cancer and the function of HECTD3 in cancer cells in response to chemotherapeutic drugs is largely unknown. Here, we demonstrated that HECTD3 depletion in multiple cancer cell lines, such as HeLa, MCF7, MDA-MB-231, and PC-3, increased the efficacy of cisplatin (Figures 1 and W1). Furthermore, HECTD3 depletion sensitized HeLa to docetaxel and γ irradiation (Figure W2). These results suggest that HECTD3 may be a potential therapeutic target for cancer combination therapy.
The increased sensitivity to cisplatin by HECTD3 depletion depends on Caspase-8 because Caspase-8 depletion or inhibition abrogated the effect of HECTD3 depletion in apoptosis in HeLa and MCF7 (Figures 1 and W3). In our study, MALT1 was identified as an HECTD3-interacting protein (Figure 2). MALT1 is well known to play a key role in NF-κB signaling in response to T cell or B cell receptor signaling and MALT lymphoma [16]. Recurrent chromosomal translocation t(11,18)(q21;q21) in MALT lymphoma leads to the production of a fusion oncoprotein, API2-MALT1, which constitutively and strongly-actives NF-κB [16]. MALT1 is ubiquitinated by TRAF6 with K63-linked poly-Ub chains that provide docking surfaces for IKKγ/NEMO and trigger NF-κB signaling [7]. Additionally, MALT1 interacts with Caspase-8 and facilitates Caspase-8-mediated c-FLIPL cleavage [11]. There is no data available, however, on the function of MALT1 in solid tumors. While we found that MALT1 also promotes cancer cell survival from cisplatin in MCF7 and HeLa (Figures 5 and 6), the functional mechanism of MALT1 in solid tumors needs further investigations.
We also demonstrated that HECTD3 promotes MALT1 non-K48-linked polyubiquitination (Figure 3). Interestingly, the catalytic inactive HECTD3 also increased MALT1 ubiquitination, suggesting that MALT1 may not be a direct substrate of HECTD3. Potentially, HECTD3 promotes MALT1 ubiquitination by other E3s. TRAF6 is an E3 ligase that mediates MALT1 poly-Ub with K63-linked chains [7]. Although we have not directly tested this hypothesis, we observed that MALT1 poly-Ub chains mediated by HECTD3 appear to contain both K63-linked and non-K63-linked. These results indicate that HECTD3 may 1) directly ubiquitinate MALT1, 2) facilitate MALT1 ubiquitination by TRAF6, and/or 3) facilitate MALT1 ubiquitination by other E3s in addition to TRAF6. Our data suggest that MALT1 degradation depends on the proteasome system because the proteasome inhibitor MG132 blocked the degradation of MALT1. It is very likely that HECTD3-mediated non-K48-linked polyubiquitination competes with K48-linked polyubiquitination in MALT1 and, therefore, protects MALT1 from proteasomal degradation.
Nevertheless, HECTD3 increases the MALT1 protein stability by preventing its proteasomal degradation. MALT1 is actually a very stable protein with a long half-life; however, cisplatin significantly decreased MALT1 stability, especially in the absence of HECTD3 (Figure 4), although the specific mechanism by which cisplatin triggers MALT1 degradation remains unclear.
In summary, we have identified a new function of HECTD3 in chemotherapeutic drug cisplatin-induced apoptosis. By forming a complex with MALT1, HECTD3 promotes MALT1 non-degradative polyubiquitination and increases MALT1 protein stability, which conferred cisplatin resistance. Our results suggest that HECTD3 and MALT1 are novel pro-survival proteins, and inhibition of HECTD3 or MALT1 may be promising strategies for future cancer therapy.
Supplementary Material
Acknowledgments
We are grateful to Daniel Krappmann of the German National Research Center for Environment and Health for providing Flag-MALT1 constructs and James Zhijian Chen from the University of Texas Southwestern Medical Center for providing the wild-type and mutant HA-Ub constructs. We also thank the Immunology Core at the Wadsworth Center for the BIAcore measurement.
Abbreviations
- CHX
cycloheximide
- DD
death domain
- DOC
destruction of cyclin
- HECT
homologous to the E6-associated protein carboxyl terminus
- HECTD3
HECT domain containing 3
- IP
immunoprecipitation
- MALT
mucosa-associated lymphoid tissue
- SPR
surface plasmon resonance
- SRB
sulforhodamine B
Footnotes
This study was supported by the National Key Basic Research Program of China (No. 2013CB910900), National Nature Science Foundation of China (81072162, 81120108019, and U1132605), Top Talents Program of Yunnan Province, China (2010CI114) and National Cancer Institute (R01CA154625) to Junran Zhang.
This article refers to supplementary materials, which are designated by Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of e3 ubiquitin ligases in human breast cancer. Mol Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 2.Woelk T, Sigismund S, Penengo L, Polo S. The ubiquitination code: asignalling problem. Cell Div. 2007;2:11. doi: 10.1186/1747-1028-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pal M, Varga K, Nagy O, Deak P. Characterization of the Apc10/Doc1 subunit of the anaphase promoting complex in Drosophila melanogaster. Acta Biol Hung. 2007;58(suppl):51–64. doi: 10.1556/ABiol.58.2007.Suppl.5. [DOI] [PubMed] [Google Scholar]
- 4.Kaustov L, Lukin J, Lemak A, Duan S, Ho M, Doherty R, Penn LZ, Arrowsmith CH. The conserved CPH domains of Cul7 and PARC are protein-protein interaction modules that bind the tetramerization domain of p53. J Biol Chem. 2007;282:11300–11307. doi: 10.1074/jbc.M611297200. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Lan J, Zhu Y, Li X, Lai X, Xue Y, Jin C, Huang H. The E3 ubiquitin ligase HECTD3 regulates ubiquitination and degradation of Tara. Biochem Biophys Res Commun. 2008;367:805–812. doi: 10.1016/j.bbrc.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Kang L, Bond W, Zhang N. Interaction between syntaxin 8 and HECTd3, a HECT domain ligase. Cell Mol Neurobiol. 2009;29:115–121. doi: 10.1007/s10571-008-9303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D. Malt1 ubiquitination triggers NF-κB signaling upon T-cell activation. EMBO J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 9.Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, Guzzardi M, Decaillet C, Grau M, Dorken B, et al. Malt1-dependent RelB cleavage promotes canonical NF-κB activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci USA. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P, Gevaert K, Beyaert R. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawadler H, Gantz MA, Riley JL, Yang X. The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol Cell. 2008;31:415–421. doi: 10.1016/j.molcel.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15:1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 13.Qiu L, Dhe-Paganon S. Oligomeric structure of the MALT1 tandem Ig-like domains. PLoS One. 2011;6:e23220. doi: 10.1371/journal.pone.0023220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Zhou Y, Guo Y, Li Z, Eisele L, Mourad W. Zinc induces dimerization of the class II major histocompatibility complex molecule that leads to cooperative binding to a superantigen. J Biol Chem. 2007;282:5991–6000. doi: 10.1074/jbc.M608482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121:2834–2841. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 16.Hosokawa Y. Anti-apoptotic action of API2-MALT1 fusion protein involved in t(11;18)(q21;q21) MALT lymphoma. Apoptosis. 2005;10:25–34. doi: 10.1007/s10495-005-6059-6. [DOI] [PubMed] [Google Scholar]
- 17.Blanc C, Deveraux QL, Krajewski S, Janicke RU, Porter AG, Reed JC, Jaggi R, Marti A. Caspase-3 is essential for procaspase-9 processing and cisplatin-induced apoptosis of MCF-7 breast cancer cells. Cancer Res. 2000;60:4386–4390. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.