Abstract
The hepatocyte growth factor receptor (c-Met) and a constitutively active mutant of the epidermal growth factor receptor (ΔEGFR/EGFRvIII) are frequently overexpressed in glioblastoma (GBM) and promote tumorigenesis. The mechanisms underlying elevated hepatocyte growth factor (HGF) production in GBM are not understood. We found higher, coordinated mRNA expression levels of HGF and c-Met in mesenchymal (Mes) GBMs, a subtype associated with poor treatment response and shorter overall survival. In an HGF/c-Met-dependent GBM cell line, HGF expression declined upon silencing of c-Met using RNAi or by inhibiting its activity with SU11274. Silencing c-Met decreased anchorage-independent colony formation and increased the survival of mice bearing intracranial GBM xenografts. Consistent with these findings, c-Met activation by ΔEGFR also elevated HGF expression, and the inhibition of ΔEGFR with AG1478 reduced HGF levels. Interestingly, c-Met expression was required for ΔEGFR-mediated HGF production, anchorage-independent growth, and in vivo tumorigenicity, suggesting that these pathways are coupled. Using an unbiased mass spectrometry-based screen, we show that signal transducer and activator of transcription 3 (STAT3) Y705 is a downstream target of c-Met signaling. Suppression of STAT3 phosphorylation with WP1193 reduced HGF expression in ΔEGFR-expressing GBM cells, whereas constitutively active STAT3 partially rescued HGF expression and colony formation in c-Met knockdown cells expressing ΔEGFR. These results suggest that the c-Met/HGF signaling axis is enhanced by ΔEGFR through increased STAT3-dependent HGF expression and that targeting c-Met in Mes GBMs may be an important strategy for therapy.
Introduction
Hepatocyte growth factor receptor (c-Met), a receptor tyrosine kinase, is typically expressed on epithelial cells and activated in a paracrine manner by its mesenchymal-derived ligand, hepatocyte growth factor (HGF) [1,2]. However in glioblastoma (GBM), which is the most common and aggressive form of adult brain cancer [3], c-Met and HGF are frequently coexpressed and function in an autocrine signaling loop [4–6]. Moreover, the coexpression of c-Met and HGF in GBM accrues with tumor grade [7,8]. When c-Met or HGF are inhibited in vivo, a dramatic reduction in GBM tumor formation and growth occurs, underscoring the importance of the c-Met/HGF axis in GBMs [9]. In addition to activating c-Met, HGF increases the transcription of the c-Met gene in GBM cells [10]. This feed-forward loop of c-Met/HGF dysregulation most likely contributes to c-Met overexpression.
For GBM patients, shorter overall survival is associated with high-level c-Met expression [11], raising the question of how this fits with the recently identified prognostic GBM subtypes that have been identified using gene expression classifiers [3,12]. Of these, the mesenchymal (Mes) GBM subtype is associated with aggressive disease, a poor prognosis [3,13], and chemotherapy resistance [14]. Interestingly, recurrent tumors shift their molecular profiles toward Mes signatures, which include signal transducer and activator of transcription 3 (STAT3) expression [3], a transcription factor required for c-Met signaling and tumorigenesis [15], and prompting our investigation of direct links between GBM subtype and c-Met pathway activity.
Not only is c-Met overexpressed in GBM [5], but it is also often hyperactivated in other cancers [16]. It has been shown that transactivation of c-Met by the epidermal growth factor receptor (EGFR) is an important contributing factor to aberrant c-Met signaling [17–19] and depends on the direct association with active EGFR [20]. In GBMs, approximately 40% of tumors overexpressing wild-type EGFR coexpress a 2- to 7-exon deletion mutant of the EGFR, known as the ΔEGFR or EGFRvIII [21]. This cancer-specific mutant signals constitutively at a low level in a ligand-independent manner, owing to inefficient receptor dimerization [22–24], internalization, and down-regulation [25,26]. ΔEGFR is a key mediator of apoptotic resistance through increased BCL-XL expression [27,28], which significantly enhances the tumorigenicity of GBM cells in vivo [25,28,29]. In the clinical setting, ΔEGFR expression has also been associated with poor patient survival [30–32]. Recent studies have shown that that the phosphorylation of Y1234, a requirement of c-Met activity, is highly responsive to titrated levels of ΔEGFR in glioma cells [33]. Notably, c-Met Y1234 is markedly increased in ΔEGFR-overexpressing cells compared to cells expressing kinase-inactive ΔEGFR, wild-type EGFR, or wild-type EGFR stimulated with EGF [34]. These reports highlight the significance of cross talk between receptor tyrosine kinases as one of the major mechanisms for their dysregulation in cancers [16].
Biologic processes that lead to the deregulation of c-Met expression and activation in tumors have been extensively investigated [16]. However, mechanisms governing aberrant HGF upregulation in GBM have not yet been identified. In our study, we show that c-Met and HGF expression is upregulated and coexpressed in Mes GBMs. We found that ΔEGFR regulates the expression of HGF through c-Met in GBM cells and that c-Met was not only critical for HGF production but also for ΔEGFR-mediated tumorigenicity. Further, we identified STAT3 as one of the downstream modulators of c-Met-mediated HGF expression in GBM cells.
Materials and Methods
Cell Culture
U87 and LN18 human GBM cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured as previously described [34]. MDCK cells [gift from Dr Zhimin Lu, University of Texas MD Anderson Cancer Center (UTMDACC)], human embryonic kidney 293FT (HEK 293FT) cells (gift from Dr Howard Colman, UTMDACC), and GP2-293 cells (Clontech, Mountain View, CA) were cultured in Dulbecco's modified Eagle's medium (10% FBS) at 5% CO2 and 37°C.
Antibodies and Reagents
The following primary antibodies were used: anti-c-Met, anti-pc-Met (Y1234/Y1235); anti-EGFR, anti-pEGFR (Y1173), anti-STAT3, and anti-pSTAT3 (Y705) (Cell Signaling Technology, Danvers, MA); anti-β-actin-HRP (Sigma-Aldrich, St Louis, MO); anti-HGF (R&D Systems, Minneapolis, MN); anti-mouse secondary antibody (Fisher Scientific, Pittsburgh, PA); anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories Inc, West Grove, PA). The following reagents were used: recombinant human HGF (rhHGF; Chemicon, Billerica, MA); SU11274 and AG1478 (Calbiochem, San Diego, CA); hygromycin B, G418, and puromycin (Fisher Scientific); WP1193 (gift from Dr Waldemar Priebe; UTMDACC).
Cell Lysate Preparation and Western Blot Analysis
Cell lysates were prepared as previously described [34]. For Western blot analysis, 20 to 30 µg of protein was separated on 4% to 12% Bis-Tris NuPage gels (Invitrogen, Carlsbad, CA), except for HGF analysis that used 150 to 200 µg of protein lysate.
Transfection or Viral Transduction
HEK 293FT cells were transfected with short hairpin RNA (shRNA) targeting c-Met or nontargeting scrambled shRNA (pLK0.1), pCMV-dR8.2dvpr, and pCMV-VSVG (gifts from Dr Ta-Jen Liu, UTMDACC) using Fugene HD (Roche, Indianapolis, IN) according to the manufacturer's instructions. Filtered viral supernatant was applied to U87 cells with 8 µg/ml hexadimethrine bromide, and sh-c-Met clones were made by limiting dilution during selection (1 µg/ml puromycin). c-Met shRNA hairpin sequences (Open Biosystems, now Thermo Fisher Scientific, Waltham, MA) are given as follows: TRCN0000009850 (sh-c-Met#A) and TRCN0000040047 (sh-c-Met#B); sh-c-Met#A: sense, 5′-CAGAATGTCATTCTACATGAG-3′; sh-c-Met#A: antisense, 5′-CTCATGTAGAATGACATTCTG-3′; sh-c-Met#B: sense, 5′-GCCAGCCTGAATGATGACATT-3′; sh-c-Met#B: antisense, 5′-AATGTCATCATTCAGGCTGGC-3′.
pLRNL-ΔEGFR (gift from Dr H-J Huang, The University of California, San Diego) was packaged into viral particles using GP2 cells, pCMV-VSVG, and Lipofectamine 2000 (Invitrogen). Viral supernatant was used to infect U87 GBM cells and later selected with G418 (2.5 µg/ml).
Constitutively active STAT3 (STAT3-CA) in pcDNA3.1/Hygro(+) (gift from Dr Robert Arceci, John Hopkins University School of Medicine) or pcDNA3.1/Hygro(+) empty vector (gift from Dr Suyun Huang, UTMDACC) was used to transfect U87 ΔEGFR or U87 sh-c-Met#B2 ΔEGFR using Fugene HD (Roche) and then selected (50 µg/ml hygromycin B).
Quantitative Real-Time Polymerase Chain Reaction
mRNA was extracted from cells using Qiagen's RNeasy Mini Kit. Reverse transcription was performed using Bio-Rad's iScript cDNA Synthesis Kit. Quantitative real-time polymerase chain reaction (PCR) was performed using FastStart SYBR Green Master reagent (Roche) with the following primers: HGF (forward): 5′-CTCACACCCGCTGGGAGTAC-3′; HGF (reverse): 5′-TCCTTGACCTTGGATGCATTC-3′; β2-microglobulin (forward): 5′-ATCCATCCGACATTGAAGTT-3′; β2-microglobulin (reverse): 5′-GGCAGGCATACTCATCTTTT-3′. Data were normalized to internal β2-microglobulin.
HGF ELISA
Conditioned media (CM) was collected from 80% confluent 24-hour serum-starved cells. Plates were coated with mouse anti-human HGF monoclonal antibody (0.5 µg/ml; R&D Systems) or isotype control antibody. Dried wells were washed with phosphate-buffered saline containing 0.05% Tween 20, blocked with 50 mM Tris (pH8.0) containing 0.14 M NaCl, 1% BSA, and 0.05% Tween 20, and rewashed. CM (concentrated using Millipore 30,000-kD cellulose ultra-filtration membranes) or HGF standard was serially diluted in TBS containing 0.1% BSA and 0.05% Tween 20 (pH 7.3) and applied to plate wells. After washing, goat anti-human HGF polyclonal antibody (0.5 µg/ml; R&D Systems) was added. Washed wells were incubated with HRP-conjugated bovine anti-goat IgG (40 ng/ml; Jackson ImmunoResearch Laboratories Inc). QuantaBlu Fluorogenic Peroxidase Substrate (Thermo Fisher Scientific) was applied to washed wells, and the reactions were terminated using QuantaBlu Stop Solution (Thermo Fisher Scientific). Fluorescence (excitation = 325 and emission = 420) was measured using a SpectraMax Gemini (Molecular Probes, now Invitrogen) fluorescent plate reader and SOFTmax Pro (v.3.0).
Anchorage-Independent Growth Assays
For anchorage-independent growth assays, 7.5 x 102 cells/well (24-well plate; Figure 3A) or 1.5 x 103 cells/well (12-well plate; Figure 7C) were cultured three-dimensionally, and colony numbers were counted as described previously [35].
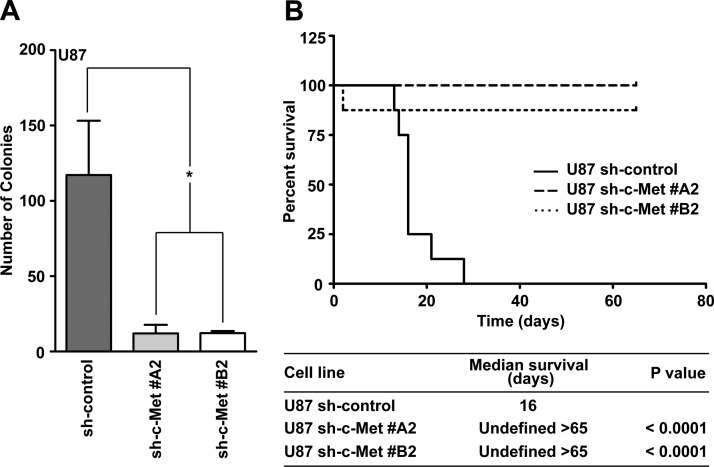
Figure 3.
Biologic effects of c-Met knockdown in U87 cells. (A) Anchorage-independent growth of U87 sh-control cells compared with U87 sh-c-Met clones (t test: *P < .05; n = 3 ± SEM; at least triplicate samples per experiment). (B) Survival curves of nude mice injected intracranially with 2 x 105 U87 sh-control or U87 sh-c-Met clones. Median survival and significant differences are shown (log-rank test of U87 sh-control versus U87 sh-c-Met clones).
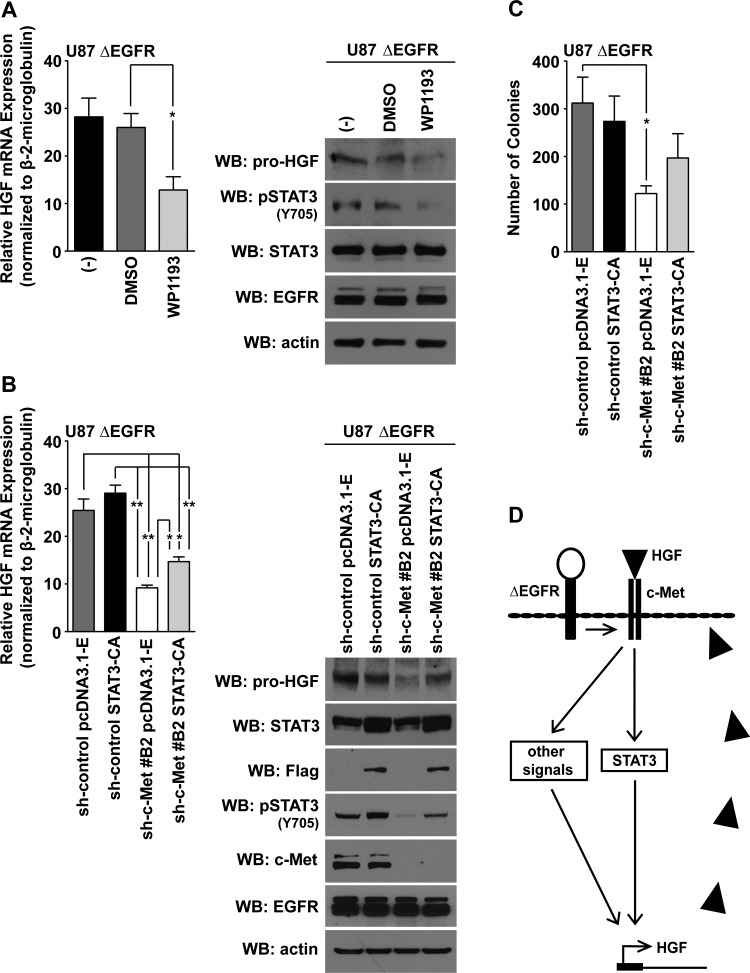
Figure 7.
STAT3 partially rescues HGF expression and anchorage-independent growth of U87 c-Met knockdown cells. (A) Left panel: Quantitative real-time PCR measured HGF mRNA levels in U87 ΔEGFR cells treated with WP1193 (2.5 µM; 16 hours; 10% FBS-containing media). Untreated (-) and 0.1% DMSO-treated cells were used as negative controls (t test compared with the 0.1% DMSO-treated cells: *P < .05; n = 3 ± SEM; at least duplicate samples per experiment). Right panel: Western blot analysis showing that WP1193 inhibited pSTAT3 Y705 phosphorylation and HGF expression in cells processed for quantitative real-time PCR analysis. (B) Left panel: Quantitative real-time PCR analysis of HGF mRNA in U87 ΔEGFR cells expressing combinations of the sh-control, pcDNA3.1-Empty, sh-c-Met#B2, or STAT3-CA (t test: *P < .05, **P < .005; n = 3 ± SEM; at least duplicate samples per experiment). Right panel: Western blot analysis of HGF and pSTAT3 (Y705) levels in cell lines represented in the left panel. (C) Anchorage independence of cell lines represented in B (t test: *P < .05; n = 3 ± SEM; at least duplicate samples per experiment). (D) Proposed model of c-Met signaling regulating HGF expression through STAT3 or other signaling mechanisms in ΔEGFR-expressing cells.
Xenograft Studies
Survival curves were generated after 2 x 105 cells per 5 µl were stereotactically injected into the right frontal lobe of 10-week-old nu/nu mice; animal experiments were performed on the same day. One U87 sh-c-Met#A2 mouse died within 48 hours of injection because of procedural stress. The maintenance and care of mice were conducted in accordance with Laboratory Animal Resources Commission standards under an approved protocol (100712131) in MD Anderson's Animal Facility.
Immunoprecipitation Assays
c-Met immunoprecipitation and subsequent immunoblot analysis techniques were performed as previously described [34].
Mass Spectrometry
Samples for mass spectrometry (MS) analysis were prepared and analyzed as previously described [34]. Liquid chromatography (LC)-MS analysis was performed with Agilent's 6340 Ion Trap System with electron transfer dissociation capability, where fragmentation alternated between collision-induced dissociation and electron transfer dissociation modes.
MS/MS spectra were extracted using Bruker CompassXport to “.mzxml” files and converted to “.mgf” files for database searches using Trans-Proteomic Pipeline (Seattle Proteome Center, Seattle, WA). Mascot search engine (v.2.3.02) searched human Swiss-Prot database's proteins to identify peptides and modifications. Phosphorylation site assignments were manually confirmed. Ideal-Q [36] software aligned the runs based on retention time, and phosphopeptide peak areas were manually calculated. Values were normalized to the total ion current of the whole run; phosphopeptide mean peak areas were then calculated.
The Cancer Genome Atlas Analyses
Level 3 gene expression data (Agilent 244K custom gene expression chip) were downloaded as log10 ratios to a Universal Human Reference RNA (Stratagene, La Jolla, CA) from 495 GBM tumors from The Cancer Genome Atlas (TCGA) data portal (http://tcgadata.nci.nih.gov/tcga/tcgaHome2.jsp) on 15 July 2011.
Gene lists defined by Verhaak et al. [12] for each GBM subtype were used to calculate average GBM subtype metagene scores for each tumor. The highest z-score normalized average metagene score was used for GBM subtype assignment.
Statistical Analysis
Data significance was analyzed using GraphPad Prism 5.03 software. For specific tests of significance, please refer to figure legends. All t tests were unpaired and two-tailed.
Results
c-Met and HGF Coexpression in Mes GBM
Analysis of a large data set of 495 GBM tumors (gene expression data from the Agilent platform) from the TCGA Network database [37] showed that c-Met and HGF transcripts are frequently coexpressed (Figure 1A), validating previous smaller studies [5,6,8].
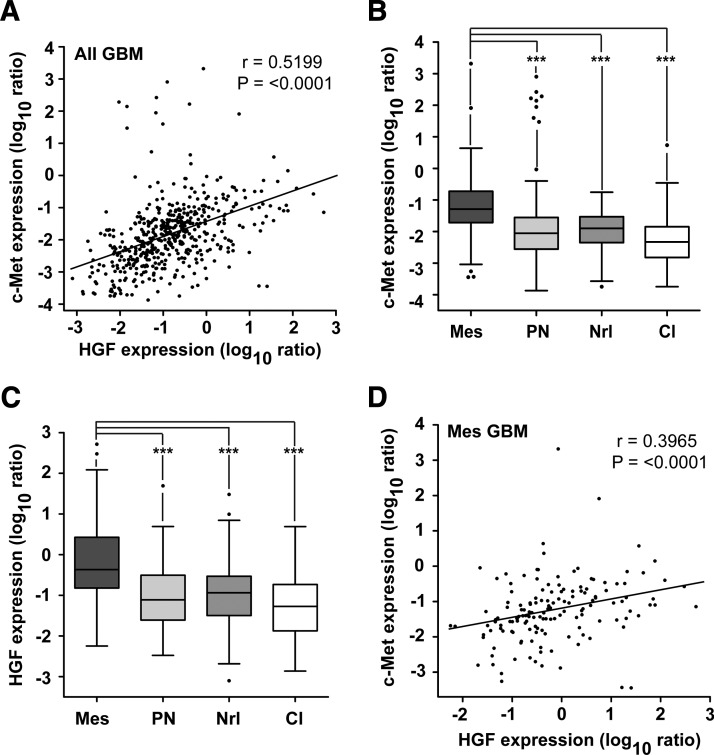
Figure 1.
Enhanced HGF and c-Met expression associate with Mes GBM. (A) HGF and c-Met mRNA expression correlate in GBMs (n = 495; Spearman correlation: r = 0.5199; P < .0001). (B) GBMs were assigned to a GBM subtype, from gene lists detailed by Verhaak et al. [12], based on highest average z-score normalized metagene scores. c-Met mRNA expression was then documented per tumor [n = 495; Tukey box plot; t tests: ***P < .0001; mesenchymal (Mes); proneural (PN); neural (Nrl); classical (Cl)]. (C) HGF mRNA expression was reported for tumors in A (n = 495; Tukey box plot; t tests: ***P < .0001). (D) HGF and c-Met mRNA expression correlate in Mes GBM tumors (n = 148; Spearman correlation: r = 0.3965; P ≤ .0001).
To assess GBM subtype-specific c-Met and HGF mRNA expression, we stratified the GBM tumors from the TCGA database according to their highest z-score corrected subtype-specific metagene scores. These scores were calculated using centroid-based gene lists that have previously been described for GBM subtypes [12]. We found that both c-Met (Figure 1B) and HGF (Figure 1C) mRNA expression were significantly higher in Mes GBMs when compared with proneural (PN), neural (Nrl), and classical (Cl) GBM subtypes (P < .0001). No discernible differences were found between the PN, Nrl, and Cl subtypes. Consistently, in 200 GBMs that Verhaak et al. [12] had assigned to subtypes, we found that c-Met and HGF mRNA expression were highest in Mes GBMs (Figure W1, A and B). Furthermore, we found that the expression of HGF and c-Met correlated significantly in the 148 Mes GBM tumors (Figure 1D; Spearman correlation; r = 0.3965; P < .0001). These data suggest that c-Met and HGF may be co-regulated in Mes GBM and contribute to their biology.
To validate our findings in an independent data set, we used Verhaak et al.'s [12] subtype-specific gene expression classifier lists to assign GBMs in the Repository for Molecular Brain Neoplasia Data (REMBRANDT) database (Affymetrix gene expression platform) to a particular GBM subtype. After determining HGF and c-Met mRNA expression levels per GBM, we found a similar trend of increased c-Met and HGF expression in Mes GBMs (Figure W1, C and D).
Phillips et al. assigned GBMs to specific subtypes based on prognostic gene expression lists [3]. Patients survived longer if their tumors had a PN signature but had the worst prognosis if their tumors were classified as Mes. Using gene lists for GBM subtype assignment (n = 495) of Phillips et al. [3], we found that c-Met and HGF expression was lower in PN tumors when compared with the more aggressive Mes or proliferative tumors, suggesting that the survival of GBM patients may be impacted with enhanced c-Met/HGF expression (Figure W1, E and F).
c-Met Modulates HGF Expression
The coincident elevation of HGF and c-Met mRNA in GBM may be caused by a positive regulatory loop that functions to enhance tumorigenesis. Because c-Met activation by HGF has been reported to induce c-Met expression in GBM [10], we hypothesized that c-Met may also positively modulate the expression of its own ligand.
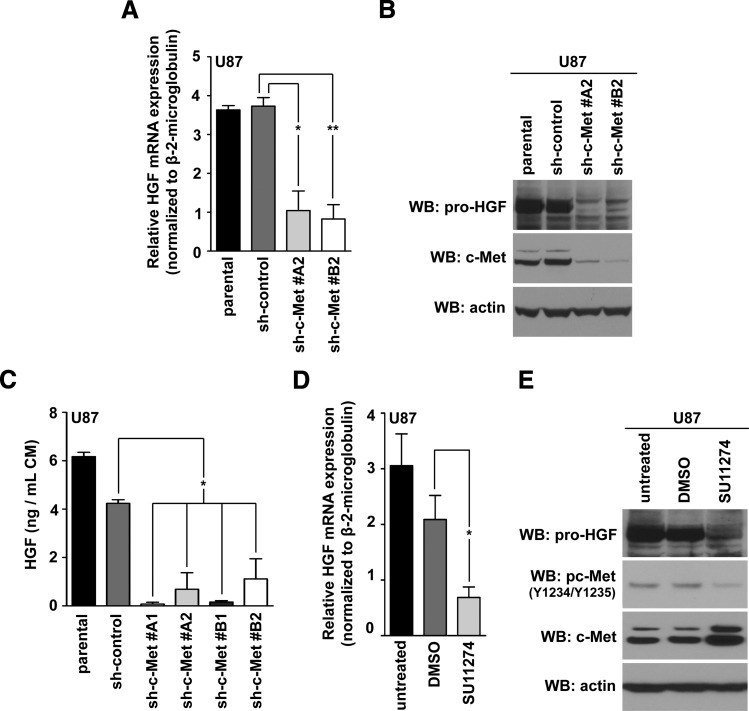
The human U87 GBM cell line coexpresses c-Met and HGF and is dependent on c-Met signaling for proliferation and survival [38,39]. shRNA-mediated knockdown of c-Met in U87 cells decreased steady-state amounts of HGF mRNA (Figure 2A). This correlated with reduced levels of HGF protein by Western blot analysis (Figure 2B) as well as attenuated levels of secreted HGF as evidenced by ELISA (Figure 2C).
Figure 2.
c-Met activity modulates HGF expression. (A) Quantitative real-time PCR measured HGF mRNA levels in U87 clones expressing two different c-Met lentiviral shRNA; 10% FBS-containing media (t test compared with sh-control: **P < .005, *P < .05; n = 3 ± SEM; at least duplicate samples per experiment). (B) Western blot analysis of HGF expression in cells detailed in A. (C) ELISA quantification of HGF in CM from control versus U87 sh-c-Met clones (one-way analysis of variance; Dunnett multiple comparison test: *P < .05; triplicate technical repeats above background analyzed per experiment; n = 2 ± SEM). (D) HGF quantitative real-time PCR of U87 cells treated with 10 µM SU11274 (16 hours); 10% FBS-containing media (t test compared with 0.1% DMSO control: *P < .05; n = 4 ± SEM; at least duplicate samples per experiment). (E) Western blot analysis of HGF and c-Met activity after 0.1% DMSO or 10 µM SU11274 treatment (16 hours); 10% FBS-containing media.
Given that HGF expression is regulated by c-Met, we asked whether c-Met's kinase activity was necessary for this. Treatment of two c-Met-dependent GBM cell lines, U87 [39] and LN18 [5], with SU11274, a specific c-Met inhibitor [40], decreased HGF mRNA and protein amounts in U87 (Figure 2, D and E) and LN18 cells (Figure W2 and Table W1). Taken together, these results suggest a feedback mechanism in which c-Met can regulate HGF expression through activated c-Met signaling.
c-Met Is Required for Anchorage-Independent Growth and Tumorigenicity of U87 Cells
We then characterized the biologic implications of c-Met silencing in U87 cells. c-Met knockdown significantly inhibited anchorage-independent growth of U87 cells (Figure 3A). To test the tumorigenic potential of these cells in vivo, we injected them intracranially into nude mice (Figure 3B). As expected, the U87 sh-control group showed a short median survival (16 days; Figure 3B). In contrast, mice that received U87 cells with c-Met knockdown did not form tumors after 65 days. The absence of tumors was verified by microscopic examination of hematoxylin and eosin (H&E)-stained serial sections from two mouse brains per group (data not shown). Taken together, these data suggest that c-Met is critical for in vitro colony formation and the tumorigenicity of U87 cells.
ΔEGFR Increases HGF Expression in a c-Met Activity-Dependent Manner
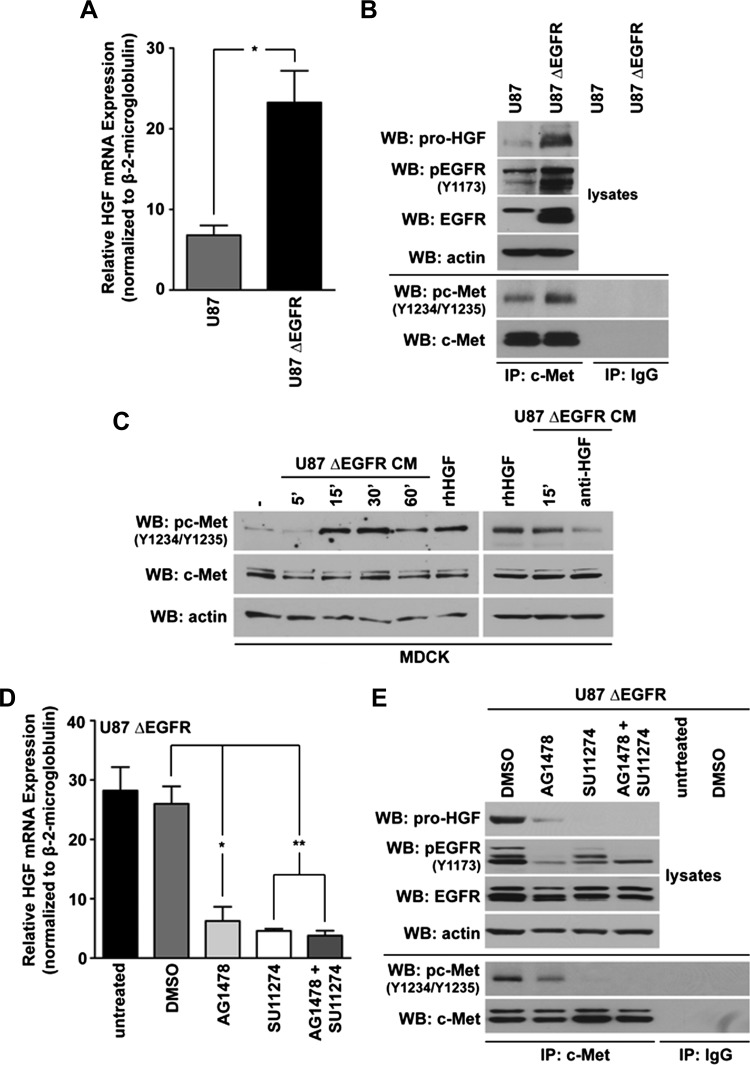
Using shotgun phosphoproteomics, we [34] and others [33] identified c-Met (Y1234) as a target of ΔEGFR signaling in GBM cells. Therefore, we examined whether ΔEGFR could regulate HGF levels through c-Met. Using quantitative real-time PCR, we show that ΔEGFR upregulated HGF mRNA levels by more than two-fold relative to parental U87 cells (Figure 4A). Similarly, Western blot analysis revealed that HGF protein levels increased with ΔEGFR expression in U87 cells (Figure 4B). As expected, the activity of c-Met increased in the presence of ΔEGFR (Figure 4B).
Figure 4.
ΔEGFR regulates HGF expression in an activity-dependent manner. (A) Quantitative real-time PCR analysis of HGF mRNA levels in U87 cells versus those expressing ΔEGFR (10% FBS-containing media); t test: *P < .05; n = 3 ± SEM; at least duplicate samples per experiment). (B) Western blots monitored HGF and immunoprecipitated c-Met for pc-Met (Y1234/Y1235) levels in U87 or U87 ΔEGFR cells (1% FBS-containing media; 20 hours). (C) CM from U87 ΔEGFR cells (1% FBS-containing media; 20 hours) were transferred to 4-hour serum-starved MDCK cells for the indicated amounts of time, and the levels of c-Met phosphorylation were detected by Western blot. Additionally, 1% serum-containing media with or without (-) 50 ng/ml rhHGF, or CM that had been pretreated with anti-HGF (0.6 µg/ml) for 2 hours, were transferred to MDCK cells for 15 minutes. (D) Quantitative real-time PCR analysis of U87 ΔEGFR cells that were either untreated (-) or treated with 0.1% DMSO, 10 µM AG1478, 10 µM SU11274, or a combination of both SU11274 and AG1478, for 16 hours in 10% FBS-containing media (t test when compared with the vehicle-treated control: **P < .005, *P < .05; n = 3 ± SEM; samples were analyzed at least in duplicate per experiment). (E)Western blot analysis of HGF, pEGFR (Y1173), and immunoprecipitated c-Met for pc-Met (Y1234/Y1235) in U87 ΔEGFR cells treated with tyrosine kinase inhibitors as in D; 10% FBS-containing media (16 hours).
To determine whether the HGF produced by GBM cells was secreted and functionally active, we examined the ability of CM from U87 ΔEGFR cells to activate c-Met (Y1234/Y1235) in HGF-responsive MDCK cells. The addition of CM resulted in acute stimulation of c-Met in a time-dependent manner in MDCK cells, which was comparable to stimulation of MDCK cells with rhHGF. Pre-neutralization of U87 ΔEGFR CM with an anti-HGF antibody blocked c-Met activation (Figure 4C).
To test whether the kinase activity of ΔEGFR modulates HGF expression, we treated U87 ΔEGFR-expressing cells with the EGFR/ΔEGFR inhibitor AG1478 [33]. Treatment with AG1478 reduced HGF mRNA levels. Using maximal doses of AG1478 and SU11274 that suppress the activities of ΔEGFR and c-Met, respectively, we found that AG1478 was as effective at reducing HGF mRNA levels as SU11274. This suggests that ΔEGFR and c-Met are likely part of the same pathway regulating HGF expression (Figure 4D).
These results were mirrored when cellular HGF protein amounts were investigated after treatment of ΔEGFR-expressing U87 cells with AG1478, SU11274, or in combination (Figure 4E). The levels of HGF protein correlated with the activity of c-Met, as measured by phosphorylation of Y1234/Y1235. As expected, c-Met's tyrosine kinase activity diminished with AG1478 treatment, which was accompanied by a reduction in HGF protein levels. A greater reduction in both c-Met's activity and HGF levels were obtained with SU11274 treatment. These results are suggestive of a positive feedforward relationship between c-Met's activity levels and HGF induction.
ΔEGFR is predominantly expressed in Cl GBMs; however, it is also expressed in Mes and PN GBMs [12]. As previously shown, the Cl GBM subtype expresses lower levels of c-Met and HGF transcripts to that of Mes GBMs (Figure 1, B and C, respectively). Given the importance of the ΔEGFR in Cl GBMs, we wanted to determine whether c-Met and HGF mRNA expression would correlate in this subtype. Using Spearman correlation, we found that their coordinated expression was significant (Figure W3; n = 141; r = 0.4562; P < .0001).
ΔEGFR Does Not Rescue c-Met Knockdown Phenotypes in U87 Cells
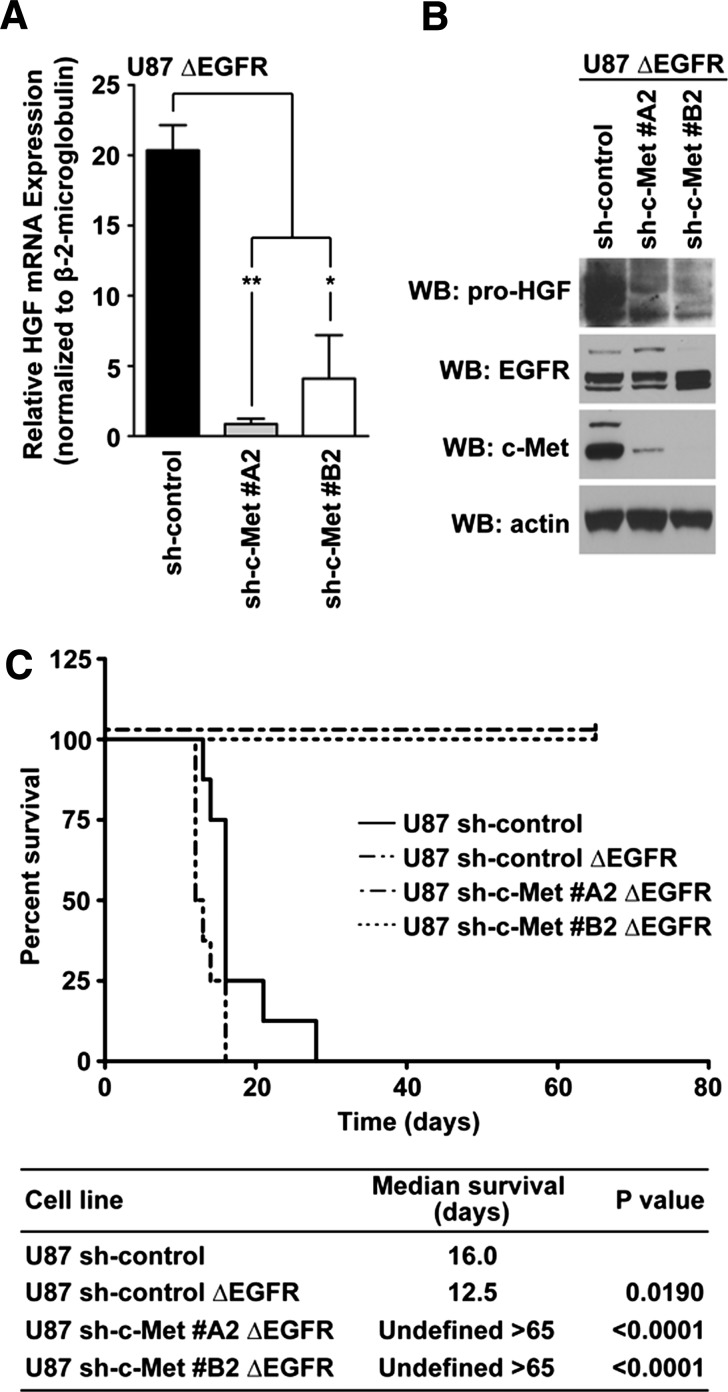
To determine whether ΔEGFR is capable of sustaining HGF expression in the absence of c-Met, we overexpressed ΔEGFR in U87 sh-c-Met clones. We found that the loss of c-Met caused a significant reduction in HGF mRNA (Figure 5A) and protein (Figure 5B) levels, suggesting that there is an absolute requirement for c-Met in ΔEGFR-mediated HGF regulation in GBM cells.
Figure 5.
c-Met is required by ΔEGFR to modulate HGF expression and tumorigenicity. (A) Quantitative real-time PCR measured HGF mRNA amounts in clonal populations of U87 cells expressing different c-Met shRNA that also expressed ΔEGFR (10% FBS-containing media; t test: **P < .005, *P < .05; n = 3 ± SEM; at least duplicate samples per experiment). (B) Western blot analysis of HGF levels present in all cells detailed in A; 10% FBS-containing media. (C) Kaplan-Meier curves of mice that were intracranially injected with 2 x 105 U87 sh-control, U87 sh-control ΔEGFR, or U87 sh-c-Met clones expressing ΔEGFR. Mice were sacrificed after 65 days. Log-rank tests determined significant differences between all survival curves compared with the U87 sh-control group; median survival per group of mice was recorded.
ΔEGFR enhances the tumorigenicity of U87 GBM xenografts [25,29], raising the possibility that its expression could overcome the diminished tumorigenicity that we had observed following c-Met knockdown in U87 cells. To test this, we injected U87 sh-control, U87 sh-control ΔEGFR, U87 sh-c-Met#A2 ΔEGFR, or U87 sh-c-Met#B2 ΔEGFR cells intracranially into nude mice and measured their survival over 65 days (Figure 5C). As expected, ΔEGFR decreased the median survival of nude mice when compared with those injected with sh-control cells (Figure 5C). Strikingly, c-Met knockdown abrogated the tumorigenicity of U87 xenografts even when expressing ΔEGFR. H&E staining of two mouse brains per group confirmed the absence of tumors in these animals (data not shown). These data suggest that c-Met loss significantly suppresses the tumorigenicity of c-Met-dependent GBM cells and that ΔEGFR is unable to rescue this phenotype.
STAT3 Y705 Is Responsive to the c-Met Signal
To identify changes in signaling that might be responsible for these observations, we examined the tyrosine phosphoproteome of U87 sh-control, U87 sh-control ΔEGFR, U87 sh-c-Met#B2, and U87 sh-c-Met#B2 ΔEGFR cells using an MS approach (for a complete list of peptides that were identified, see Table W2).
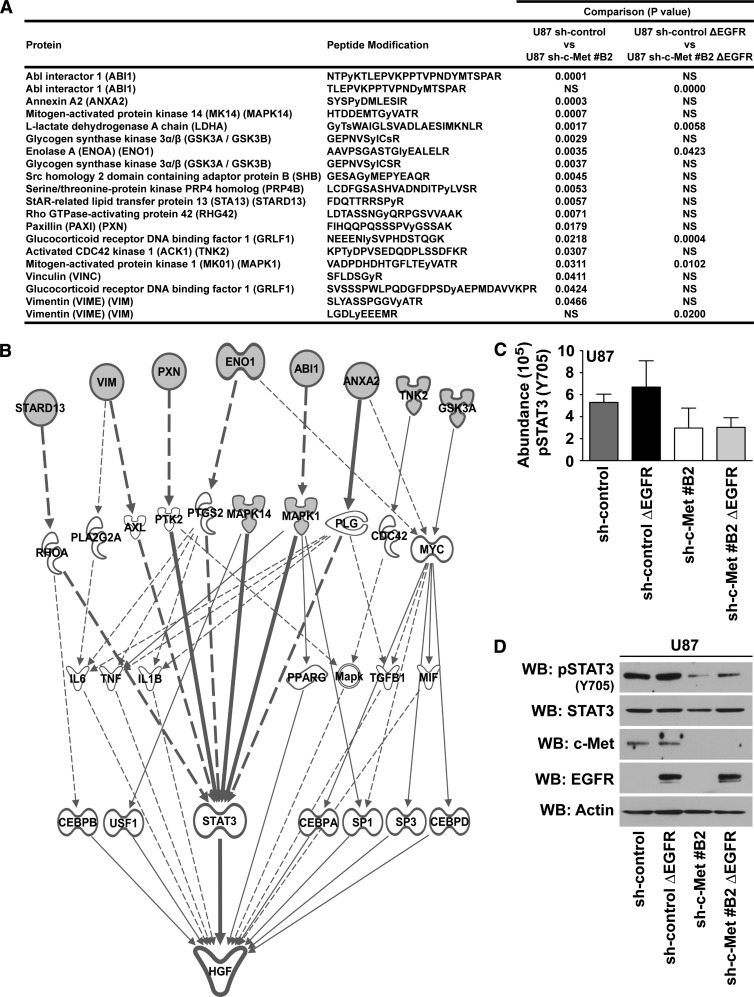
The peptides showing significant decreases (P < .05) in tyrosine phosphorylation in U87 sh-c-Met#B2 cells compared to U87 shcontrol cells, and in U87 sh-c-Met#B2 ΔEGFR cells compared to U87 sh-control ΔEGFR cells, are listed in Figure 6A alongside their modified tyrosine phosphorylation sites. Interestingly, all of the proteins identified as being responsive to the c-Met signal in ΔEGFR-expressing cells were found to change significantly with c-Met knockdown in parental cells. Using Ingenuity Pathway Analysis (www.ingenuity.com) to identify signaling connections of the c-Met-responsive proteins identified here with established regulators of HGF expression within Ingenuity Knowledge Base, we identified STAT3 as a potential central node modulating HGF expression in GBM cells (Figure 6B).
Figure 6.
Identification of STAT3 as responsive to the c-Met signal in U87 GBM cells. (A) PhosphoScan identified phosphotyrosine-enriched peptides by MS in U87 sh-control, U87 sh-c-Met#B2, U87 sh-control ΔEGFR, and U87 sh-c-Met#B2 ΔEGFR cells. Significant peptides that differed with c-Met knockdown in parental U87 sh-control cells, and in U87 sh-control ΔEGFR cells, are listed (t test of matched cells with or without c-Met knockdown: P < .05; n = 2; duplicate samples per experiment). (B) Biologic relationships were discovered from the list of proteins in A with Ingenuity Knowledge Base's known modulators of HGF expression. (C) Average abundance of STAT3 Y705 PhosphoScan analysis signal. (D) Western blot validation of pSTAT3 Y705 phosphorylation in samples used for Phospho-Scan analysis.
Analysis of our MS data for phosphorylation intensities confirmed that STAT3 Y705 decreased with c-Met knockdown in U87 cells and in c-Met knockdown cells that also expressed ΔEGFR (Figure 6C), findings that were confirmed by Western blot analysis (Figure 6D). Additionally, HGF stimulation of U87 and U87 ΔEGFR-expressing cells lead to increased STAT3 activation (Figure W4, A and B, respectively). These data show that the activity of STAT3 is also responsive to the c-Met signal in U87 GBM cells and that it may serve as a key node regulating HGF.
STAT3 Activity Partially Rescues Loss of c-Met-Dependent Phenotypes in ΔEGFR-Expressing GBM Cells
Given the strong association between c-Met expression and the potential regulation of HGF expression by STAT3, we investigated whether STAT3 activity was necessary for HGF expression. To address this, we suppressed STAT3 activity in U87 ΔEGFR cells with WP1193, a STAT3 phosphorylation inhibitor [41], and found that HGF mRNA and protein amounts were attenuated (Figure 7A). Next, we tested whether STAT3 activity can rescue the reduction of HGF expression in U87 ΔEGFR cells resulting from c-Met knockdown. STAT3-CA [42] partially rescued HGF expression in ΔEGFR-expressing cells lacking c-Met expression both at the mRNA and protein levels (Figure 7B). To test whether these events had an impact at the biologic level, we asked whether STAT3-CA could rescue the suppression of in vitro colony formation associated with c-Met knockdown in ΔEGFR-expressing cells. Anchorage-independent growth assays demonstrated that colony formation was suppressed with c-Met knockdown and that STAT3-CA was not able to rescue anchorage-independent growth of these cells (Figure 7C). These results support our earlier phosphoproteomic and bioinformatic analyses that STAT3 is a downstream effector of HGF expression in ΔEGFR-expressing GBM cells. STAT3 likely works in concert with additional pathways to maximally upregulate HGF expression (Figure 7D).
Discussion
In GBM, autocrine c-Met/HGF signaling contributes to tumor progression [26]. We found that the elevated expression of HGF and c-Met correlated well in a large data set of GBMs, and this was found predominantly in Mes GBMs. Among the GBM subtypes, Mes GBMs are associated with a poorer overall survival, with GBM recurrence [3] and treatment resistance [14], and their gene expression signatures can predict for this outcome [43,44], allowing for the possibility that the HGF/c-Met signaling axis is making a contribution to their aggressive phenotype. Although we found that HGF and c-Met mRNA expression were lower in Cl when compared to Mes tumors, we determined that their coexpression also correlated significantly in the Cl subtype. Interestingly, Verhaak et al. [12] identified that the expression of the ΔEGFR was most frequently found in Cl GBMs (5 of 22 Cl GBMs) and, to a lesser extent, in Mes and PN GBMs (1 of 38 Mes GBMs and 1 of 37 PN GBMs), suggesting that in Cl GBMs the connection of these two pathways is most critical. Overall, these findings suggest that the c-Met/HGF axis is an important contributing factor to the oncogenicity of both Cl and Mes GBM subtypes, with some variations on the exact triggers for its elevation.
Because both HGF and c-Met are located on chromosome 7, chromosomal duplication partially account for their increased dosage in GBM [5]. However, their high expression levels are not likely accounted for by chromosome 7 trisomy alone. We found that c-Met signaling increases the expression of its own ligand. c-Met activation has previously been reported to transcriptionally activate the c-Met gene [10], highlighting the importance of self-regulation of this signaling axis in GBM. The biologic significance of this pathway is shown by the impact of reducing c-Met activity on U87 in vitro colony formation and tumorigenicity in xenografts. In agreement with our results, decreased tumorigenicity occurs when c-Met/HGF-dependent xenografts are treated with antagonists of c-Met [38] or HGF [9,39]. Taken together, these data suggest that targeting either c-Met or HGF in tumors that rely heavily on their autocrine signal for growth and tumorigenicity may be an effective target for therapeutic intervention, particularly because of the autonomous autocrine loop that we describe here. Interestingly, autonomous regulation of the autocrine signal has also been described for other receptor/ligand pairs, such as for the EGFR [45] and its cognate ligands, heparin-binding EGF-like growth factor (HB-EGF), epiregulin, and amphiregulin [46].
In GBM, c-Met is a preferential target of the ΔEGFR [33,34,39]. Our results indicate that the lateral activation of c-Met by ΔEGFR enhances HGF production, suggesting that targeting HGF could be beneficial in combination with ΔEGFR-targeted therapies. Studies have found that antibody-mediated HGF antagonism is effective in parental U87 or wild-type EGFR-expressing U87 xenografts and not against U87 xenografts expressing ΔEGFR [39]. This could be due to the higher levels of HGF expression in ΔEGFR-expressing U87 xenografts that cannot be completely neutralized with anti-HGF treatment alone. This idea is supported by a significant reduction in tumor growth when ΔEGFR and HGF antagonists are used in combination to treat ΔEGFR-expressing U87 xenografts [39]. We found that c-Met was crucial for ΔEGFR to maintain not only enhanced HGF levels but also for ΔEGFR-mediated oncogenicity of U87 cells. Therefore, our data indicate that c-Met may be an important driver of tumorigenicity for ΔEGFR-expressing GBMs that are addicted to c-Met/HGF signaling, suggesting that the addition of therapeutics targeting c-Met to ΔEGFR regimes may prove to be a more effective treatment strategy.
STAT3 is a master transcriptional regulator of the Mes phenotype in GBM [47]. STAT3 signaling is important for HGF/c-Met-mediated anchorage-independent growth and tumorigenicity [15] and is a driver of HGF expression in various cancer cell lines [2,48]. Interestingly, we did not find that HGF promoter activity was attenuated with c-Met knockdown in U87 cells when we evaluated the first -1029 bp in the 5′ flanking region of the HGF gene, which contains a cis-acting STAT3 binding element [48] (data not shown).
Another novel finding from our study is that STAT3 signaling regulated HGF expression in U87 ΔEGFR cells. Interestingly, STAT3 expression predicts poorer outcomes for GBM patients [49], correlating closely with GBM aggressiveness [49]. Although we found that STAT3 Y705 phosphorylation does not change significantly in response to ΔEGFR, it is required by ΔEGFR for cellular transformation, proliferation, and viability [50]. This suggests that ΔEGFR recruits the activity of STAT3 through signaling intermediates, such as is the case with c-Met for HGF production, to maintain strengthened ΔEGFR-mediated tumorigenicity. Our data also suggested that additional signaling effectors may be required by STAT3 to maximally upregulate HGF expression in HGF/c-Met-dependent GBM cells expressing ΔEGFR.
In summary, our data highlight the importance of c-Met signaling for GBM tumorigenesis, the manner in which c-Met upregulates HGF expression in GBM, and the contribution of ΔEGFR for perpetuation of the HGF/c-Met signal in GBM. Our data show that STAT3 is an important component necessary for enhanced c-Met-mediated HGF expression in ΔEGFR-expressing cells. Additionally, we have shown that the c-Met/HGF axis is significantly upregulated in Mes GBMs, indicating that these signals are most important in this GBM subtype, and by implication represents an opportunity for therapy of these tumors.
Supplementary Material
Acknowledgments
We would like to thank Michelle Barton, Zhimin Lu, and Dihua Yu for helpful discussions. We thank Laura Gibson for DNA finger-printing the cell lines and Verlene Henry and Lindsay Holmes (UTMDACC) for carrying out the animal experiments.
Abbreviations
- Cl
classical
- CM
conditioned media
- c-Met
hepatocyte growth factor receptor
- EGFR
epidermal growth factor receptor
- GBM
glioblastoma
- HGF
hepatocyte growth factor
- Mes
mesenchymal
- Nrl
neural
- PN
proneural
- rhHGF
recombinant human HGF
- shRNA
short hairpin RNA
- STAT3
signal transducer and activator of transcription 3
- STAT3-CA
constitutively active STAT3
- TCGA
The Cancer Genome Atlas
Footnotes
These studies were supported in part by grants from the National Cancer Institute of the National Institutes of Health [RO1CA108500 (O.B.) and P50CA127001 (O.B.)] and through The University of Texas MD Anderson's Cancer Center Support grant CA016672. None of the authors report any conflict of interests.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 2.Wojcik EJ, Sharifpoor S, Miller NA, Wright TG, Watering R, Tremblay EA, Swan K, Mueller CR, Elliott BE. A novel activating function of c-Src and Stat3 on HGF transcription in mammary carcinoma cells. Oncogene. 2006;25:2773–2784. doi: 10.1038/sj.onc.1209306. [DOI] [PubMed] [Google Scholar]
- 3.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Abounader R, Laterra J. HGF/c-Met signaling and targeted therapeutics in brain tumors. In: van Meir EG, editor. CNS Cancer: Cancer Drug Discovery and Development. New York, NY: Humana Press; 2009. pp. 933–952. [Google Scholar]
- 5.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Q, Bradley R, Kang L, Koeman J, Ascierto ML, Worschech A, De Giorgi V, Wang E, Kefene L, Su Y, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci USA. 2012;109:570–575. doi: 10.1073/pnas.1119059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koochekpour S, Jeffers M, Rulong S, Taylor G, Klineberg E, Hudson EA, Resau JH, Vande Woude GF. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- 8.Moriyama T, Kataoka H, Kawano H, Yokogami K, Nakano S, Goya T, Uchino H, Koono M, Wakisaka S. Comparative analysis of expression of hepatocyte growth factor and its receptor, c-Met, in gliomas, meningiomas and schwannomas in humans. Cancer Lett. 1998;124:149–155. doi: 10.1016/s0304-3835(97)00469-2. [DOI] [PubMed] [Google Scholar]
- 9.Abounader R, Ranganathan S, Lal B, Fielding K, Book A, Dietz H, Burger P, Laterra J. Reversion of human glioblastoma malignancy by U1 small nuclear RNA/ribozyme targeting of scatter factor/hepatocyte growth factor and c-Met expression. J Natl Cancer Inst. 1999;91:1548–1556. doi: 10.1093/jnci/91.18.1548. [DOI] [PubMed] [Google Scholar]
- 10.Abounader R, Ranganathan S, Kim BY, Nichols C, Laterra J. Signaling pathways in the induction of c-Met receptor expression by its ligand scatter factor/hepatocyte growth factor in human glioblastoma. J Neurochem. 2001;76:1497–1508. doi: 10.1046/j.1471-4159.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- 11.Kong DS, Song SY, Kim DH, Joo KM, Yoo JS, Koh JS, Dong SM, Suh YL, Lee JI, Park K, et al. Prognostic significance of c-Met expression in glioblastomas. Cancer. 2009;115:140–148. doi: 10.1002/cncr.23972. [DOI] [PubMed] [Google Scholar]
- 12.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducray F, de Reyniès A, Chinot O, Idbaih A, Figarella-Branger D, Colin C, Karayan-Tapon L, Chneiweiss H, Wager M, Vallette F, et al. An ANOCEF genomic and transcriptomic microarray study of the response to radiotherapy or to alkylating first-line chemotherapy in glioblastoma patients. Mol Cancer. 2010;9:234. doi: 10.1186/1476-4598-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y-W, Wang L-M, Jove R, Vande Woude GF. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21:217–226. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 16.Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009;15:2207–2214. doi: 10.1158/1078-0432.CCR-08-1306. [DOI] [PubMed] [Google Scholar]
- 17.Bergström JD, Westermark B, Heldin NE. Epidermal growth factor receptor signaling activates met in human anaplastic thyroid carcinoma cells. Exp Cell Res. 2000;259:293–299. doi: 10.1006/excr.2000.4967. [DOI] [PubMed] [Google Scholar]
- 18.Fischer OM, Giordano S, Comoglio PM, Ullrich A. Reactive oxygen species mediate Met receptor transactivation by G protein-coupled receptors and the epidermal growth factor receptor in human carcinoma cells. J Biol Chem. 2004;279:28970–28978. doi: 10.1074/jbc.M402508200. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto N, Mammadova G, Song RX-D, Fukami Y, Sato K. Tyrosine phosphorylation of p145met mediated by EGFR and Src is required for serum-independent survival of human bladder carcinoma cells. J Cell Sci. 2006;119:4623–4633. doi: 10.1242/jcs.03236. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal S, Zerillo C, Kolmakova J, Christensen JG, Harris LN, Rimm DL, Digiovanna MP, Stern DF. Association of constitutively activated hepatocyte growth factor receptor (Met) with resistance to a dual EGFR/Her2 inhibitor in non-small-cell lung cancer cells. Br J Cancer. 2009;100:941–949. doi: 10.1038/sj.bjc.6604937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang PH, Cavenee WK, Furnari FB, White FM. Uncovering therapeutic targets for glioblastoma: a systems biology approach. Cell Cycle. 2007;6:2750–2754. doi: 10.4161/cc.6.22.4922. [DOI] [PubMed] [Google Scholar]
- 22.Chu CT, Everiss KD, Wikstrand CJ, Batra SK, Kung H-J, Bigner DD. Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII) Biochem J. 1997;324:855–861. doi: 10.1042/bj3240855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang Y, Chumbalkar V, Latha K, Bogler O. Forced dimerization increases the activity of ΔEGFR/EGFRvIII and enhances its oncogenicity. Mol Cancer Res. 2011;9:1199–1208. doi: 10.1158/1541-7786.MCR-11-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ymer SI, Greenall SA, Cvrljevic A, Cao DX, Donoghue JF, Epa VC, Scott AM, Adams TE, Johns TG. Glioma specific extracellular missense mutations in the first cysteine rich region of epidermal growth factor receptor (EGFR) initiate ligand independent activation. Cancers. 2011;3:2032–2049. doi: 10.3390/cancers3022032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt MH, Furnari FB, Cavenee WK, Bogler O. Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc Natl Acad Sci USA. 2003;100:6505–6510. doi: 10.1073/pnas.1031790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang H-JS. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 28.Cavenee WK. Genetics and new approaches to cancer therapy. Carcinogenesis. 2002;23:683–686. doi: 10.1093/carcin/23.5.683. [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa R, Ji X-D, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang H-JS. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 31.Pelloski CE, Ballman KV, Furth AF, Zhang L, Lin E, Sulman EP, Bhat K, McDonald JM, Yung WK, Colman H, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 32.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 33.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chumbalkar V, Latha K, Hwang Y, Maywald R, Hawley L, Sawaya R, Diao L, Baggerly K, Cavenee WK, Furnari FB, et al. Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of ΔEGFR. J Proteome Res. 2011;10:1343–1352. doi: 10.1021/pr101075e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajiwara Y, Panchabhai S, Levin VA. A new preclinical 3-dimensional agarose colony formation assay. Technol Cancer Res Treat. 2008;7:329–334. doi: 10.1177/153303460800700407. [DOI] [PubMed] [Google Scholar]
- 36.Tsou CC, Tsai CF, Tsui YH, Sudhir PR, Wang YT, Chen YJ, Chen JY, Sung TY, Hsu WL. IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol Cell Proteomics. 2010;9:131–144. doi: 10.1074/mcp.M900177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network, author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martens T, Schmidt N-O, Eckerich C, Fillbrandt R, Merchant M, Schwall R, Westphal M, Lamszus K. A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo. Clin Cancer Res. 2006;12:6144–6152. doi: 10.1158/1078-0432.CCR-05-1418. [DOI] [PubMed] [Google Scholar]
- 39.Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, Scott AM, Johns TG. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia. 2009;11:448–458. doi: 10.1593/neo.09230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Le P, Liang C, Chan J, Kiewlich D, Miller T, Harris D, Sun L, Rice A, Vasile S, et al. Potent and selective inhibitors of the Met [hepatocyte growth factor/scatter factor (HGF/SF) receptor] tyrosine kinase block HGF/SF-induced tumor cell growth and invasion. Mol Cancer Ther. 2003;2:1085–1092. [PubMed] [Google Scholar]
- 41.Kong LY, Gelbard A, Wei J, Reina-Ortiz C, Wang Y, Yang EC, Hailemichael Y, Fokt I, Jayakumar A, Qiao W, et al. Inhibition of p-STAT3 enhances IFN-α efficacy against metastatic melanoma in a murine model. Clin Cancer Res. 2010;16:2550–2561. doi: 10.1158/1078-0432.CCR-10-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning A-Q, Li J, McGuinness M, Arceci RJ. STAT3 activation is required for Asp816 mutant c-Kit induced tumorigenicity. Oncogene. 2001;20:4528–4536. doi: 10.1038/sj.onc.1204590. [DOI] [PubMed] [Google Scholar]
- 43.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sulman EP, Aldape K. The use of global profiling in biomarker development for gliomas. Brain Pathol. 2011;21:88–95. doi: 10.1111/j.1750-3639.2010.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seth D, Shaw K, Jazayeri J, Leedman PJ. Complex post-transcriptional regulation of EGF-receptor expression by EGF and TGF-α in human prostate cancer cells. Br J Cancer. 1999;80:657–669. doi: 10.1038/sj.bjc.6690407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu EK, Foley JS, Cheng J, Patel AS, Drazen JM, Tschumperlin DJ. Bronchial epithelial compression regulates epidermal growth factor receptor family ligand expression in an autocrine manner. Am J Respir Cell Mol Biol. 2005;32:373–380. doi: 10.1165/rcmb.2004-0266OC. [DOI] [PubMed] [Google Scholar]
- 47.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomida M, Saito T. The human hepatocyte growth factor (HGF) gene is transcriptionally activated by leukemia inhibitory factor through the Stat binding element. Oncogene. 2004;23:679–686. doi: 10.1038/sj.onc.1207190. [DOI] [PubMed] [Google Scholar]
- 49.Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neurooncol. 2010;100:339–343. doi: 10.1007/s11060-010-0195-8. [DOI] [PubMed] [Google Scholar]
- 50.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.