Abstract
Background
Percentage of free-to-total prostate-specific antigen (%fPSA) is an independent predictor of risk for prostate cancer among men with modestly elevated level of total PSA (tPSA) in blood. Physiological and pathological factors have been shown to influence the %fPSA value and diagnostic accuracy.
Materials/Methods
To evaluate genetic determinants of %fPSA, we conducted a genome-wide association study of serum %fPSA by genotyping 642,584 single nucleotide polymorphisms (SNPs) in 3192 men of European ancestry, each with a tPSA level of 2.5 to 10 ng/ml, that were recruited in the REduction by DUtasteride of Prostate Cancer Events study. Single nucleotide polymorphisms (SNPs) with P < 10-5 were further evaluated among the controls of a population-based case-control study in Sweden (2899 prostate cancer cases and 1722 male controls), including 464 controls having tPSA levels of 2.5 to 10 ng/ml.
Results
We identified two loci that were associated with %fPSA at a genome-wide significance level (P <5 x 10-8). The first associated SNP was rs3213764 (P = 6.45 x 10-10), a nonsynonymous variant (K530R) in the ATF7IP gene at 12p13. This variant was also nominally associated with tPSA (P = .015). The second locus was rs1354774 (P = 1.25 x 10-12), near KLK2 at 19q13, which was not associated with tPSA levels, and is separate from the rs17632542 locus at KLK3 that was previously associated with tPSA levels and prostate cancer risk. Neither rs3213764 nor rs1354774 was associated with prostate cancer risk or aggressiveness.
Conclusions
These findings demonstrate that genetic variants at ATF7IP and KLK2 contribute to the variance of %fPSA.
Introduction
Serum prostate-specific antigen (PSA) test is widely used for prostate cancer screening before diagnosis in Western countries. The introduction of PSA screening for prostate cancer has considerably increased the detection of early-stage cancer. The results from large randomized trials show that PSA-based screening reduces prostate cancer mortality among men who would not otherwise be screened in Europe [1,2], and which has not been feasible to evaluate in the United States [3]. As a consequence of PSA screening, overdiagnosis and complications of treatment for prostate cancer, including urinary, sexual, and bowel dysfunction, have also been a concern [4,5]. Nevertheless, serum PSA levels are currently still the most important noninvasive indicator for the emergence and progression of prostate cancer.
Although the PSA level in blood is strongly associated both with risk of diagnosis and long-term outcome of prostate cancer, it has low to modest specificity for prostate cancer diagnosis at a modestly elevated PSA level in blood [6]. Most of the abnormally elevated PSA results are false positives in terms of prostate cancer. For example, 75% of men with PSA levels in the range of 4.0 to 10 ng/ml have a negative prostate biopsy. In contrast, about 15% of men whose PSA levels were <4.0 ng/ml, have prostate cancer on biopsy [7]. Importantly, about 15% of these cancers detected at a PSA < 4.0 ng/ml were also shown to be high-grade disease and, as such, are likely to progress [7]. To improve the diagnostic performance of the PSA test, numerous approaches have been proposed, including measuring PSA velocity (change over time), levels of free and protein-bound PSA, PSA density (the PSA level divided by the prostate volume), and the use of cutoff values for PSA levels that are specific to the individual's age, race, or ethnic group [8].
Levels of free PSA (fPSA) can be detected and compared to total PSA (tPSA), yielding the percentage of fPSA (%fPSA, also known as ratio PSA). Using %fPSA can improve specificity over tPSA alone, especially in men with intermediate levels of serum PSA, and reduce the proportion of unnecessary biopsies [9]. %fPSA has already been used as an assisting clinical parameter in the screening and diagnosis of prostate cancer for men with intermediate tPSA levels. However, physiological and pathological factors, including hereditary factors, have been shown to influence %fPSA value and diagnostic accuracy [10]. To date, the exact genetic determinants of %fPSA are largely unknown. We now report on a genome-wide association study (GWAS) and replication study among men with intermediate PSA levels, to identify genetic variants associated with %fPSA and to assess their relationship with prostate cancer risk.
Materials and Methods
Study Subjects for GWAS and Replication Stage
For the GWAS portion of our study, we utilized samples collected from subjects that were previously recruited for the REduction by DUtasteride of Prostate Cancer Events (REDUCE) study. Details of the REDUCE study design and implementation have been described elsewhere [11,12]. Briefly, the REDUCE study is a multicenter, randomized, double-blind, placebo-controlled clinical trial, which was designed to evaluate the clinical value of Dutasteride at a dose of 0.5 mg daily, a dual 5α-reductase inhibitor, in reducing the risk of incident prostate cancer. Of 3239 men of European descent who consented for genetic studies in REDUCE (Table W1), 3206 subjects having a baseline tPSA level between 2.5 and 10 ng/ml were used for the GWAS of %fPSA. For analysis of associations between single nucleotide polymorphisms (SNPs) and prostate cancer risk, the study subjects were restricted to the placebo group using case and non-case status after 4 years of follow-up (410 of 1654 men in the placebo group developed prostate cancer within this follow-up period). Among these 410 incident prostate cancer patients, associations of SNPs with aggressive disease were further examined by defining aggressiveness as men (n = 124) who developed prostate cancer with a Gleason score of 7 or higher, stage T3b or higher, and/or were lymph node or metastasis positive (N+ or M+, respectively).
The replication study subjects were from a population-based case-control study in Sweden, termed Cancer Prostate in Sweden (CAPS). The study sample was described in detail elsewhere [13]. In brief, prostate cancer patients were identified between July 2001 and October 2003 and recruited from four of the six regional cancer registries in Sweden and the National Prostate Cancer Register. Control subjects were randomly selected from the Swedish Population Registry, without a diagnosis of prostate cancer, by frequency matching to the cases on age (groups of 5-year intervals) and geographic region. DNA samples from blood were available for 2899 patients and 1722 control subjects after informed consent. As shown in Table W1, 464 control subjects with a tPSA level in the range of 2.5 to 10 ng/ml were used for the replication study to confirm GWAS results of %fPSA in REDUCE. In the CAPS case-control study, 2899 cases and 1722 controls were employed to evaluate the association of SNPs with prostate cancer risk. Associations of SNPs with prostate cancer aggressiveness were tested in a case-only study including 1231 aggressive and 1619 nonaggressive cases. Patients were classified as having aggressive disease if their tumors had a clinical stage of T3/T4, N+, M+, Gleason score of 8 or higher, or a serum PSA level of >50 ng/ml; otherwise, the patients were classified as nonaggressive cases.
The study was approved by the research ethical committees of the Karolinska Institute, Umea University, and Wake Forest University School of Medicine.
Measurement of tPSA and fPSA Levels
In REDUCE, serum levels of tPSA and fPSA were determined with an enzyme immunoassay at Quest Diagnostics (Van Nuys, CA and Heston, Middlesex, United Kingdom). In CAPS, the levels of tPSA and fPSA in EDTA-anticoagulated plasma were measured in the laboratory of Dr Hans Lilja in the Department of Laboratory Medicine, Lund University, Skane University Hospital (Malmo, Sweden). A dual-label assay (DELFIA Prostatus PSA F/T; PerkinElmer Life Science, Waltham, MA) [14] was used to simultaneously measure fPSA and tPSA.
Genotyping and Imputation for GWAS in REDUCE
GWAS genotyping was performed on the Illumina HumanOmni-Express BeadChip platform at the Center for Cancer Genomics, Wake Forest University School of Medicine. A total of 729,755 SNPs were genotyped in 3225 samples (14 samples were not genotyped because of poor DNA quality). All of the samples had a genome-wide call rate of ≥95% with an overall call rate of 99.7%. After excluding 33 individuals with tPSA levels less than 2.5 ng/ml or more than 10 ng/ml, 3192 subjects were included in the GWAS analysis. The following quality control (QC) criteria were used to exclude SNPs from further analysis: minor allele frequency (MAF) < 0.01 (n = 75,170), genotype call rate < 95% (n = 6961), and P < .001 (n = 8589) for the Hardy-Weinberg Equilibrium test. After exclusions, genotype data on 642,584 SNPs were used for the final genome-wide association analysis.
Using the combined data of the 1000 Genomes low-coverage pilot project and HapMap3 data as reference haplotype maps, imputation was performed to infer genotypes of SNPs that were not genotyped by the IMPUTE computer program [15]. A posterior probability of >0.90 was applied to call imputed genotypes. The same QC procedures for genotyped SNPs were also applied to imputed SNPs.
SNP Selection and Genotyping for Replication in CAPS Samples
SNPs associated with %fPSA with a P value less than 1 x 10-5 were selected as candidate loci for replication. The SNPs with the lowest P values among multiple SNPs in linkage disequilibrium (LD) at an r2 > 0.50 were chosen to be genotyped in CAPS samples. As shown in Table W2, a total of five SNPs were genotyped in CAPS samples. Genotyping was performed using the MassARRAY iPLEX (Sequenom, Inc, San Diego, CA) at the Center for Cancer Genomics, Wake Forest University. Duplicates and negative controls were included in each 384-well plate to ensure QC. Genotyping was performed by technicians blinded to sample status. The average concordance rate was >98%.
Statistical Analysis
A linear regression model was used to analyze the association of each SNP with quantitative traits, assuming an additive genetic model, which was implemented in the PLINK software package. Quantitative associations of %fPSA and tPSA were performed after log transformation to limit potential bias because of deviation from normality. We estimated the population stratification using a principal component approach implemented by EIGENSTRAT software [16]. A common P of 5 x 10-8 was used as a cutoff for genome-wide significance. For the regions that met the statistical criteria of genome-wide association with %fPSA, ungenotyped SNPs were then imputed and conditional analysis was then applied to test the independence of SNPs, using the originally significant SNPs as covariates for the subsequent analysis. The effects of identified SNPs on prostate cancer risk or aggressiveness were further evaluated in the placebo group of REDUCE and in the case controls of CAPS using logistic regression models.
We adopted the methods reported by Gudmundsson et al. [17] to calculate a personalized %fPSA cutoff value corresponding to the commonly used uniform cutoff of 25%. In brief, this was done by multiplying the value of 25%with the estimated relative genetic effect for the two %fPSA associated SNPs (rs3213764 and rs1354774). Personalized cutoff values for PSA were calculated for each subject. First, a linear regression model was fitted where log transformed %fPSA were treated as outcomes and the number of alleles associated with higher level of biomarkers was treated as a covariate. Genetic effects were calculated for noncarriers (aa), one carrier (Aa), and two carriers (AA) of the allele associated with elevated values, using the fitted values. Relative allelic effects (% of increase per allele) were calculated by dividing the fitted values of aa and Aa. The relative risk to the population for each of the three genotypes (aa, Aa, and AA) were then computed on the basis of the relative allelic effect and genotypic frequency. Second, assuming a multiplicative model, the combined genetic relative effect was calculated by multiplying the relative genotypic effects for each SNP relative to the general population. Third, the personalized cutoff of %fPSA was generated by multiplying a uniform cutoff (i.e., 25%) by the combined genetic relative effect for each subject.
Results
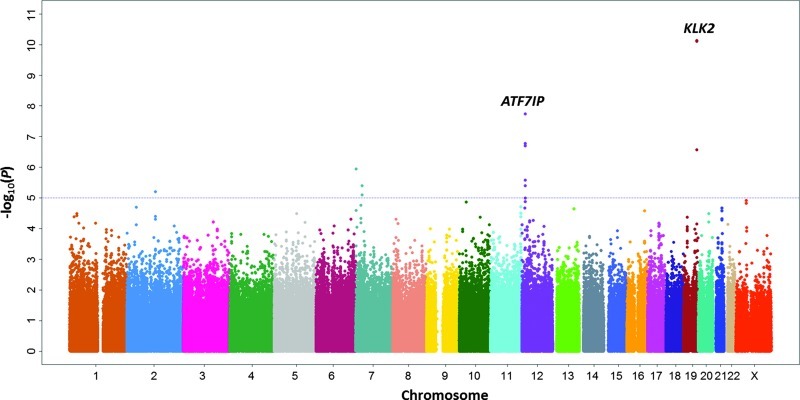
After QC filters, a total of 642,584 genotyped SNPs were analyzed for association with %fPSA among 3192 individuals who had a tPSA level between 2.5 and 10 ng/ml. Principal component analysis showed that all of the subjects were clustered around European descent, and the quantile-quantile (Q-Q) plot showed no obvious evidence of genetic stratification among subjects (genomic inflation factor λ = 1.009; Figure W1). Thus, the reported P values are not corrected for genomic inflation. Throughout the genome (Figure 1), two signals, one at 19q13 (P = 7.39 x 10-11 and 7.90 x 10-11 for rs1354774 and rs16987929, respectively) and another at 12p13 (P = 1.85 x 10-8 for rs3213764), were associated with %fPSA at a genome-wide significance level (P < 5 x 10-8). These lead SNPs, rs16987929 and rs1354774 at 19q13, are in moderate LD (r2 = 0.591).
Figure 1.
Manhattan plot of the strength of associations (-log10 (P) values; Y-axis) between SNPs (X-axis by chromosome and chromosomal position) and %fPSA.
The most significant SNP at each locus that was associated with % fPSA with a P value less than 1 x 10-5 at the GWAS stage was then fast-tracked for further genotyping in a replication analysis using an independent sample set consisting of 464 control subjects with tPSA levels between 2.5 and 10 ng/ml in CAPS. Among these five SNPs, two were also consistently associated with %fPSA in the replication among CAPS controls, with intermediate tPSA levels (2.5–10 ng/ml; rs3213764 at 12p13: P = 9.65 x 10-3; and rs1354774 at 19q13: P = 4.71 x 10-3; Tables W2 and 1). After pooling the GWAS and replication results, we observed a genome-wide significant association of rs3213764 (P = 6.45 x 10-10) and rs1354774 (P = 1.25 x 10-12) with %fPSA (Table 1). When we extended the replication analysis to all controls of the CAPS study population (Table W3), a more significant association was observed between rs1354774 and %fPSA (P = 1.05 x 10-11), resulting in a combined P value of 6.48 x 10-20. For association between rs3213764 and %fPSA, similar results were observed between controls with intermediate tPSA levels and all of the controls in CAPS (Pcombined = 1.97 x 10-9).
Table 1.
Summary Results for SNPs Associated with %fPSA at Genome-Wide Significance Level (P 5 x 10-8).
| Chromosome | SNP | Gene (Location) | Stage | MAF | Mean* | β | SE | P† | Pcombined‡ |
| 12p13 | rs3213764 (A/G)§ | ATF7IP (K530R) | GWAS (N = 3192) | 0.467 | 15.73/16.61/17.34 | 0.049 | 0.009 | 1.85 x 10-8 | 6.45 x 10-10 |
| Replication (N = 464) | 0.504 | 23.85/26.22/27.15 | 0.065 | 0.025 | 9.65 x 10-3 | ||||
| 19q13 | rs1354774 (A/G)§ | KLK2 (∼9 kb at 3′-flank) | GWAS (N = 3192) | 0.343 | 15.64/17.14/17.45 | 0.058 | 0.009 | 7.39 x 10-11 | 1.25 x 10-12 |
| Replication (N = 464) | 0.309 | 23.98/27.82/27.05 | 0.075 | 0.026 | 4.71 x 10-3 |
Mean values of %fPSA (%) by genotypes (major homozygote/heterozygote/minor homozygote).
P values were from linear regression models adjusted for age.
P values were estimated using inverse variance-weighted meta-analyses according to regression coefficients (β) and corresponding SEs across studies.
Major/minor allele.
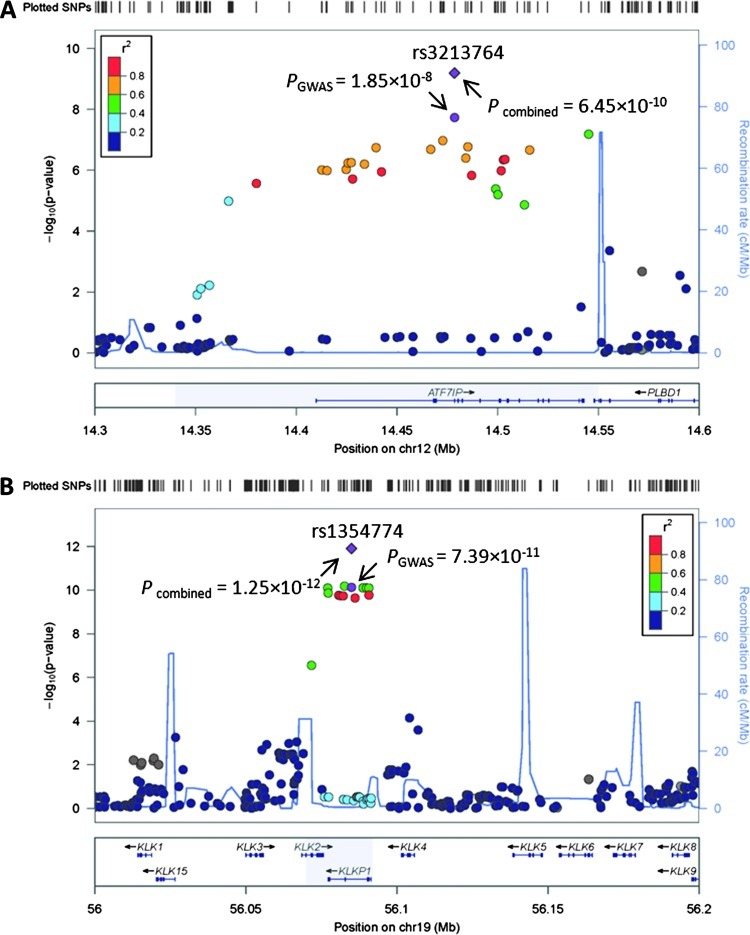
We imputed ungenotyped SNPs at 12p13 in a 300-kb window and tested their associations with %fPSA (Figure 2A). The SNPs with P < 10-5 are presented in Table W4, all of which are located within a 210-kb region of LD, incorporating the entire ATF7IP gene and 3′-end of the PLBD1 gene. The most significant SNP was rs3213764, a missense variant at codon 530 of the ATF7IP gene. This was the only SNP at 12p13 that showed a genome-wide significant association with %fPSA. There were no other SNPs at 12p13 that were associated with %fPSA at P < .01 after conditioning on rs3213764.
Figure 2.
Regional plots of the associations between 12p13 (A) and 19q13 (B) and %fPSA. Associations of individual SNPs have been plotted as -log10 P against the chromosomal position. Results of both genotyped and imputed SNPs are shown. Colors indicate the LD strength between themost significant SNPs (shown at the top of each plot with a point in purple) and the other SNPs assessed. The right Y-axis shows the recombination rate estimated from the 1000 Genomes Utah residents with Northern and Western European ancestry (CEU) population.
For chromosome 19q13, imputation was performed within a 200-kb window. As shown in Figure 2B, multiple SNPs associated with %fPSA less than 5 x 10-8 are centralized in a 22-kb region between two recombination hotspots. This region contains the entire KLKP1 gene and a majority of the KLK2 gene, except exon 1 and intron 1. This locus is distinct from the region containing KLK3 (the gene encoding PSA) and the tPSA-associated locus at 19q13 (rs17632542) [17]. As shown in Table W4, all of the SNPs at 19q13 with P < 10-5 for %fPSA were in LD with rs1354774, with r2 values ranging from 0.519 to 1. None of the associations for these SNPs reached a significance level of 0.01 after conditioning on rs1354774.
To evaluate whether the %fPSA-associated loci at 12p13 and 19q13 were also associated with tPSA (Table 2), we tested the associations of rs3213764 and rs1354774 with levels of tPSA. The SNP rs3213764 at 12p13 was suggestively associated with tPSA (Pcombined = .015). However, no association was observed between rs1354774 at 19q13 and tPSA levels (Pcombined = .904). We further evaluated whether these two loci were also associated with prostate cancer risk in case-control studies (REDUCE: 410 new cases and 1234 non-cases in a four-year follow-up of the placebo group; CAPS: 2899 cases and 1722 controls) or prostate cancer aggressiveness in case-only studies (REDUCE: 124 aggressive and 281 nonaggressive cases; CAPS: 1231 aggressive and 1619 nonaggressive cases). As shown in Table 3, no associations were observed for prostate cancer risk (Pcombined = .214 and .722, respectively) or aggressiveness (Pcombined = .737 and .716, respectively).
Table 2.
Relationships of the Loci ATF7IP at 12p13 and KLK2 at 19q13 with tPSA Levels.
| Locus | SNP (Minor Allele) | REDUCE (N = 3192) | CAPS (N = 464) | Pcombined* | ||||
| Mean† | β | P‡ | Mean† | β | P‡ | |||
| 12p13 (ATF7IP) | rs3213764 (G) | 6.00/5.86/5.81 | -0.015 | 0.069 | 4.88/4.41/4.37 | -0.053 | 0.027 | 0.015 |
| 19q13 (KLK2) | rs1354774 (G) | 5.89/5.85/6.00 | 0.002 | 0.846 | 4.58/4.49/4.31 | -0.024 | 0.342 | 0.904 |
Pcombined values were estimated using inverse variance-weighted meta-analyses according to regression coefficients (β) and corresponding SEs across studies.
Mean values of tPSA (%) by genotypes (major homozygote/heterozygote/minor homozygote).
P values were from linear regression models adjusted for age.
Table 3.
Relationships of the Loci ATF7IP at 12p13 and KLK2 at 19q13 with Prostate Cancer Risk (Case-Control Study) or Aggressiveness (Case-Only Study).
| Locus | SNP (Minor Allele) | Case-Control Study | Case-Only Study | ||||||
| MAFcase/control | OR (95% CI)* | P* | MAFagg/nonagg | OR (95% CI)* | P* | ||||
| REDUCE (N = 410/1234) | CAPS (N = 2899/1722) | REDUCE (N = 124/281) | CAPS (N = 1231/1619) | ||||||
| ATF7IP | rs3213764 (G) | 0.445/0.465 | 0.519/0.499 | 1.05 (0.97–1.13) | 0.214 | 0.456/0.441 | 0.524/0.517 | 1.02 (0.92.1.13) | 0.737 |
| KLK2 | rs1354774 (G) | 0.345/0.341 | 0.340/0.335 | 1.01 (0.94–1.10) | 0.722 | 0.367/0.336 | 0.332/0.344 | 0.98 (0.88.1.09) | 0.716 |
Odds ratios (ORs), 95% confidence intervals (CIs), and P values were estimated using inverse variance-weighted meta-analyses according to regression coefficients and corresponding SEs across REDUCE and CAPS studies.
We further calculated personalized %fPSA cutoff values to interpret the effects of the two identified variants on the variation of % fPSA levels. We adopted the methods reported by Gudmundsson et al. [17] and calculated a personalized%fPSA cutoff value corresponding to the commonly used cutoff of 25%. As shown in Figure W2, in contrast to the uniform %fPSA cutoff value of 25%, a personalized cutoff may range from 21.3% to 27.9% for an individual after genetic correction.
Discussion
PSA circulates within the serum in unbound (free) form or bound to one of several antiproteases, most predominantly α-1-antichymotrypsin (ACT/SERPINA3) [18,19]. fPSA represents 10% to 40% of tPSA. Previous studies suggest that %fPSA predicts risk of prostate cancer independent of tPSA and contributes modest diagnostic enhancements above and beyond tPSA alone amongmen in the “diagnostic gray zone” [9]. Numerous studies have explored methods of optimizing the diagnostic accuracy of %fPSA [10]. In the current study, we focused on the influence of genetic factors on %fPSA using a GWAS approach, leading to the identification of two loci at 12p13 and 19q13 that are associated with %fPSA, especially among men with intermediate tPSA levels. These findings may be useful for improving the performance of %fPSA in distinguishing malignant from benign prostate disease. The possible added value of incorporating these SNPs within existing PSA screening practices needs to be evaluated in future studies, as this may help avoid significant numbers of unnecessary and invasive biopsies.
Our results indicate that an SNP 1354774 at 19q13 is associated with %fPSA but not with tPSA. This SNP is located in the 3′-flanking region of KLK2 and an intron of KLKP1. As separated by a recombination hotspot (Figure 2), our lead SNP (rs1354774) is independent of SNPs in the KLK3 locus at 19q13 previously reported to be associated with tPSA and prostate cancer risk [17,20]. Specifically, rs1354774 is not in LD with either rs17632542 (r2 = 0.000 and 0.049 for REDUCE GWAS and 1000 Genomes, respectively) or rs2735839 (r2 = 0.001 and 0.002 for REDUCE GWAS and 1000 Genomes, respectively). In contrast to the complex associations observed between rs1763542 (or rs2735839) and tPSA and prostate cancer risk, rs1354774 was not associated with prostate cancer risk and aggressiveness. Interestingly, Nam et al. [21,22] and our group [23] have shown that this locus (defined by rs198977) was also associated with blood KLK2 levels. The SNP rs198977, showing moderate LD with rs1354774 (r2 = 0.505 and 0.547 for REDUCE GWAS and 1000 Genomes, respectively), was also associated with %fPSA (P = 4.77 x 10-8 in REDUCE) and fPSA (P = 6.55 x 10-7 in REDUCE) in this study. Because multiple SNPs in LD with rs135774 at 19q13 were associated with %fPSA and fPSA, we cannot dissect whether any of these variants are independently functional and how these variants at 19q13 might affect individual % fPSA and fPSA values at this stage. The KLK2 gene product has been shown to convert the inactive PSA zymogen to catalytically active PSA, which may influence the ratio (%fPSA) in the blood, and act as a physiologic PSA processing protease in the conversion of pro-PSA into active PSA [24–26]. %fPSA was decreased when KLK3 and KLK2-expressing cells were co-inoculated subcutaneously and double transgenic mice expressing both KLK2 and KLK3 in the prostate produced more active PSA compared to single transgenic animals [27]. Taken together, these multiple lines of evidence suggest that genetic variants in 19q13 may modify fPSA and%fPSA by influencing the biologic function of the KLK2 gene product.
We also identified a locus at 12p13 that was associated with %fPSA at a genome-wide significance level. The most significant SNP, rs3213764, results in an amino acid substitution (K530R) in ATF7IP, which encodes for the activating transcription factor 7 interacting protein [also known as MBD1-containing chromatin-associated factor 1 (MCAF1)]. Of interest, ATF7IP has been identified as a susceptibility gene for testicular germ cell tumor (TGCT) in a recent GWAS [28]. The reported SNP rs2900333 in TGCT GWAS is in moderate LD with missense variant rs3213764 (r2 = 0.534 and 0.553 for REDUCE GWAS and 1000 Genomes, respectively). The TGCT-related SNP rs2900333 was also associated with %fPSA in our REDUCE GWAS (P = 6.56 x 10-8). These results suggest that %fPSA shares the same genetic locus in ATF7IP as that for TGCT susceptibility. ATF7IP was found to regulate telomerase activity by acting on the promoter of TERT and TERC in an Sp1-dependent manner [29]. Overexpression of ATF7IP was also frequently observed in cancers [29]. The locus at TERT on 5p15 (defined by rs2736098) regulates telomerase activity and has been associated with tPSA and risk of multiple cancers, including prostate cancer [30] and TGCT [28]. In addition, we observed a suggestive association of rs3213764 in ATF7IP with circulating levels of tPSA (P = .015) but not with prostate cancer risk (P = .214) or aggressiveness (P = .737). However, these results were primarily because of the limited sample size, and further studies with larger sample sizes are needed to further clarify the relationships. Nevertheless, the exact mechanism by which genetic variants in ATF7IP may have an effect on PSA regulation is not known. It is possible that ATF7IP may regulate key genes in PSA processing as it does in TERT [29] and thus mediates the overall proportion of fPSA in circulation.
The relatively small sample size in the replication set is a limitation of this study, especially in the analyses restricted to individuals with a tPSA level range of 2.5 to 10 ng/ml. This may have resulted in false-negative findings because of limited statistical power. The moderate sample size also did not provide sufficient power to more extensively replicate additional suggestive signals (e.g., P values ranging from 10-3 to 10-5) observed in the REDUCE GWAS. In addition, the different levels of %fPSA between REDUCE and CAPS may also result in falsenegative results. However, this is not a major concern since we successfully replicated our findings in the CAPS population. Nevertheless, the two identified variants could explain only 2% of variance of %fPSA, which may lead to limited clinical utility. The identification of additional genetic loci that contribute to the level of %fPSA is needed in order to define a more differentiated personal cutoff value. The clinical value of personalized PSA test also needs to be evaluated in other independent and large prospective studies.
Conclusions
Two loci at 12p13 (rs3213764 in ATF7IP) and 19q13 (rs1354774 near KLK2) are identified to be associated with %fPSA. These findings may provide insight into individual variance of %fPSA and may be clinically useful for improving the predictive accuracy of %fPSA in PSA screening.
Supplementary Material
Acknowledgments
We thank the patients enrolled in REDUCE who provided consent and genetic samples that enabled this study and the clinicians who contributed their expertise in recruiting study patients for the REDUCE clinical study. Dave Pulford, Jennifer Aponte, Jon Charnecki, and Mary Ellyn Volk participated in consent reconciliation and sample management to enable genetic sample selection for inclusion and genotype determination. Karen King provided data management support for this project. We appreciate the assistance of Lauren Marmor in coordinating the support of the Avodart Collaborative Research Team. Editorial assistance was provided by Aubrey Turner.
Footnotes
This study was supported in part by a National Cancer Institute RC2 grant (CA148463) to J. Xu, a research contract by GlaxoSmithKline (GSK) to J. Xu, National Cancer Institute (grants R33 CA 127768-02 and P50-CA92629), Swedish Cancer Society (3455), Swedish Research Council (Medicine) (20095), the Sidney Kimmel Center for Prostate and Urologic Cancers, and David H. Koch through the Prostate Cancer Foundation to H. Lilja. J. Xu and J. Sun certify that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (e.g., employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: H. Lilja holds patents for free PSA, intact PSA, and hK2 assays. L. D. Condreay was a GSK employee during this study and holds stock in GSK.
This article refers to supplementary materials, which are designated by Tables W1 to W4 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hugosson J, Carlsson S, Aus G, Bergdahl S, Khatami A, Lodding P, Pihl CG, Stranne J, Holmberg E, Lilja H. Mortality results from the Göteborg randomised population-based prostate-cancer screening trial. Lancet. 2010;Oncol 11:725–732. doi: 10.1016/S1470-2045(10)70146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andriole GL, Crawford ED, Grubb RL, III, Buys SS, Chia D, Church TR, Fouad MN, Isaacs C, Kvale PA, Reding DJ, et al. Prostate cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: mortality results after 13 years of follow-up. J Natl Cancer Inst. 2012;104:125–132. doi: 10.1093/jnci/djr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilt TJ, MacDonald R, Rutks I, Shamliyan TA, Taylor BC, Kane RL. Systematic review: comparative effectiveness and harms of treatments for clinically localized prostate cancer. Ann Intern Med. 2008;148:435–448. doi: 10.7326/0003-4819-148-6-200803180-00209. [DOI] [PubMed] [Google Scholar]
- 6.Vickers AJ, Cronin AM, Bjork T, Manjer J, Nilsson PM, Dahlin A, Bjartell A, Scardino PT, Ulmert D, Lilja H. Prostate specific antigen concentration at age 60 and death or metastasis from prostate cancer: case-control study. BMJ. 2010;341:c4521. doi: 10.1136/bmj.c4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson IM, Ankerst DP, Chi C, Lucia MS, Goodman PJ, Crowley JJ, Parnes HL, Coltman CA., Jr Operating characteristics of prostatespecific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005;294:66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 8.Greene KL, Albertsen PC, Babaian RJ, Carter HB, Gann PH, Han M, Kuban DA, Sartor AO, Stanford JL, Zietman A, et al. Prostate specific antigen best practice statement: 2009 update. J Urol. 2009;182:2232–2241. doi: 10.1016/j.juro.2009.07.093. [DOI] [PubMed] [Google Scholar]
- 9.Roddam AW, Duffy MJ, Hamdy FC, Ward AM, Patnick J, Price CP, Rimmer J, Sturgeon C, White P, Allen NE. Use of prostate-specific antigen (PSA) isoforms for the detection of prostate cancer in men with a PSA level of 2–10 ng/ml: systematic review and meta-analysis. Eur Urol. 2005;48:386–399. doi: 10.1016/j.eururo.2005.04.015. discussion 398–389. [DOI] [PubMed] [Google Scholar]
- 10.Stephan C, Jung K, Lein M, Sinha P, Schnorr D, Loening SA. Molecular forms of prostate-specific antigen and human kallikrein 2 as promising tools for early diagnosis of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:1133–1147. [PubMed] [Google Scholar]
- 11.Andriole G, Bostwick D, Brawley O, Gomella L, Marberger M, Tindall D, Breed S, Somerville M, Rittmaster R. Chemoprevention of prostate cancer in men at high risk: rationale and design of the reduction by dutasteride of prostate cancer events (REDUCE) trial. J Urol. 2004;172:1314–1317. doi: 10.1097/01.ju.0000139320.78673.2a. [DOI] [PubMed] [Google Scholar]
- 12.Andriole GL, Bostwick DG, Brawley OW, Gomella LG, Marberger M, Montorsi F, Pettaway CA, Tammela TL, Teloken C, Tindall DJ, et al. Effect of dutasteride on the risk of prostate cancer. N Engl J Med. 2010;362:1192–1202. doi: 10.1056/NEJMoa0908127. [DOI] [PubMed] [Google Scholar]
- 13.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Balter K, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 14.Mitrunen K, Pettersson K, Piironen T, Bjork T, Lilja H, Lovgren T. Dual-label one-step immunoassay for simultaneous measurement of free and total prostate-specific antigen concentrations and ratios in serum. Clin Chem. 1995;41:1115–1120. [PubMed] [Google Scholar]
- 15.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Gudmundsson J, Besenbacher S, Sulem P, Gudbjartsson DF, Olafsson I, Arinbjarnarson S, Agnarsson BA, Benediktsdottir KR, Isaksson HJ, Kostic JP, et al. Genetic correction of PSA values using sequence variants associated with PSA levels. Sci Transl Med. 2010;2:62ra92. doi: 10.1126/scitranslmed.3001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensson A, Laurell CB, Lilja H. Enzymatic activity of prostatespecific antigen and its reactions with extracellular serine proteinase inhibitors. Eur J Biochem. 1990;194:755–763. doi: 10.1111/j.1432-1033.1990.tb19466.x. [DOI] [PubMed] [Google Scholar]
- 19.Lilja H, Christensson A, Dahlen U, Matikainen MT, Nilsson O, Pettersson K, Lovgren T. Prostate-specific antigen in serum occurs predominantly in complex with alpha 1-antichymotrypsin. Clin Chem. 1991;37:1618–1625. [PubMed] [Google Scholar]
- 20.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 21.Nam RK, Zhang WW, Klotz LH, Trachtenberg J, Jewett MA, Sweet J, Toi A, Teahan S, Venkateswaran V, Sugar L, et al. Variants of the hK2 protein gene (KLK2) are associated with serum hK2 levels and predict the presence of prostate cancer at biopsy. Clin Cancer Res. 2006;12:6452–6458. doi: 10.1158/1078-0432.CCR-06-1485. [DOI] [PubMed] [Google Scholar]
- 22.Nam RK, Zhang WW, Trachtenberg J, Diamandis E, Toi A, Emami M, Ho M, Sweet J, Evans A, Jewett MA, et al. Single nucleotide polymorphism of the human kallikrein-2 gene highly correlates with serum human kallikrein-2 levels and in combination enhances prostate cancer detection. J Clin Oncol. 2003;21:2312–2319. doi: 10.1200/JCO.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Klein RJ, Hallden C, Cronin AM, Ploner A, Wiklund F, Bjartell AS, Stattin P, Xu J, Scardino PT, Offit K, et al. Blood biomarker levels to aid discovery of cancer-related single-nucleotide polymorphisms: kallikreins and prostate cancer. Cancer Prev Res (Phila) 2010;3:611–619. doi: 10.1158/1940-6207.CAPR-09-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Mikolajczyk SD, Goel AS, Millar LS, Saedi MS. Expression of pro form of prostate-specific antigen by mammalian cells and its conversion to mature, active form by human kallikrein 2. Cancer Res. 1997;57:3111–3114. [PubMed] [Google Scholar]
- 25.Lovgren J, Rajakoski K, Karp M, Lundwall A, Lilja H. Activation of the zymogen form of prostate-specific antigen by human glandular kallikrein 2. Biochem Biophys Res Commun. 1997;238:549–555. doi: 10.1006/bbrc.1997.7333. [DOI] [PubMed] [Google Scholar]
- 26.Takayama TK, Fujikawa K, Davie EW. Characterization of the precursor of prostate-specific antigen. Activation by trypsin and by human glandular kallikrein. J Biol Chem. 1997;272:21582–21588. doi: 10.1074/jbc.272.34.21582. [DOI] [PubMed] [Google Scholar]
- 27.Williams SA, Xu Y, DeMarzo AM, Isaacs JT, Denmeade SR. Prostate-specific antigen (PSA) is activated by KLK2 in prostate cancer ex vivo models and in prostate-targeted PSA/KLK2 double transgenic mice. Prostate. 2010;70:788–796. doi: 10.1002/pros.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Ishihara K, Ichimura T, Fujita N, Hino S, Tomita S, Watanabe S, Saitoh N, Ito T, Nakao M. MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem. 2009;284:5165–5174. doi: 10.1074/jbc.M807098200. [DOI] [PubMed] [Google Scholar]
- 30.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.