Abstract
The CCAAT box is one of the most common cis-elements present in eukaryotic promoters and is bound by the transcription factor NUCLEAR FACTOR Y (NF-Y). NF-Y is composed of three subunits, NF-YA, NF-YB, and NF-YC. Unlike animals and fungi, plants have significantly expanded the number of genes encoding NF-Y subunits. We provide a comprehensive classification of NF-Y genes, with a separation of closely related, but distinct, histone fold domain proteins. We additionally review recent experiments that have placed NF-Y at the center of many developmental stress-responsive processes in the plant lineage.
INTRODUCTION
NUCLEAR FACTOR Y (NF-Y) transcription factors (also known as Heme-associated proteins [HAPs] and CCAAT box binding factors [CBFs]) are rapidly emerging as important regulators of numerous plant developmental and stress-induced responses. NF-Ys are sequence-specific transcription factors with histone-like subunits, with the unique characteristic that they bind DNA at CCAAT sites as heterotrimeric complexes composed of single subunits from each of three protein families: NF-YA, NF-YB, and NF-YC. In the plant lineage, each subunit type is encoded by a family of ∼10 genes, both in dicots and monocots, which differentiates them from animal systems, where there is typically only one or two genes encoding each subunit type. The expansion of NF-Y families in plants, combined with their heterotrimeric nature, means that many possible NF-Y complexes can form. This leads to the formation of a flexible, combinatorial system of transcription factors that may allow subtle adjustments to many different environmental conditions.
Because of overlapping functionality in the plant NF-Y families, our knowledge with respect to their functions has lagged somewhat compared with animal and fungal systems. Nevertheless, in recent years, numerous reports have emerged demonstrating the roles of individual subunits in many important processes. In this review, we comprehensively examine progress in understanding NF-Y functions in the plant lineage, providing an entrée for the nonexpert and a broad review for the aficionados. Additionally, to avoid further confusion with acronyms and the related, but functionally distinct, histone fold domain (HFD) proteins, we propose a reclassification of the plant NF-Y genes. For additional information/perspectives, we recommend the recent NF-Y review by Laloum et al. (2012).
CLASSIFICATION
NF-Ys and HAPs
The initial reports of NF-Y genes in plants date back to the 1990s (Li et al., 1992b; Albani and Robert, 1995; Edwards et al., 1998; Lotan et al., 1998; Kusnetsov et al., 1999). The comprehensive search for NF-Y genes in Arabidopsis thaliana (Gusmaroli et al., 2001, 2002; Yang et al., 2005; Siefers et al., 2009) was followed by similar ones in other plant species, including rice (Oryza sativa), wheat (Triticum aestivum), and Brachypodium distachyon (Stephenson et al., 2007; Thirumurugan et al., 2008; Cao et al., 2011b). It became clear that each plant NF-Y gene family has undergone significant amplification (e.g., seven NF-YC genes in rice [Thirumurugan et al., 2008] and 14 in wheat [Stephenson et al., 2007]).
Identification of members of each NF-Y family is based on the presence of highly conserved domains. NF-YA proteins have a conserved ∼56–amino acid domain that is composed of two helices (A1 and A2) with separate functions: A1 is required for subunit interactions and A2 for sequence specificity to bind DNA at CCAAT boxes. NF-YB and NF-YC each share HFDs with the core histones H2B and H2A, respectively. Mutagenesis studies in yeast and mammals confirmed that the HFD is important for NF-Y subunit interactions and DNA binding (Romier et al., 2003 and references therein). Currently, the various NF-Y subunits of rice (and other species, although less fully adopted) are often referred to by the yeast HAP2/3/5 nomenclature (homologs of NF-YA, NF-YB, and NF-YC, respectively; Miyoshi et al., 2003; Yazawa and Kamada, 2007; Thirumurugan et al., 2008; Xie et al., 2008; Yu et al., 2011). This has created confusion and difficulty following the literature in three principal ways: (1) To the best of our knowledge, plants are devoid of homologs of HAP4, which is required for transcriptional activation in fungi (McNabb and Pinto, 2005; Kato, 2005), and they form a mammalian-like trimer. (2) Most genetic experiments and two thorough biochemical studies were performed on Arabidopsis, adhering to the NF-Y terminology (Gusmaroli et al., 2001, 2002; Siefers et al., 2009). (3) The phylogenetically unrelated HAPLESS mutants of Arabidopsis are also known as hap (Johnson et al., 2004), which precludes changing the Arabidopsis nomenclature. Thus, where the HAP terminology is used, we propose to reclassify these genes following the At-NF-Y nomenclature (Table 1).
Table 1. Classification of Rice NF-Y Genes.
| Os-NF-YA Family | ||
|---|---|---|
| NF-Y Name | Locus Name | Other Names |
| NF-YA1 | Os03g07880 | HAP2C |
| NF-YA2 | Os03g29760 | HAP2E |
| NF-YA3 | Os03g44540 | HAP2H |
| NF-YA4 | Os03g48970 | HAP2D |
| NF-YA5 | Os07g06470 | HAP2J |
| NF-YA6 | Os07g41720 | HAP2G |
| NF-YA7 | Os08g09690 | HAP2A |
| NF-YA8 | Os10g25850 | HAP2I |
| NF-YA9 | Os12g41880 | HAP2B |
| NF-YA10 | Os12g42400 | HAP2F |
| Os-NF-YB Family | ||
| NF-Y Name | Locus Name | Other Names |
| NF-YB1 | Os02g49410 | HAP3K |
| NF-YB2 | Os01g61810 | HAP3A |
| NF-YB3 | Os05g38820 | HAP3B |
| NF-YB4 | Os05g49780 | HAP3C |
| NF-YB5 | Os01g70880 | HAP3J |
| NF-YB6 | Os01g70890 | HAP3G |
| NF-YB7 | Os02g49370 | HAP3E, L1L |
| NF-YB8 | Os03g29970 | HAP3I |
| NF-YB9 | Os06g17480 | HAP3D |
| NF-YB10 | Os07g41580 | HAP3F |
| NF-YB11 | Os08g07740 | HAP3H, Hd5, DTH8, Ghd8 |
| Os-NF-YC Family | ||
| NF-Y Name | Locus Name | Other Names |
| NF-YC1 | Os02g07450 | HAP5A |
| NF-YC2 | Os03g14669 | HAP5C |
| NF-YC3 | Os04g58680 | HAP5G |
| NF-YC4 | Os06g45640 | HAP5B |
| NF-YC5 | Os08g10560 | HAP5F |
| NF-YC6 | Os08g38780 | HAP5D |
| NF-YC7 | Os09g30310 | HAP5E |
We also propose to maintain use of the current nomenclature on Arabidopsis LEAFY COTYLEDON1 (At-LEC1) and LEC1-LIKE (L1L; At-NF-YB9 and At-NF-YB6, respectively). This suggestion comes from the fact that LEC1 and L1L were identified before the original Arabidopsis classification and many studies refer to this nomenclature (Zhang et al., 2002; Yazawa et al., 2004; Alemanno et al., 2008; Schellenbaum et al., 2008; Maillot et al., 2009; Salvini et al., 2012). We suggest that reporting on these proteins include mention of the NF-Y names to assist nonexpert readers in understanding the family connection.
Relationship between NF-Y, NC2, and Dpb3/4 Genes
A second level of misunderstanding has been generated by the inclusion of plant homologs of NC2 and Dpb3/Dpb4 within the NF-Y gene family. These genes collectively form a subfamily of H2A/H2B-like genes, with high primary sequence identity, 30/35%, reflected in structural similarities (Kamada et al., 2001; Romier et al., 2003; Hartlepp et al., 2005). It is therefore understandable that plant NC2s and Dpb3/4 might be included in a common family. In fact, these genes are more closely related to NF-Ys than other HFD proteins. Nevertheless, it has been shown that some of the classified Arabidopsis, wheat, and B. distachyon genes are outliers in the phylogenetic analyses of NF-Y proteins (Stephenson et al., 2007; Siefers et al., 2009; Cao et al., 2011b). This point is relevant, as the two related HFD groups NC2 and Dpb3/4 are not functionally overlapping with NF-Y; NC2 associates with TBP to bind TATA boxes in core eukaryotic promoters (Kamada et al., 2001), whereas Dpb3/4 are involved in complexes with DNA Pol ε and the chromatin remodeling complex Chrac (Hartlepp et al., 2005).
In summary, we propose a return to the original classification of 29 bona fide At-NF-Y proteins with the addition of At-NF-YC12 (Gusmaroli et al., 2001, 2002; Siefers et al., 2009). This nomenclature also serves to close a gap, as both NC2 and Dpb3/Dpb4 genes are highly conserved from yeast to mammals. It is now clear that At-NC2 genes were duplicated and At-Dpb3/4 were not expanded in plants (Table 2). Thus, plant NF-Y genes serve a far greater role in gene regulatory processes, leading to variable plant developmental pathways.
Table 2. Reclassification of Plant HFD Proteins.
| Former NF-Y Name | New Name | Locus ID |
|---|---|---|
| At-NF-YC11 | At-NC2α | AT3G12480 |
| At-NF-YB12 | At-NC2β1 | AT5G08190 |
| At-NF-YB13 | At-NC2β2 | AT5G23090 |
| Bd-NF-YC4 | Bd-NC2α1 | Bradi2g21290 |
| Bd-NF-YC11 | Bd-NC2α2 | Bradi4g16840 |
| Bd-NF-YB16 | Bd-NC2β1 | Bradi3g34930 |
| Ta-NF-YC6 | Ta-NC2α1 | TC233433 |
| Ta-NF-YC8 | Ta-NC2α2 | TC241235 |
| Ta-Dr1A | Ta-NC2β1 | AF464903_1 |
| Ta-Dr1B | Ta-NC2β2 | TC416575 |
| At-NF-YC10 | At-Dpb3-1 | AT1G07980 |
| At-NF-YC13 | At-Dpb3-2 | AT5G43250 |
| At-NF-YB11 | At-Dpb4 | AT2G27470 |
| Ta-NF-YC13 | Ta-Dpb3-1 | BJ308764 |
| Ta-NF-YC14 | Ta-Dpb3-2 | TC270995 |
| Bd-NF-YC1 | Bd-Dpb3 | Bradi1g01300 |
| Bd-NF-YB3 | Bd-Dpb4 | Bradi1g43470 |
FUNCTIONAL STUDIES OF NF-Y GENES
Expression of NF-Y Subunits
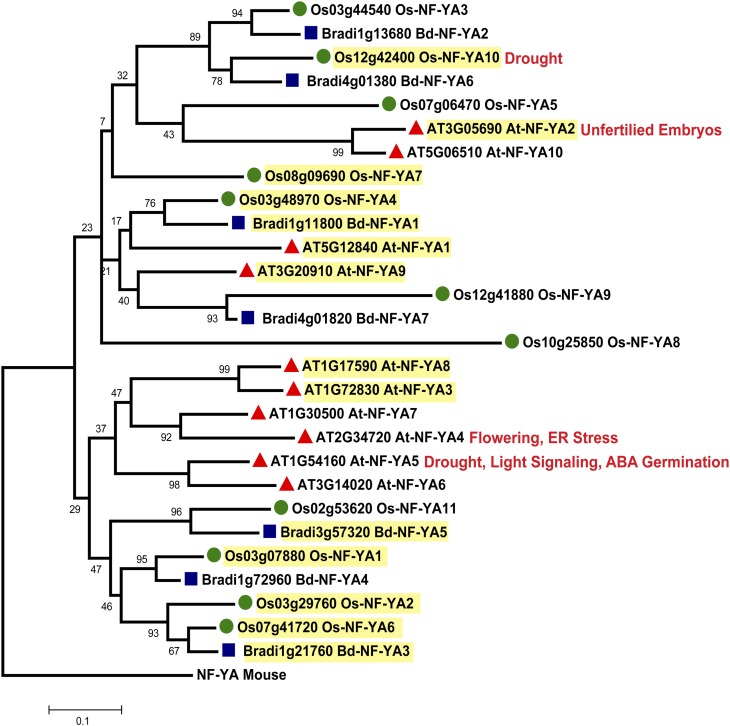
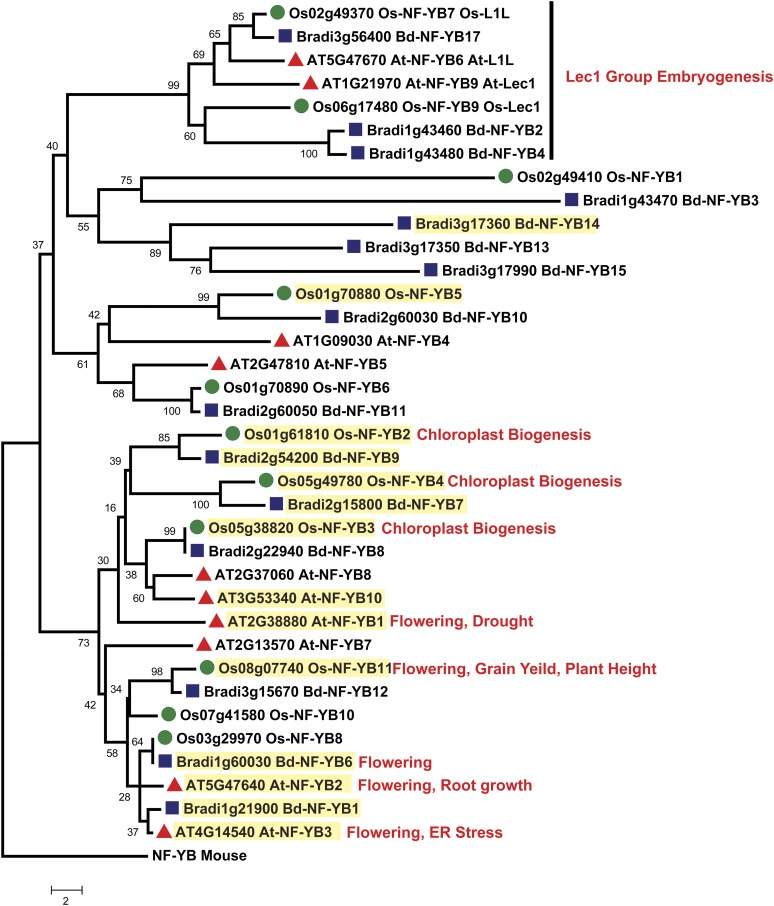
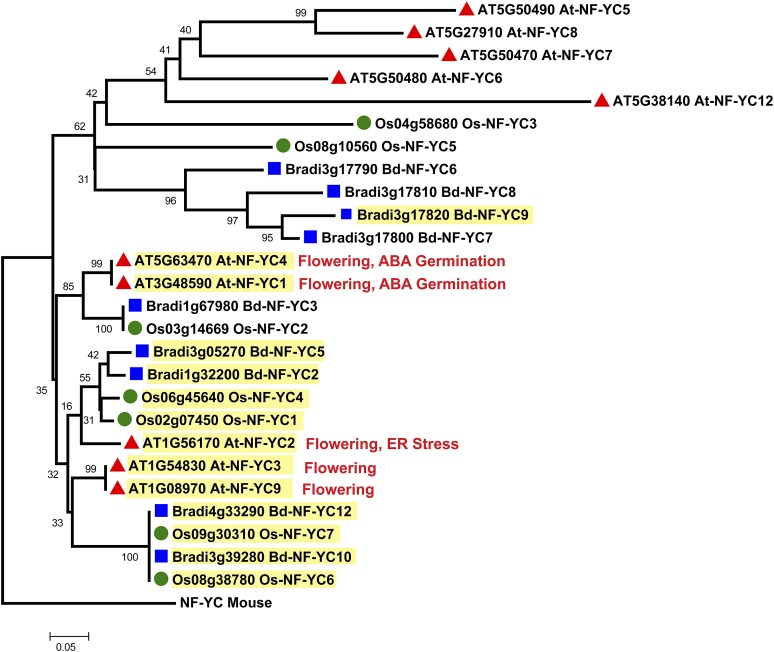
The expansion of plant NF-Y gene families has led to the intimidating task of identifying biologically relevant NF-Y complexes. To begin sorting through the many possible NF-Y heterotrimers, several groups have characterized NF-Y mRNA expression patterns for diverse plant species. Organ-specific mRNA expression patterns and phylogenetic relationships have been systematically established for Arabidopsis, B. distachyon, rice, and wheat (Gusmaroli et al., 2001, 2002; Stephenson et al., 2007; Thirumurugan et al., 2008; Cao et al., 2011b), although the Triticum phylogenetic classifications and database annotations require further input to make the published analyses broadly useful. Here, we developed phylogenetic trees for each NF-Y family comparing Arabidopsis, B. distachyon, and rice full-length proteins (Figure 1, Figure 2, and Figure 3; Supplemental Data Sets 1, 2, and 3 online; Laloum et al. [2012] provide trees comparing Arabidopsis, soybean [Glycine max], Medicago truncatula, and rice). Due to the high amino acid identity among family members, even across the dicot/monocot split, the bootstrap support for some clades is quite low (<70%); thus, these trees are not necessarily accurate for lineage inferences. Nevertheless, we opted not to collapse these branches because they provide potentially useful information for functional inferences. In fact, despite the lack of strong bootstrap support, similar trees have been useful for identifying likely NF-Y candidates involved in flowering (Kumimoto et al., 2010; Cao et al., 2011b). Based on these data, no strong patterns connecting NF-Y phylogenetics and reported expression patterns emerge, but NF-Y expression can be broken down into two very general classes: genes that are broadly expressed (found in nearly all tissues tested) and genes with more restricted or organ-specific expression patterns (see highlighting on Figure 1, Figure 2, and Figure 3). To date, most NF-Y phenotypes have been associated with the broadly expressed subunits, with a notable exception being the narrowly expressed LEC class (Figure 2).
Figure 1.
Phylogenetic Tree of Arabidopsis, B. distachyon, and Rice NF-YA Subunits.
For each family of NF-Y proteins, multiple sequence alignments were generated using ClustalW (Thompson et al., 2002) on full-length proteins as implemented by Molecular Evolutionary Genetics Analysis (MEGA) software, version 5.0 (Tamura et al., 2011; note that multiple alignments using MUSCLE [Edgar, 2004] generated essentially identical phylogenetic trees). Phylogenetic trees were constructed by neighbor joining with complete deletions as implemented by MEGA. Reliability values at each branch represent bootstrap samples (5000 replicates). Bootstrap values below 70% are not considered reliable for phylogenetic inferences, although trees are not collapsed at this value (see explanation in main text). The mouse NF-YA subunit was used to root the tree. Yellow highlighted genes are broadly expressed in all RT-PCR experiments. Documented phenotypes for a given gene are in red text.
Figure 2.
Phylogenetic Tree of Arabidopsis, B. distachyon, and Rice NF-YB Subunits.
See Figure 1 legend.
Figure 3.
Phylogenetic Tree of Arabidopsis, B. distachyon, and Rice NF-YC Subunits.
See Figure 1 legend.
In addition to RT-PCR data, tissue-specific expression has also been examined for each of the At-NF-Y genes using promoter:β-glucuronidase (pNF-Y:GUS) reporter gene fusions (Siefers et al., 2009). These tissue-specific expression patterns have proven valuable for identifying NF-Y involved in photoperiod-dependent flowering, which is regulated by interactions between NF-Y subunits and the floral promoting protein CONSTANS (CO) (Kumimoto et al., 2010). Because CO function in flowering time requires vascular expression in leaves (An et al., 2004), the pNF-Y:GUS reporter lines were instrumental for identifying three vascular expressed At-NF-YC involved in the control of photoperiod-dependent flowering (Kumimoto et al., 2010; see below).
Adding to surveys of organ- and tissue-specific expression patterns, several studies have systematically examined NF-Y responses to various environmental conditions and stresses (Stephenson et al., 2007, 2010, 2011; Hackenberg et al., 2012), and numerous studies have implicated NF-Y in regulating photosynthesis and drought responses (Kusnetsov et al., 1999; Miyoshi et al., 2003; Nelson et al., 2007; Li et al., 2008; Stephenson et al., 2010, 2011). Analysis of public microarray data also points to possible NF-Y functions related to photosynthesis, flowering time, and drought. Analysis of cell-specific transcriptome data from rice revealed the CCAAT box element is associated with cell-specific transcripts in both leaf (mesophyll, vein, and primordia) and dehydration-response data sets (Jiao et al., 2009). Finally, it was recently shown that upregulation of Arabidopsis NF-Y is correlated with leaf senescence (Breeze et al., 2011). Together, these expression data sets, along with publicly available microarrays, serve as a basic entrée point to NF-Y analyses and can be used to narrow down the number of candidates acting in a particular developmental stage or tissue type.
Embryo and Plant Development
Our initial knowledge on developmental processes regulated by NF-Y genes came from studies on the embryogenesis regulators At-LEC1 and L1L (At-NF-YB9 and NF-YB6, respectively; Meinke, 1992; West et al., 1994; Parcy et al., 1997; Lotan et al., 1998; Kwong et al., 2003; Gaj et al., 2005; Braybrook and Harada, 2008; Junker et al., 2012). Orthologs of Arabidopsis LEC and L1L have since been identified and studied in several monocot and dicot species, as well as conifers, where they consistently regulate the transition from embryo to adult status: lec1 mutants display pleiotropic phenotypes, including abnormal development of trichomes on cotyledons, desiccation intolerance, abnormal suspensors, and seed defects in starch, protein, and lipid accumulation (Zhang et al., 2002; Yazawa et al., 2004; Fambrini et al., 2006; Alemanno et al., 2008; Mu et al., 2008; Schellenbaum et al., 2008; Maillot et al., 2009; Shen et al., 2010; Cao et al., 2011b; Tan et al., 2011; Uddenberg et al., 2011; Salvini et al., 2012). Recent evidence suggests that LEC1 control extends beyond the embryo to etiolation responses in young seedlings (Junker et al., 2012). Furthermore, using a combination of dexamethasone-inducible LEC1, chromatin immunoprecipitation, and microarray analyses, these authors implicated LEC1 as a general integrator of light and hormone signaling during embryogenesis. To our knowledge, this article is also the first to show a clear enrichment for CCAAT boxes as targets of NF-Y transcriptional action in plants, a basic and long accepted expectation from the animal literature.
From an agronomic standpoint, overexpression of LEC1 or L1L in various species can result in significant changes in seed lipids/oils (Mu et al., 2008; Shen et al., 2010; Tan et al., 2011). The specific genes involved are currently unknown, but there is a surprising parallel with the mammalian system, in which sterol regulatory element binding proteins, the master transcription factor regulators of fatty acids and cholesterol metabolisms, team up with NF-Y to activate hundreds of genes that fine-tune lipid levels (Reed et al., 2008). It will be interesting to assess whether these pathways are conserved in plants. Interestingly, LEC1 and L1L, but not several other tested At-NF-YB subunits, can activate seed specific promoters via interactions with the abscisic acid response element binding transcription factor bZIP67 (Yamamoto et al., 2009). Finally, in rice, Os-NF-YB7/L1L has roles during both vegetative and floral meristem development. Os-NF-YB7/L1L1–overexpressing plants have a dwarf phenotype and erected leaves, as well as a dense panicle, abnormal rachis, and double flowers (Ito et al., 2011).
In addition to At-LEC1/NF-YB9 and At-L1L/NF-YB6, other NF-Y genes play roles in vegetative and reproductive development (Table 3). Recent evidence suggests overlapping roles for At-NF-YA1, 5, 6, and 9 during numerous stages of embryogenesis (Mu et al., 2012), and NF-YA loss of function can be embryo lethal (Pagnussat et al., 2005). Overexpression of At-NF-YB2 enhances primary root elongation due to a faster cell division and/or elongation (Ballif et al., 2011). In the model legume Medicago truncatula, Mt-HAP2-1 is expressed in the root nodule meristematic zone and is essential for the differentiation of root nodule cells (Combier et al., 2006). Spatial and temporal expression of this gene (and NF-YA subunits in general across plant lineages) is controlled by microRNA 169 (miR169; Jones-Rhoades and Bartel, 2004) as well as a small peptide encoded by its own 5′ leader sequence (Combier et al., 2008). In Arabidopsis, miR169 is also strongly downregulated in response to nitrogen starvation, and this is correlated with the concomitant induction of multiple NF-YA family members, changes that are at least correlated to nitrogen sensitivity through the analysis of transgenic plants overexpressing miR169 (Zhao et al., 2011). Studies of Pv-NF-YC1 overexpression and RNA silencing further implicate NF-Ys in nodule organogenesis for the legume Phaseolus vulgaris (Zanetti et al., 2010). Additionally, overexpression of Pv-NF-YC1 is linked to the selection of Rhizobium etli strains during nodulation. An NF-YA from Brassica napus (Bn-CBF-B) has a function in reproductive tissues: Plants expressing an antisense Bn-CBF-B (NF-YA) in the anthers show reduced quantity of viable pollen due to degeneration of the tapetal cell layer and reduced female fertility (Lévesque-Lemay et al., 2003). Finally, Yu et al. (2011) showed a novel function of an NF-YC gene from Picea wilsonii that has a role in regulation of pollen tube growth orientation.
Table 3. Summary of Genotype/Phenotype Correlations.
| Gene | Other Name | Organism | Function | References |
|---|---|---|---|---|
| Dicots | ||||
| At-NF-YA5 | Arabidopsis | Drought resistance | Li et al. (2008) | |
| At-NF-YB1 | Arabidopsis | Drought-related stress tolerance | Nelson et al. (2007) | |
| At-NF-YB2 | At-HAP3b | Arabidopsis | Promotion of flowering, root growth | Chen et al. (2007); Kumimoto et al. (2008); Ballif et al. (2011) |
| At-NF-YB3 | Arabidopsis | Promotion of flowering | Chen et al. (2007); Kumimoto et al. (2008) | |
| At-L1L | At-NF-YB6 | Arabidopsis | Embryo development | Kwong et al. (2003); Lee et al. (2003); Yamamoto et al. (2009); Le et al. (2010) |
| Vv-L1L | Vitis vinifera | Somatic embryogenesis | Schellenbaum et al. (2008); Maillot et al. (2009) | |
| Tc-L1L | Theobroma cacao | Partial rescue of lec1 mutant | Alemanno et al. (2008) | |
| Ha-L1L | Helianthus | Zygotic and in somatic | Fambrini et al. (2006); Chiappetta et al. (2009); Salvini et al. (2012) | |
| annuus | embryogenesis | |||
| At-LEC1 | At-NF-YB9 | Arabidopsis | Early and late embryogenesis | Meinke et al. (1992); Kagaya et al. (2005); Casson et al. (2006); Mu et al. (2008); Junker et al. (2012) |
| Dc-C-LEC1 | Daucus carota | Embryo development; complements lec1 mutant | Yazawa et al. (2004); Yazawa and Kamada. (2007) | |
| Bn-LEC1 Bn-L1L | B. napus | Seed oil production | Tan et al. (2011) | |
| At-NF-YC1 | Arabidopsis | Floral induction | Hackenberg et al. (2012) | |
| At-NF-YC2 | Arabidopsis | Floral induction, ER stress | Liu and Howell (2010); Hackenberg et al. (2012); | |
| At-NF-YC4 | Arabidopsis | Germination | Warpeha et al. (2007 Liu and Howell (2010) | |
| At-NF-YC3 | Arabidopsis | Flowering | Kumimoto et al. (2010 Liu and Howell (2010) | |
| At-NF-YC4 | ||||
| At-NF-YC9 | ||||
| Mt-HAP2-1 | M. truncatula | Root nodule development | Combier et al. (2006); Liu and Howell (2010) | |
| Bn-CBF-B | B. napus | Development of tapetal cell layer of anthers and female fertility | Lévesque-Lemay et al. (2003); Liu and Howell (2010) | |
| Sl-THAP5a | S. lycopersicum | Flowering | Ben-Naim et al. (2006) | |
| Pv-NF-YC1 | P. vulgaris | Root nodule development | Zanetti et al. (2010) | |
| Monocots | ||||
| Zm-NF-YB2 | Z. mays | Drought tolerance in maize | Nelson et al. (2007) | |
| Zm-LEC1 | Z. mays | Embryogenesis and seed oil production | Zhang et al. (2002); Shen et al. (2010) | |
| Os-NF-YB2 | Os-HAP3A | O. sativa | Chloroplast biogenesis | Miyoshi et al. (2003) |
| Os-NF-YB3 | Os-HAP3B | |||
| Os-NF-YB4 | Os-HAP3C | |||
| Os-NF-YB11 | Os-HAP3H, | O. sativa | Photoperiodic flowering | Wei et al. (2010) |
| DTH8, Ghd8 | ||||
| Os-L1L | Os-HAP3E | O. sativa | Floral meristem identity | Ito et al. (2011) |
| Ta-NF-YB2 | T. aestivum | Drought adaptation | Stephenson et al. (2007) | |
| Ta-NF-YB3 | T. aestivum | Positive regulation of photosynthesis genes | Stephenson et al. (2011) | |
| Hv-NF-YB1 | H. vulgare | Overexpression results in earlier flowering in Arabidopsis | Liang et al. (2012) | |
| Bd-NF-YB6 | B. distachyon | Overexpression results in earlier flowering in Arabidopsis and rescue of the late-flowering phenotype in nf-yb2 nf-yb3 mutants | Cao et al. (2011b) | |
| Conifers | ||||
| Pw-HAP5 | P. wilsonii | Pollen tube development and control of tube orientation | Yu et al. (2011) | |
| Pa/Ps-HAP3A, HAP3B | Picea abies, Pinus silvestris | Embryogenesis | Uddenberg et al. (2011) | |
Flowering Time
An important discovery is the involvement of NF-Y genes in the control of photoperiod-dependent flowering time (Ben-Naim et al., 2006; Wenkel et al., 2006). At-NF-YB2 and At-NF-YB3 are necessary for the promotion of flowering in response to inductive long-day photoperiodic conditions (Cai et al., 2007; Kumimoto et al., 2008). At-NF-YB2 and At-NF-YB3 act through the activation of the key floral regulator FLOWERING LOCUS T (FT), a gene responsible for the vegetative to floral meristem conversion (Samach et al., 2000; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007). At-NF-YB2 and At-NF-YB3 interact in vivo with At-NF-YC3, At-NF-YC4, and At-NF-YC9, which are also required for flowering (Kumimoto et al., 2010). Additionally, At-NF-YC1 and At-NF-YC2 can activate flowering when overexpressed, which is additionally correlated with increased FT transcript levels (Hackenberg et al., 2012). Importantly, NF-YB and NF-YC subunits physically interact with CO, a key regulator of photoperiod-induced flowering time, via the conserved CCT (for CO, CO-Like, and TOC1) domain (Ben-Naim et al., 2006; Wenkel et al., 2006; see section 3). The interaction between NF-YC and CO-like proteins is conserved in tomato (Solanum lycopersicum), and overexpression of THAP5a (an NF-YC protein) in Arabidopsis accelerates flowering time (Ben-Naim et al., 2006).
In the absence of NF-Y, overexpressed CO is either unable or strongly reduced in its ability to activate FT and drive early flowering (Kumimoto et al., 2010). The converse is also true; in the absence of CO, overexpressed NF-Ys do not effectively activate flowering (Tiwari et al., 2010), but recent work showed that overexpression of At-NF-YB2 fused to a novel activation domain can activate flowering in a co null mutant (Tiwari et al., 2012). These data support a model in which NF-Y complexes provide a DNA binding component to CO, although this simple interpretation does not address the recent finding that CO can bind DNA directly at TGTG(N2-3)ATG sites (Tiwari et al., 2010). Blackman and Michaels (2010) proposed a mixed model where CO can interact with the FT promoter via direct DNA interactions and/or through interactions with NF-Y complexes (i.e., NF-Ys act as a binding platform). These possibilities are supported by the presence of multiple CCAAT and TGTG(N2-3)ATG sites throughout the functionally defined minimal FT promoter (Adrian et al., 2010).
NF-Y function in flowering time has also been reported in monocots. In particular, rice NF-YB11 (also called DTH8/Ghd8/LHD1) probably plays a suppressive role in the signal network of photoperiodic flowering by downregulating the expression of several floral regulators during noninductive long-day conditions (rice is a short day flowering plant; Wei et al., 2010; Yan et al., 2011; Dai et al., 2012). It was also demonstrated that overexpression of B. distachyon NF-YB6 (Cao et al., 2011b) and barley (Hordeum vulgare) NF-YB1 (Liang et al., 2012), orthologs of At-NF-YB2/3, resulted in significantly earlier flowering in Arabidopsis. Furthermore, Bd-NF-YB6 could rescue late-flowering nf-yb2 nf-yb3 mutants (Cao et al., 2011b). Finally, in wheat, interactions between the CCT domains of VRN2 and CO with NF-Ys integrate seasonal vernalization and photoperiod signals, providing a flexible combinatorial system to connect multiple developmental and environmental signals in the regulation of flowering initiation in cereals (Li et al., 2011).
Plant–Environment Interactions
NF-Ys have been identified as regulators of drought tolerance in different plant species. Transgenic Arabidopsis and maize (Zea mays) plants overexpressing At-NF-YB1 and the maize ortholog Zm-NF-YB2, respectively, have improved performance and survival under drought conditions (Nelson et al., 2007). Microarray data suggest At-NF-YB1 does not transcriptionally regulate the dehydration-responsive element binding proteins or the abscisic acid (ABA)–dependent drought tolerance pathway, suggesting it may act through a novel drought resistance pathway. Nevertheless, no alternative mechanism has been proposed, and we note increasing evidence for NF-Y/bZIP interactions and the fact that bZIP proteins are well known to be involved in ABA signaling.
At-NF-YA5 is also involved in drought resistance; its expression is strongly induced by drought, as well as osmotic and salt stresses. At-NF-YA5 is transcriptionally regulated through an ABA-dependent mechanism and posttranscriptionally regulated by miR169. miR169 is downregulated by drought stress through an ABA-dependent pathway. This dual regulation is consistent with the critical importance of At-NF-YA5 for drought resistance. In stomatal guard cells, At-NF-YA5 expression regulates aperture size. In nonguard cells, At-NF-YA5 is likely important for dehydration tolerance via activating stress-responsive genes, such as genes involved in oxidative stress responses (Li et al., 2008). In rice, an NF-YA gene (Os-HAP2E) has been identified as a target gene of miR169, which is induced by high salinity and is probably regulated by an ABA-dependent pathway, since ABA response elements have been found in its promoter (Zhao et al., 2009).
Drought-related expression has been systematically examined for NF-Y from wheat and Arabidopsis (Stephenson et al., 2007; Hackenberg et al., 2012), as well a more limited study of five NF-YB genes in barley (Liang et al., 2012). Furthermore, in addition to its role in flowering time, At-NF-YB2 is also upregulated by osmotic stress (Chen et al., 2007). While many At-NF-Ys were upregulated by drought stress, none were downregulated (Hackenberg et al., 2012). The opposite is true in wheat (Stephenson et al., 2007), pointing to a possible functional divergence between monocots and dicots. The NF-YA family is targeted by the stress-responsive miR169 (Reinhart et al., 2002; Kidner and Martienssen, 2005; Jones-Rhoades et al., 2006; Reynoso et al., 2012; Leyva-González et al., 2012). In rice, miR169 family members were shown to be upregulated under drought and salt stress conditions (Zhao et al., 2007, 2009). This was in contrast with Arabidopsis, where miR169 members were downregulated (Li et al., 2008), further supporting the possible functional divergence of NF-Y in monocots and dicot during stress responses.
The endoplasmic reticulum (ER) stress response is phylogenetically conserved. In mammals, gene expression programs are promoted by several transcription factors, including ATF6 (Bailey and O’Hare, 2007; Yoshida, 2007), a bZIP transcription factor that binds to ER stress–responsive elements. ATF6 recruitment requires previous binding of NF-Y to a nearby CCAAT box (Kabe et al., 2005). In Arabidopsis, NF-YC2, NF-YA4, and NF-YB3 form a transcriptional complex with bZIP28 that upregulates ER stress-induced genes (Liu and Howell, 2010). At-NF-YC2 is also strongly induced in response to photodynamic, light, oxidative, heat, and drought stresses, and a tobacco (Nicotiana tabacum) NF-YC was found to be inducible by photooxidative stress. Despite the stress induction, At-nf-yc2 mutants and At-NF-YC2 overexpressors did not show phenotypic differences compared with wild-type seedlings in response to photooxidative stress, possibly due to the compensatory potential of other members of the At-NF-YC family (Hackenberg et al., 2012).
Several studies have suggested that the NF-Y family is involved in light-mediated gene regulation (Kusnetsov et al., 1999; Miyoshi et al., 2003; Warpeha et al., 2007). Kusnetsov et al. (1999) demonstrated that assembly of the NF-Y complex at the CCAAT box in the spinach (Spinacia oleracea) AtpC promoter is regulated by light and rice plants with antisense or RNA interference constructs of Os-HAP3A (Os-NF-YB2) have reduced leaf chlorophyll content and degenerated chloroplasts. Furthermore, three NF-YB proteins (Os-HAP3A-C/Os-NF-YB2/3/4) regulate a number of photosynthesis genes, including chlorophyll a/b binding protein (CAB) and the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (Miyoshi et al., 2003). In Arabidopsis, NF-YA5 and LEC1 (NF-YB9) are also involved in the regulation of CAB expression in response to blue light and ABA (Warpeha et al., 2007). Transgenic wheat lines constitutively overexpressing Ta-NF-YB3 have a significant increase in leaf chlorophyll content, photosynthesis rate, and early growth rate (Stephenson et al., 2011). Additionally, many Ta-NF-YCs are regulated by light (Ta-NF-YC3,Ta-NF-YC5, Ta-NF-YC8, Ta-NF-YC9, Ta-NF-YC11, and Ta-NF-YC12), and Ta-NF-YC11 is important in the regulation of photosynthesis-related genes (Stephenson et al., 2010).
In summary, in addition to the original role in embryogenesis discovered for the LEC1/L1L branch of NF-YBs, genetic experiments are pinpointing several other NF-Y genes specifically involved in single pathways. In parallel, biochemical work is starting to unveil the possible molecular mechanisms.
BIOCHEMISTRY
Interactions of NF-Y Subunits
The presence of 30 NF-Y genes in the Arabidopsis genome could result in the formation of ∼1000 alternative heterotrimeric combinations, and an obvious question is whether there is specificity in those interactions and in DNA binding.
A first important concept is that NF-YA binds only to a preformed HFD dimer: Selected HFD interactions have been initially reported (Warpeha et al., 2007; Yazawa and Kamada, 2007; Thirumurugan et al., 2008; Kumimoto et al., 2010; Liu and Howell, 2010), and two recent systematic studies employed yeast two-hybrid (Y2H) assays (Hackenberg et al., 2011; Calvenzani et al., 2012). These studies agreed on most interactions, with some limited preferences and reduced affinities for some of the heterodimers. In general, yeast assays should be interpreted with great caution, particularly because of the presence of endogenous yeast HAPs, shown to be able to interact with mammalian and plant NF-Ys (Chodosh et al., 1988; Hooft van Huijsduijnen et al., 1990; Ben-Naim et al., 2006; Yazawa and Kamada, 2007; Kumimoto et al., 2008).
Importantly, in accordance with the new classification proposed here, At-NF-YC12 interacts with all At-NF-YBs, but not with At-Dpb4 (formerly NF-YB11), while At-Dpb3-2 (formerly At-NF-YC13) does not interact with any of the At-NF-YBs (Hackenberg et al., 2011). Note that At-NC2α (formerly At-NF-YC11) shows interactions with At-NF-YB2/3 (Hackenberg et al., 2011), and Y2H library screens using At-NF-YB2 and At-NF-YB3 as bait occasionally isolate At-NC2α1/2 (B.F. Holt and R.W. Kumimoto, unpublished data), so there may remain some potential for cross-reactivity between families.
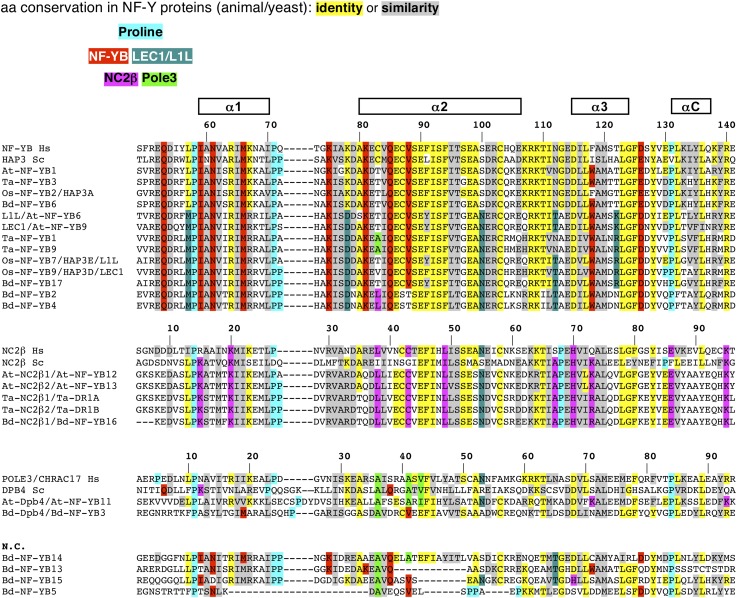
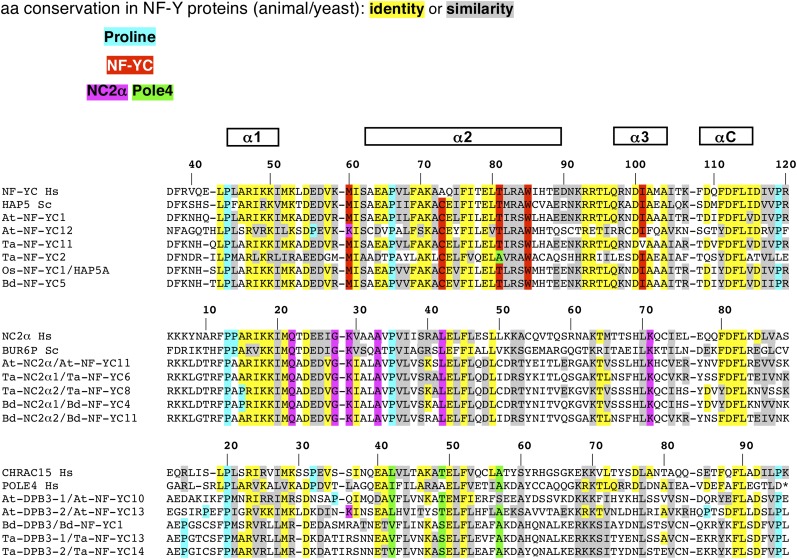
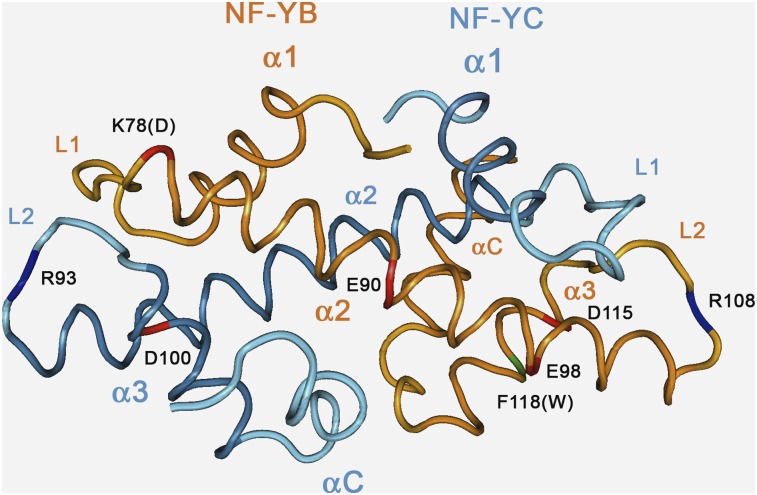
It is safe to conclude that At-NF-YBs and At-NF-YCs can interact promiscuously with each other, a result expected from our knowledge of the mouse HFD dimer crystal structure (Romier et al., 2003). Important hydrophobic amino acid contacts in helices α2 of the two HFDs are well conserved, as is the α1 helix of NF-YBs, whose hydrophobic interactions are stacked against the perfectly conserved Trp-85 of NF-YC helix α2 (Figure 4, Figure 5, and Figure 6). Additionally, bidentate salt bridges between Arg-108 and Asp-115 of NF-YB and Arg-93 and Asp-100 of NF-YC, which are perfectly conserved in all plant genes, are absent in the other HFDs (Figure 4, Figure 5, and Figure 6 ). The HFD side of the trimerization surface relies on selected residues in the α2 helix of NF-YB and in the αC helix of NF-YC. Glu-90 and Glu-98 of mouse NF-YB are important for NF-YA binding and conserved in all At-NF-YBs. In NF-YC, some members have notable modifications in the crucial αC helix, such as Arg-109 (At-NF-YC5), two Asp residues at 111-112 and Ile-113 (At-NF-YC8); At-NF-YC7 has a four amino acids addition in the α3 helix, which extends it for an additional turn, thus displacing the loop (LC) and αC helix. Note that these proteins interact with At-NF-YA6 in vitro (Calvenzani et al., 2012).
Figure 4.
Protein Sequence Alignments of Conserved Domains of NF-YB, NC2β, and Dpb4.
Protein sequence alignments of NF-YB and putative NF-YB, NC2β, and Dpb4 conserved domains. Alignments were performed using the ClustalW program (Thompson et al., 2002). Amino acid residues conserved among listed proteins are shaded and gaps (dashes) were introduced for maximum matching. Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; At, Arabidopsis; Bd, B. distachyon; Ta, T. aestivum; Os, O. sativa.
Figure 5.
Protein Sequence Alignments of Conserved Domains of NF-YC, NC2α, and Dpb3.
Protein sequence alignments of the conserved domains as in Figure 4.
Figure 6.
Structure of the Human NF-Y HFD Subunit Dimer.
Ribbon representation of human NF-YB/NF-YC dimer structure (Protein Data Bank code: 1N1J) and location of selected residues, determined by Romier et al. (2003). NF-YB and NF-YC main chains traces are represented in orange and cyan color, respectively. Secondary structure elements are labeled, and main chain location of selected relevant residues described in the text (human numbering) is highlighted in blue (positive) or red (negative) for side-chain charge. NF-YB Lys-78 (Asp in LEC1/L1L proteins) is colored in red. Human NF-YB Phe-118 (Trp in plants) is highlighted in green.
A second level of variability is represented by formation of the trimer. The NF-YA side of the trimerization surface is in the A1 helix. Many At-NF-YAs are able to interact with HFD subunits (Liu and Howell, 2010; Hackenberg et al., 2011; Calvenzani et al., 2012). At-NF-YA4 showed no interaction with the At-L1L/ NF-YC2 dimer in both ABA-mediated recruitment assays and electrophoretic mobility shift assays (EMSAs; Yamamoto et al., 2009; Calvenzani et al., 2012), but a positive interaction was reported from EMSA with At-NF-YB3/At-NF-YC2 (Liu and Howell, 2010). Further biochemical work on At-NF-YA4 is required to verify its trimerization and DNA binding specificity. Overall, the lack of impediment to subunit association, valid for the HFD dimers, should also apply for trimerization. In summary, highly selective NF-Y complexes are very likely the exception rather than the rule.
DNA Binding
Another fundamental question is whether all NF-Y trimers bind DNA with the same specificity as their mammalian and yeast orthologs (Mantovani R., 1998). In general, many of these genes are so similar to mammalian NF-Y, it seems highly likely that they also exhibit DNA binding. The HFD dimer contributes to DNA binding through the α1 helices and the L1 and L2 loops (Romier et al., 2003, and references therein), with residues equivalent to those in H2A/H2B that contact the DNA phosphate backbone in the nucleosome (Luger et al., 1997). Specifically, all the “right” residues are found in the right place in all At-NF-YBs with two possible exceptions, LEC1/NF-YB9 and L1L/NF-YB6. Experiments with chimeric constructs demonstrated that the HFD domain is necessary and sufficient for LEC1 function in embryos, specifically pinpointing Asp-55 as a crucial residue (Lee et al., 2003; Figure 4 and Figure 6; Asp-55 corresponds to human Lys-78). L1L also has an Asp at this signature position for the subfamily. Asp-55 is located at the beginning of the α2 helix in a region that lies on the surface of the dimer; most other Arabidopsis and mammalian NF-YBs have a Lys (or Arg), as in H2B, predicted to be involved in protein–DNA interactions based on the nucleosome model (Luger et al., 1997; Romier et al., 2003). In theory, the change in charge might abolish DNA binding, but this is not the case, since LEC1/ NF-YC3 and L1L/ NF-YC3 bind to CCAAT with efficiencies that are marginally lower than the mammalian homolog (Calvenzani et al., 2012). As for At-NF-YCs, only two members were negative in CCAAT-based EMSAs, NF-YC7 and NF-YC8, possibly because of a lack of NF-YA interactions (discussed above) or DNA binding peculiarities. Finally, EMSAs indicate that the majority of At-NF-YAs bind to CCAAT (Calvenzani et al., 2012), with the exception of NF-YA2 and NF-YA4 (discussed above). Slight variations in sequence specificities, or in the purification procedure/tags, might influence these in vitro results. With such complex interfamily interactions, combined genetic and biochemical approaches will be necessary to solve the many NF-Y puzzles.
Interactions with CCT Proteins
Tomato NF-YC proteins were shown to physically interact with a tomato CO-Like protein through the conserved CCT domain (Ben-Naim et al., 2006). This discovery was extended to Arabidopsis, where it was shown that CO can bind numerous HFD subunits in Y2H assays (At-NF-YB1/2/5/LEC1/L1L and At-NF-YC1/C2/C3/C5/C6/C7/C9; Wenkel et al., 2006). Although there remains a minor controversy over the existence of direct association of NF-YB and CO (Kumimoto et al., 2010; Cao et al., 2011a), these interactions are relevant as (1) higher order mutants in both NF-YB and NF-YC families phenocopied the late flowering of co mutants (Kumimoto et al., 2008, 2010), and (2) NF-YB and NF-YC were at least partially required for CO action in vivo (Kumimoto et al., 2010; Tiwari et al., 2010). More recently, work from Li et al. (2011) confirmed and extended these data in wheat by showing that the CCT domain–containing VRN2 proteins ZCCT1 and ZCCT2 interact with selected NF-YB and NF-YC family members. Furthermore, they can compete with CO for this behavior (Li et al., 2011).
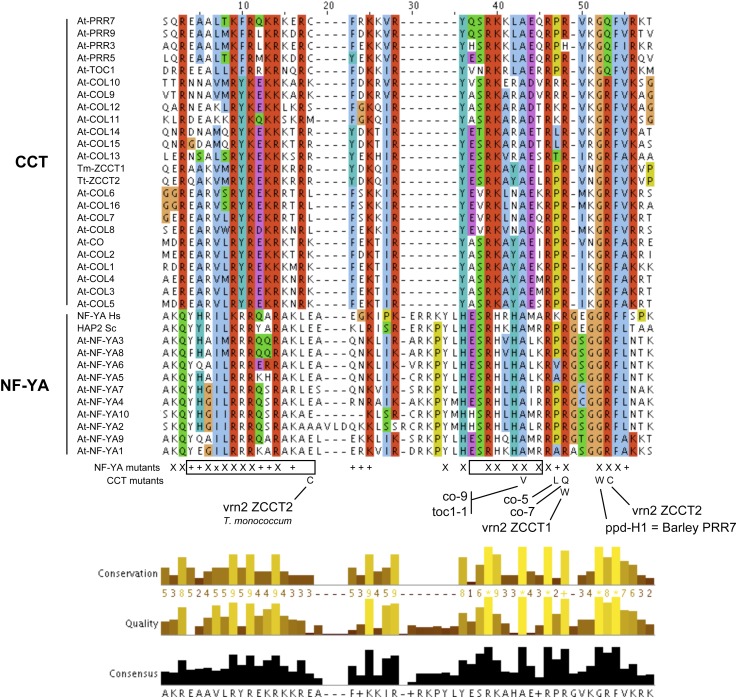
CO and VRN2 belong to a large family of plant specific regulators containing the CCT domain (Putterill et al., 1995; Yan et al., 2004), a 43–amino acid stretch conserved across species, sharing homology with the conserved domain of NF-YA (Figure 7). The homology is striking in the DNA binding A2 helix. Indeed, known mutations that affect CO (co-9, co-5, co-7, and ppd-H1), VRN2, and TOC1 activity in vivo alter amino acids within the A2 homology region (Wenkel et al., 2006; Distelfeld et al., 2009). The N-terminal part of CCTs does not superimpose as well with the corresponding region of NF-YA, but both are predicted to form an α-helical structure (Wenkel et al., 2006). As with NF-YAs, it is rich in basic amino acids potentially responsible for association to a highly negatively charged domain of the HFD dimer (Romier et al., 2003). These similarities between NF-YA and CCT proteins led to the hypothesis that they may compete for interactions with HFD dimers (Wenkel et al., 2006; Distelfeld et al., 2009), which was then formally demonstrated as a possibility in yeast three-hybrid assays (Li et al., 2011). Two key issues to address are (1) whether CO and other CCT proteins need to form complexes with additional proteins (e.g., NF-Ys) to bind DNA (Blackman and Michaels, 2010; Tiwari et al., 2010) and (2) whether the HFDs/CO trimers bind DNA with variations of the CCAAT sequence. Now that the minimal FT promoter has been defined (Adrian et al., 2010), future experiments should focus on in vivo binding assays for both CO and NF-Y proteins in conjunction with directed promoter mutagenesis to address the nature of NF-Y/CO DNA binding during flowering.
Figure 7.
Amino Acid Alignments of the A2 Helix of NF-YA and CCT Proteins.
Alignments were performed using the ClustalW program (Thompson et al., 2002). Amino acid residues conserved among listed proteins are shaded and gaps (dashes) were introduced for maximum matching. Below the alignment, the location of mutations that affect CCT proteins CO, VRN2, TOC1, PPR7 (co-9, co-5, co-7, ppd-H1, toc1-1, and vrn2), or NF-YA/HAP2 activity in vivo (X symbols) are indicated.
Interactions between NF-Y and Unrelated Proteins
The above mentioned At-bZIP28/NF-YB3/NF-YC2/NF-YA4 complex that regulates ER stress (Liu and Howell, 2010) and At-NF-YC2/LEC1/bZIP67 activating an ABA-responsive element in the promoters of seed-specific genes (Yamamoto et al., 2009) suggest that a bZIP activator might be more widely targeted by NF-Y subunits. Intriguingly, overexpression of At-NF-YA4/5/7/9 in reporter assays interfered with gene activation by At-bZIP67/LEC1/NF-YC2, suggesting that activation is not CCAAT dependent and possibly due to a tethering phenomenon, through other DNA-bound transcription factors. At least one other study indicates that noncanonical NF-Y complexes can be formed in plants: The rice MADS box protein MADS18, alone or in combination with the natural partner MADS6, is able to interact with Os-NF-YB1 (Masiero et al., 2002).
Finally, two interacting proteins with no overt transcriptional role were described: (1) Arabidopsis Pirin1, an iron-containing member of the cupin superfamily involved in a pathway leading to an ABA-mediated delay in seed germination, binds to LEC1, potentially serving as coactivator of NF-YA5, LEC1 or L1L, and At-NF-YC1/4/9 (Warpeha et al., 2007); (2) P. wilsonii HAP5a, related to At-NF-YC3/9, interacts with FKBP12 (Yu et al., 2011), a member of a large family shown to have a multitude of cellular functions in both mammals and plants. The growing list of transcription factors and cofactors interacting with NF-Y subunits strongly suggests that a large part of the expansion might have occurred to accommodate such interactions, either at the levels of nearby DNA elements or promoter tethering mechanisms.
CONCLUSIONS AND PERSPECTIVES
We anticipate that the plant NF-Y field will further expand in several directions: (1) further systematic genetic experiments on NF-Y subunits in different plant models, including higher order mutants, which might be required to observe phenotypes of related members of the families; (2) discovery of NF-Y posttranslational modifications, which are currently poorly understood even in the mammalian system; (3) determining the structure of NF-YA, and possibly CCT proteins, in complex with HFDs and the DNA, which will give us mechanistic details on the plant variants; (4) a thorough bioinformatic analysis of plant promoters for the presence of CCAAT and CCAAT-like sequences and genome-wide analysis of association of single-plant NF-Y subunits by chromatin immunoprecipitation sequencing.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data Set 1. Alignment of NF-YA Amino Acid Sequences Used to Generate Figure 1.
Supplemental Data Set 2. Alignment of NF-YB Amino Acid Sequences Used to Generate Figure 2.
Supplemental Data Set 3. Alignment of NF-YC Amino Acid Sequences Used to Generate Figure 3.
AUTHOR CONTRIBUTIONS
K.P., R.W.K., N.G., V.C., and M.F. performed and edited alignments of the figures and Table 2. C.T., B.F.H., and R.M. contributed to writing the article.
References
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani D., Robert L.S. (1995). Cloning and characterization of a Brassica napus gene encoding a homologue of the B subunit of a heteromeric CCAAT-binding factor. Gene 167: 209–213 [DOI] [PubMed] [Google Scholar]
- Alemanno L., Devic M., Niemenak N., Sanier C., Guilleminot J., Rio M., Verdeil J.L., Montoro P. (2008). Characterization of leafy cotyledon1-like during embryogenesis in Theobroma cacao L. Planta 227: 853–866 [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bailey D., O'Hare P. (2007). Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid. Redox Signal. 9: 2305–2321 [DOI] [PubMed] [Google Scholar]
- Ballif J., Endo S., Kotani M., Macadam J., Wu Y. (2011). Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol. Biochem. 49: 579–583 [DOI] [PubMed] [Google Scholar]
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46: 462–476 [DOI] [PubMed] [Google Scholar]
- Blackman B.K., Michaels S.D. (2010). Does CONSTANS act as a transcription factor or as a co-activator? The answer may be—yes. New Phytol. 187: 1–3 [DOI] [PubMed] [Google Scholar]
- Braybrook S.A., Harada J.J. (2008). LECs go crazy in embryo development. Trends Plant Sci. 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Ballif J., Endo S., Davis E., Liang M., Chen D., DeWald D., Kreps J., Zhu T., Wu Y. (2007). A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 145: 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvenzani V., Testoni B., Gusmaroli G., Lorenzo M., Gnesutta N., Petroni K., Mantovani R., Tonelli C. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Kumimoto R.W., Siriwardana C.L., Risinger J.R., Holt B.F., III (2011b). Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS ONE 6: e21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Siriwardana C.L., Kumimoto R.W., Holt B.F., III (2011a). Construction of high quality Gateway™ entry libraries and their application to yeast two-hybrid for the monocot model plant Brachypodium distachyon. BMC Biotechnol. 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S.A., Lindsey K. (2006). The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol. 142: 526–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.Z., Zhang X.Q., Wei P.C., Chen Q.J., Ren F., Chen J., Wang X.C. (2007). AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J. Biochem. Mol. Biol. 40: 1083–1089 [DOI] [PubMed] [Google Scholar]
- Chiappetta A., Michelotti V., Fambrini M., Bruno L., Salvini M., Petrarulo M., Azmi A., Van Onckelen H., Pugliesi C., Bitonti M.B. (2006). Zeatin accumulation and misexpression of a class I knox gene are intimately linked in the epiphyllous response of the interspecific hybrid EMB-2 (Helianthus annuus × H. tuberosus). Planta. 223: 917–931 [DOI] [PubMed] [Google Scholar]
- Chodosh L.A., Olesen J., Hahn S., Baldwin A.S., Guarente L., Sharp P.A. (1988). A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell 53: 25–35 [DOI] [PubMed] [Google Scholar]
- Combier J.P., de Billy F., Gamas P., Niebel A., Rivas S. (2008). Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev. 22: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., Vernié T., Ott T., Gamas P., Crespi M., Niebel A. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dai X., Ding Y., Tan L., Fu Y., Liu F., Zhu Z., Sun X., Sun X., Gu P., Cai H., Sun C. (2012). LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J. Integr. Plant Biol. 54: 790–799 [DOI] [PubMed] [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D., Murray J.A., Smith A.G. (1998). Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrini M., Durante C., Cionini G., Geri C., Giorgetti L., Michelotti V., Salvini M., Pugliesi C. (2006). Characterization of LEAFY COTYLEDON1-LIKE gene in Helianthus annuus and its relationship with zygotic and somatic embryogenesis. Dev. Genes Evol. 216: 253–264 [DOI] [PubMed] [Google Scholar]
- Gaj M.D., Zhang S., Harada J.J., Lemaux P.G. (2005). Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222: 977–988 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Tonelli C., Mantovani R. (2001). Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 264: 173–185 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Tonelli C., Mantovani R. (2002). Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283: 41–48 [DOI] [PubMed] [Google Scholar]
- Hackenberg D., Keetman U., Grimm B. (2012). Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 13: 3458–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D., Wu Y., Voigt A., Adams R., Schramm P., Grimm B. (2011). Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 5: 876–888 [DOI] [PubMed] [Google Scholar]
- Hartlepp K.F., Fernández-Tornero C., Eberharter A., Grüne T., Müller C.W., Becker P.B. (2005). The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol. Cell. Biol. 25: 9886–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R., Li X.Y., Black D., Matthes H., Benoist C., Mathis D. (1990). Co-evolution from yeast to mouse: cDNA cloning of the two NF-Y (CP-1/CBF) subunits. EMBO J. 9: 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Thirumurugan T., Serizawa A., Hiratsu K., Ohme-Takagi M., Kurata N. (2011). Aberrant vegetative and reproductive development by overexpression and lethality by silencing of OsHAP3E in rice. Plant Sci. 181: 105–110 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2009). A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat. Genet. 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., Preuss D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Junker A., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71: 427–442 [DOI] [PubMed] [Google Scholar]
- Kabe Y., Yamada J., Uga H., Yamaguchi Y., Wada T., Handa H. (2005). NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell. Biol. 25: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Toyoshima R., Okuda R., Usui H., Yamamoto A., Hattori T. (2005). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kamada K., Shu F., Chen H., Malik S., Stelzer G., Roeder R.G., Meisterernst M., Burley S.K. (2001). Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106: 71–81 [DOI] [PubMed] [Google Scholar]
- Kato M. (2005). An overview of the CCAAT-box binding factor in filamentous fungi: Assembly, nuclear translocation, and transcriptional enhancement. Biosci. Biotechnol. Biochem. 69: 663–672 [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8: 38–44 [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Adam L., Hymus G.J., Repetti P.P., Reuber T.L., Marion C.M., Hempel F.D., Ratcliffe O.J. (2008). The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723 [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Zhang Y., Siefers N., Holt B.F., III (2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63: 379–391 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V., Landsberger M., Meurer J., Oelmüller R. (1999). The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J. Biol. Chem. 274: 36009–36014 [DOI] [PubMed] [Google Scholar]
- Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B., Harada J.J. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2012). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. (in press). [DOI] [PubMed] [Google Scholar]
- Le B.H., Cheng C., Bui A.Q., Wagmaister J.A., Henry K.F., Pelletier J., Kwong L., Belmonte M., Kirkbride R., Horvath S., Drews G.N., Fischer R.L., Okamuro J.K., Harada J.J., Goldberg R.B. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA. 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Fischer R.L., Goldberg R.B., Harada J.J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque-Lemay M., Albani D., Aldcorn D., Hammerlindl J., Keller W., Robert L.S. (2003). Expression of CCAAT-binding factor antisense transcripts in reproductive tissues affects plant fertility. Plant Cell Rep. 21: 804–808 [DOI] [PubMed] [Google Scholar]
- Leyva-González M.A., Ibarra-Laclette E., Cruz-Ramírez A., Herrera-Estrella L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7: e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Distelfeld A., Comis A., Dubcovsky J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X., Oono Y., Zhu J., He X.J., Wu J.M., Iida K., Lu X.Y., Cui X., Jin H., Zhu J.K. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y., Mantovani R., Hooft van Huijsduijnen R., Andre I., Benoist C., Mathis D. (1992b). Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 20: 1087–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Hole D., Wu J., Blake T., Wu Y. (2012). Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta 235: 779–791 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Maillot P., Lebel S., Schellenbaum P., Jacques A., Walter B. (2009). Differential regulation of SERK, LEC1-like and pathogenesis-related genes during indirect secondary somatic embryogenesis in grapevine. Plant Physiol. Biochem. 47: 743–752 [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26: 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D.S., Pinto I. (2005). Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 4: 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero S., Imbriano C., Ravasio F., Favaro R., Pelucchi N., Gorla M.S., Mantovani R., Colombo L., Kater M.M. (2002). Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J. Biol. Chem. 277: 26429–26435 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Meinke D.W. (1992). A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Ito Y., Serizawa A., Kurata N. (2003). OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 36: 532–540 [DOI] [PubMed] [Google Scholar]
- Mu J., Tan H., Hong S., Liang Y., Zuo J. (August 29, 2012). Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant. doi:10.1093/mp/sss061. [DOI] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., Yang X., Wang T., Chong K., Wang X.J., Zuo J. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.E., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Parcy F., Valon C., Kohara A., Miséra S., Giraudat J. (1997). The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Reed B.D., Charos A.E., Szekely A.M., Weissman S.M., Snyder M. (2008). Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 4: e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso M.A., Blanco F.A., Bailey-Serres J., Crespi M., Zanetti M.E. (October 11, 2012). Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J., doi: 10.1111/tpj.12033 [DOI] [PubMed] [Google Scholar]
- Romier C., Cocchiarella F., Mantovani R., Moras D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278: 1336–1345 [DOI] [PubMed] [Google Scholar]
- Salvini M., Sani E., Fambrini M., Pistelli L., Pucciariello C., Pugliesi C. (2012). Molecular analysis of a sunflower gene encoding an homologous of the B subunit of a CAAT binding factor. Mol. Biol. Rep. 39: 6449–6465 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schellenbaum P., Jacques A., Maillot P., Bertsch C., Mazet F., Farine S., Walter B. (2008). Characterization of VvSERK1, VvSERK2, VvSERK3 and VvL1L genes and their expression during somatic embryogenesis of grapevine (Vitis vinifera L.). Plant Cell Rep. 27: 1799–1809 [DOI] [PubMed] [Google Scholar]
- Shen B., Allen W.B., Zheng P., Li C., Glassman K., Ranch J., Nubel D., Tarczynski M.C. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N., Dang K.K., Kumimoto R.W., Bynum W.E., IV, Tayrose G., Holt B.F., III (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson T.J., McIntyre C.L., Collet C., Xue G.P. (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 65: 77–92 [DOI] [PubMed] [Google Scholar]
- Stephenson T.J., McIntyre C.L., Collet C., Xue G.P. (2010). TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Funct. Integr. Genomics 10: 265–276 [DOI] [PubMed] [Google Scholar]
- Stephenson T.J., McIntyre C.L., Collet C., Xue G.P. (2011). TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct. Integr. Genomics 11: 327–340 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Yang X., Zhang F., Zheng X., Qu C., Mu J., Fu F., Li J., Guan R., Zhang H., Wang G., Zuo J. (2011). Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 156: 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumurugan T., Ito Y., Kubo T., Serizawa A., Kurata N. (2008). Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genomics 279: 279–289 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Higgins D.G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2: Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., et al. (2012). The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70: 855–865 [DOI] [PubMed] [Google Scholar]
- Uddenberg D., Valladares S., Abrahamsson M., Sundström J.F., Sundås-Larsson A., von Arnold S. (2011). Embryogenic potential and expression of embryogenesis-related genes in conifers are affected by treatment with a histone deacetylase inhibitor. Planta 234: 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha K.M., Upadhyay S., Yeh J., Adamiak J., Hawkins S.I., Lapik Y.R., Anderson M.B., Kaufman L.S. (2007). The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Yee K.M., Danao J., Zimmerman J.L., Fischer R.L., Goldberg R.B., Harada J.J. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Li X., Glover B.J., Bai S., Rao G.Y., Luo J., Yang J. (2008). Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 25: 1581–1592 [DOI] [PubMed] [Google Scholar]
- Yan W.H., Wang P., Chen H.X., Zhou H.J., Li Q.P., Wang C.R., Ding Z.H., Zhang Y.S., Yu S.B., Xing Y.Z., Zhang Q.F. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4: 319–330 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Xie Z., Glover B.J. (2005). Asymmetric evolution of duplicate genes encoding the CCAAT-binding factor NF-Y in plant genomes. New Phytol. 165: 623–631 [DOI] [PubMed] [Google Scholar]
- Yazawa K., Kamada H. (2007). Identification and characterization of carrot HAP factors that form a complex with the embryo-specific transcription factor C-LEC1. J. Exp. Bot. 58: 3819–3828 [DOI] [PubMed] [Google Scholar]
- Yazawa K., Takahata K., Kamada H. (2004). Isolation of the gene encoding Carrot leafy cotyledon1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol. Biochem. 42: 215–223 [DOI] [PubMed] [Google Scholar]
- Yoshida H. (2007). Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid. Redox Signal. 9: 2323–2333 [DOI] [PubMed] [Google Scholar]
- Yu Y., Li Y., Huang G., Meng Z., Zhang D., Wei J., Yan K., Zheng C., Zhang L. (2011). PwHAP5, a CCAAT-binding transcription factor, interacts with PwFKBP12 and plays a role in pollen tube growth orientation in Picea wilsonii. J. Exp. Bot. 62: 4805–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M.E., Blanco F.A., Beker M.P., Battaglia M., Aguilar O.M. (2010). A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22: 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wong L., Meng L., Lemaux P.G. (2002). Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize (Zea mays L.). Planta 215: 191–194 [DOI] [PubMed] [Google Scholar]
- Zhao B., Ge L., Liang R., Li W., Ruan K., Lin H., Jin Y. (2009). Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., Ruan K., Jin Y. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 354: 585–590 [DOI] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J.K., Zhang F., Li W.X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]