Many plant microRNAs are deeply conserved in all land plants, from mosses to flowering plants. We carry out functional studies of two such microRNAs, miR156 and miR390, in the moss Physcomitrella patens and identify them as components of a broadly conserved gene regulatory network that controls the timing of plant development.
Abstract
microRNA156 (miR156) affects developmental timing in flowering plants. miR156 and its target relationships with members of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family appear universally conserved in land plants, but the specific functions of miR156 outside of flowering plants are unknown. We find that miR156 promotes a developmental change from young filamentous protonemata to leafy gametophores in the moss Physcomitrella patens, opposite to its role as an inhibitor of development in flowering plants. P. patens miR156 also influences accumulation of trans-acting small interfering RNAs (tasiRNAs) dependent upon a second ancient microRNA, miR390. Both miR156 and miR390 directly target a single major tasiRNA primary transcript. Inhibition of miR156 function causes increased miR390-triggered tasiRNA accumulation and decreased accumulation of tasiRNA targets. Overexpression of miR390 also caused a slower formation of gametophores, elevated miR390-triggered tasiRNA accumulation, and reduced level of tasiRNA targets. We conclude that a gene regulatory network controlled by miR156, miR390, and their targets controls developmental change in P. patens. The broad outlines and regulatory logic of this network are conserved in flowering plants, albeit with some modifications. Partially conserved small RNA networks thus influence developmental timing in plants with radically different body plans.
INTRODUCTION
Plants produce both a multicellular haploid entity (the gametophyte) and a multicellular diploid entity (the sporophyte) during their sexual life cycles. Mosses and other bryophytes have morphologically complex, photosynthetic gametophytes with attached simple nonphotosynthetic sporophytes. By contrast, other lineages, including flowering plants, have simple, microscopic gametophytes enclosed by, and nutritionally dependent upon, a morphologically complex sporophyte. The extent to which genes with ancestral roles in the development of complex gametophytes were recruited to control complex sporophytic development is unclear as both positive (Menand et al., 2007) and negative (Tanahashi et al., 2005) examples have been described. The question is particularly acute for microRNAs (miRNAs) because several miRNA families are conserved in a wide range of land plants, including both sporophyte-dominated and gametophyte-dominated species (Cuperus et al., 2011). The regulatory targets of these conserved miRNA families are, as a general rule, also conserved, and the majority of the most ancient plant miRNA families regulate mRNAs encoding transcription factors (Axtell and Bowman, 2008). The functions of many of these miRNAs and their targets have been intensively investigated in several flowering plants and frequently found to affect various aspects of sporophytic development (Axtell and Bowman, 2008). Global reduction of miRNA accumulation in DICER-LIKE 1A (DCL1a) mutants of the moss Physcomitrella patens, whose life cycle is dominated by a morphologically complex gametophyte, has pleiotropic effects on gametophytic development (Khraiwesh et al., 2010). However, the functions of specific ancient plant miRNA families in P. patens or other gametophyte-dominated basal plant lineages remain unclear.
miR156 is a master regulator of developmental transitions in flowering plants. miR156 accumulation in flowering plants is temporally regulated, with high levels in early embryos, and a progressive reduction as the plant ages (Wu and Poethig, 2006; Wang et al., 2009). Conversely, the accumulation of its SQUAMOSA PROMOTER BINDING PROTEIN- LIKE (SPL) targets generally increases with age in flowering plants (Wu and Poethig, 2006; Wang et al., 2009). SPL genes in flowering plants promote adult leaf morphology and flowering (Poethig, 2009; Huijser and Schmid, 2011). miR172-mediated regulation of APETALA2 (AP2) family genes has a reciprocal relationship with miR156/SPL: miR172 levels increase with plant age, while miR172-targeted AP2 family genes, several of which serve to repress adult traits, decrease (Huijser and Schmid, 2011). In Arabidopsis thaliana, this coordination is achieved in part by direct stimulation of miR172 transcription by miR156-targeted SPL9 and SPL10 (Wu et al., 2009). Genetic screens for Arabidopsis vegetative developmental mutants (Hunter et al., 2003, 2006; Peragine et al., 2004; Fahlgren et al., 2006) have identified a third small RNA pathway that influences vegetative developmental transitions. miR390 stimulates production of trans-acting small interfering RNAs (tasiRNAs) that target AUXIN RESPONSE FACTOR3 (ARF3)/ETTIN and ARF4 mRNAs. Loss of tasiRNA-mediated ARF3/4 regulation accelerates vegetative developmental transitions, implying that ARF3/ETTIN and ARF4 promote these changes. The miR390/tasiRNA/ARF regulatory system also influences organ polarity in flowering plants (Garcia et al., 2006; Nogueira et al., 2007). The miR156/SPL regulatory interaction appears universal among land plants (Arazi et al., 2005; Axtell et al., 2007), and the miR390/tasiRNA/ARF cascade appears nearly universal (Axtell et al., 2006) save for an apparent secondary loss in the lycophyte lineage (Banks et al., 2011), while the miR172/AP2 interaction appears to have emerged later, after the divergence of the moss and lycophyte lineages (Axtell and Bartel, 2005).
Here, we report a functional study of P. patens miR156, a miR156-targeted target SPL gene, and miR390. We find that miR156 promotes the transition from tip-growing protonemal growth to production of leafy gametophores. Interestingly, miR156-mediated promotion of this vegetative transition is contrary to the inhibitory role of miR156 upon developmental transitions in flowering plants. In addition, P. patens miR156 and miR390 both directly target the same major tasiRNA primary transcript, and perturbation of miR156 influences accumulation of tasiRNAs and their target mRNAs. Overexpression of MIR390C resulted in slower gametophore production, accompanied by reduction of tasiRNA targets. Finally, we describe a gene regulatory network in P. patens in which a set of miRNAs, tasiRNAs, and three transcription factor families converge to control developmental timing. This network is partially conserved in flowering plants, suggesting that the regulatory networks for developmental transition present in modern species descended and diversified from those present in an ancient ancestor.
RESULTS
Temporal Regulation of miR156 and Its Targets in P. patens
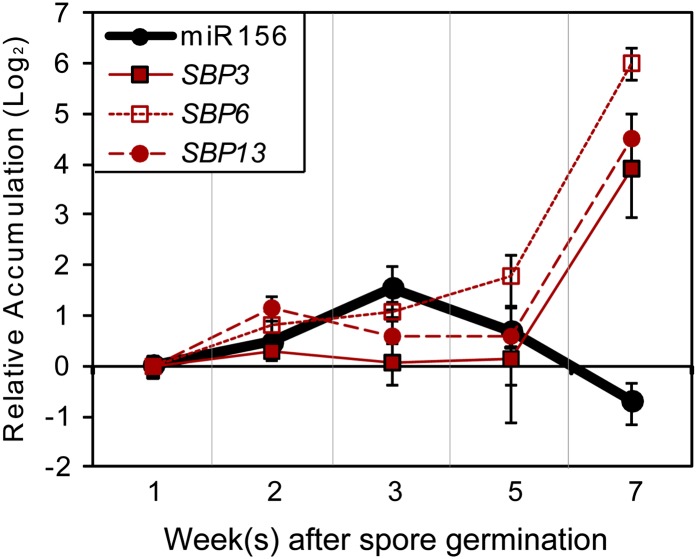
miR156 accumulation in flowering plant sporophytes is temporally regulated, with high levels in early embryos and a progressive reduction as the plant ages (Wu and Poethig, 2006; Wang et al., 2009). Conversely, the accumulation of its SPL targets generally increases with age in flowering plants (Wu and Poethig, 2006; Wang et al., 2009). Stem loop–mediated quantitative RT-PCR (qRT-PCR) (Varkonyi-Gasic et al., 2007) indicated that miR156 accumulation is also temporally regulated in P. patens gametophytes (Figure 1; see Supplemental Figure 1 online). Young plants comprised solely of tip-growing, filamentous protonemata tissues (chloronema and caulonema; 1 to 2 weeks after germination from spores; see Supplemental Figure 2 online) have relatively little miR156 accumulation. Young buds and early gametophores with one or two phylloids are most enriched in 3-week old plants (see Supplemental Figure 2 online), with patches of buds on top of the lawn of filamentous protonemata (Huijser and Schmid, 2011). Buds are the intermediate tissues between protonemata and gametophores. Thus, we define these plants as undergoing the developmental transition from young protonemata to adult leafy gametophores. In these plants, the accumulation of miR156 reaches its maximum level (Figure 1; see Supplemental Figure 1 online). miR156 levels then gradually decline from their 3-week maximum as the leafy gametophores mature (Figure 1; see Supplemental Figure 1 online).
Figure 1.
Accumulation of miR156 and Its Targets during P. patens Gametophyte Development.
Accumulation of P. patens miR156 and target SBP genes was assessed using qRT-PCR. Relative accumulation levels relative to the 1-week sample were plotted. Error bars indicate sd from three biological replicates. Analyses of significant differences are shown in Supplemental Figure 1 online.
[See online article for color version of this figure.]
The three P. patens SQUAMOSA PROMOTER BINDING PROTEIN (SBP) targets of miR156 (SBP3, SBP6, and SBP13) (Arazi et al., 2005; Riese et al., 2007; Addo-Quaye et al., 2009) also accumulated in a temporal fashion. (Note that the P. patens miR156-targeted SBP genes are in the same gene family as the miR156-targeted SPL genes in Arabidopsis and other flowering plants; to be consistent with the prior literature [Riese et al., 2007], we use the SBP acronym when referring to the P. patens members of this gene family.) Like miR156, the SBP targets were minimally expressed in 1- to 2-week-old plants. Increased accumulation of SBP targets became apparent in older plants at time points when miR156 accumulation levels were declining from their maximum values (Figure 1; see Supplemental Figure 1 online).
miR156 Promotes Gametophore Production in P. patens via SBP3
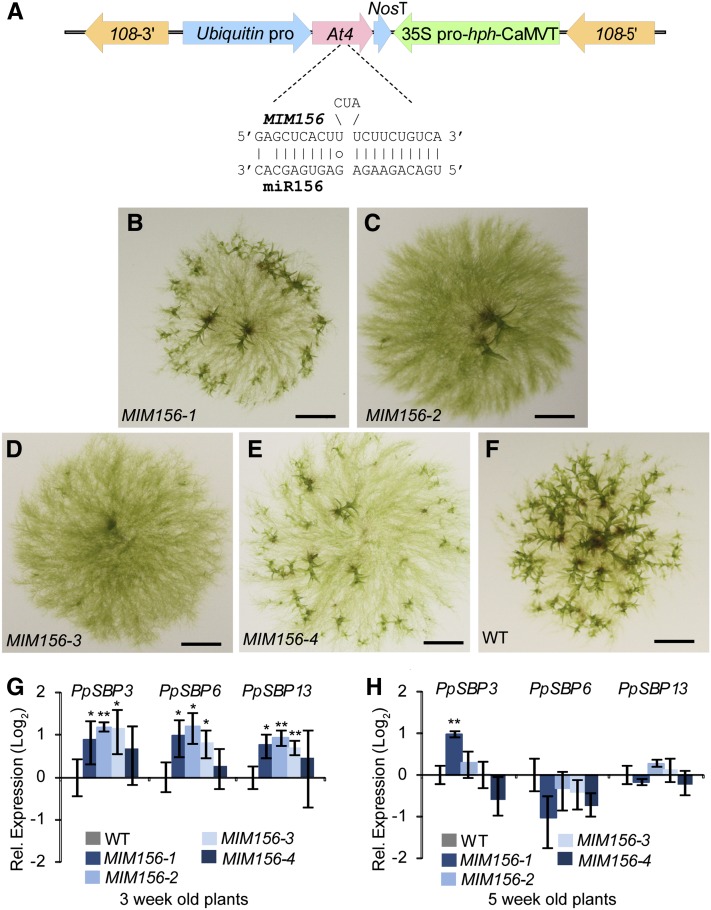
A miRNA target mimic (Franco-Zorrilla et al., 2007; Todesco et al., 2010) designed to reduce miR156 function was overexpressed in wild-type P. patens (Figure 2A). Four independent MIM156 transgenic lines had delayed bud initiation and reduced numbers of leafy buds and subsequent gametophores (Figures 2B to 2F). In general, phenotypic severity was positively correlated with the accumulation of the MIM156 target mimic RNA, with the two stronger MIM156 expressers (MIM156-2 and -3) more delayed and the two weaker expressers (MIM156-1 and -4) less delayed (see Supplemental Figure 3A online). Mature miR156 accumulation was also inhibited in MIM156 plants to varying degrees (see Supplemental Figure 3B online). We noted that the magnitude of reduction of mature miR156 accumulation was not perfectly correlated with MIM156 transcript levels nor with phenotypic severity. The MIM method is based on the sequestration of miRNA by overexpressed decoy targets, and MIM-induced reductions in mature miRNA levels have been inconsistently observed (Franco-Zorrilla et al., 2007; Todesco et al., 2010). Therefore, the amount of functional inhibition of miR156 in individual MIM156 lines should not be directly inferred from accumulation of mature miR156 alone. In 3-week-old MIM156 plants, accumulation of SBP3, SBP6, and SBP13 was elevated approximately two-fold (Figure 2G). By 5 weeks, accumulation of these three miR156 target mRNAs in MIM156 plants was indistinguishable from the wild type (Figure 2H). We noted that the magnitude of target upregulation was similar in the weak MIM156-1 line and the strong MIM156-2 and MIM156-3 lines (Figure 2G). This suggests that there are other factors besides miR156 that act as a buffer for regulating the accumulation of SBP mRNAs in P. patens. The timing of target upregulation in the MIM156 plants coincided with both the maximal accumulation of miR156 and with the maximal initiation of leafy buds in the wild type. Collectively, these data indicate that miR156 functions to promote the vegetative developmental transition from protonemata to leafy gametophores in P. patens.
Figure 2.
miR156 Promotes Bud and Leafy Gametophore Formation.
(A) Schematic of the MIM156 overexpression construct. pro, promoter; NosT and CaMVT, Nos and cauliflower mosaic virus terminator, respectively; At4, At4 gene from Arabidopsis; 108, 108 targeting locus from the P. patens genome (Schaefer and Zrÿd, 1997).
(B) to (F) Delay of bud and gametophore production in MIM156 lines. Four-week-old plants (four independent MIM156 lines [B] to [E] and the wild type (WT; [F])] are shown. Bars = 3 mm.
(G) to (H) Increased accumulation of miR156 targets in MIM156 plants at the time of maximal bud initiation. RNAs from spore-germinated 3-week-old tissues (G) and 5-week-old tissues (H) were analyzed by qRT-PCR as in Figure 1. Expression relative to the wild type was plotted. Error bars indicate sd from three biological replicates. Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
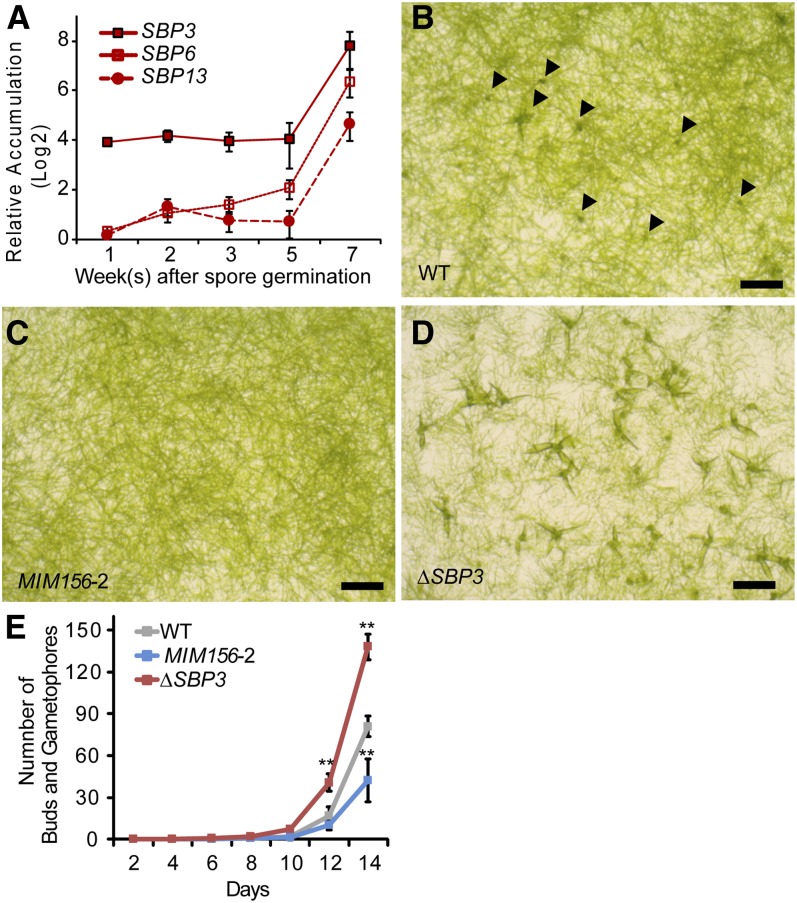
Relative accumulation of SBP3 was much higher than that of SBP6 and SBP13 in wild-type P. patens (Figure 3A; see Supplemental Figure 4 online). We therefore hypothesized that misregulation of SBP3 could be a major component of the MIM156-associated phenotypes. Deletion of the SBP3 locus was achieved using a gene replacement strategy (see Supplemental Figure 5 online). The resulting ΔSBP3 null mutant had a phenotype opposite to that seen in MIM156 plants: increased initiation and numbers of leafy buds relative to the wild type (Figures 3B to 3E). SBP3 is therefore a negative regulator of bud and leafy gametophore formation in the P. patens gametophyte. We conclude that elimination of single miR156 target, SBP3, is sufficient to produce a phenotype opposite to that caused by reduction of miR156 activity, implying that SBP3 is a miR156 target of major importance.
Figure 3.
SBP3 Represses Bud and Leafy Gametophore Formation.
(A) Accumulation of SBP transcripts during the P. patens life cycle. Expression levels relative to EF1α in a sample were plotted. Error bars indicate sd from three biological replicates. Analyses of significant differences are shown in Supplemental Figure 4 online.
(B) to (D) Opposite effects of miR156 and SBP3 upon bud formation. Blended protonemal tissues of the wild type (WT) (B), MIM156-2 (C), and ΔSBP3 (D) were plated on cellophane overlaid media and incubated for 2 weeks. Arrowheads in (B) indicate buds. Bars = 1 mm.
(E) Rates of bud and gametophore appearance. Seven-day-old protonemal tissues were inoculated on media. The numbers of buds and subsequent gametophores in four colonies were counted every 2 days. Error bars represent sd of four replicates. Significant differences from the wild type on each day were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
miR156 Suppresses Accumulation and Function of miR390-Dependent tasiRNAs
Several distinct small RNA pathways influence the rate and timing of leafy gametophore formation in P. patens. Loss of the P. patens–specific miRNA miR534a leads to elevated accumulation of two Blade on Petiole (BOP) target mRNAs and accelerated production of leafy gametophores (Saleh et al., 2011). Accumulation of the miR534a-targeted BOP mRNAs was not significantly affected in 3-week-old MIM156 plants, suggesting that the influences of miR156 and miR534a on bud formation are separate (see Supplemental Figure 6A online). P. patens ΔDCL3 mutants, in which 22- to 24-nucleotide small interfering RNAs (siRNAs) derived from intergenic/repetitive elements are lost, also have an accelerated leafy gametophore formation phenotype (Cho et al., 2008). Accumulation of SBP3, SBP6, and SBP13 was not detectably altered in 3-week-old ΔDCL3 plants, suggesting that the influence of DCL3 upon P. patens leafy bud formation is also separate from that conditioned by miR156 (see Supplemental Figure 6B online).
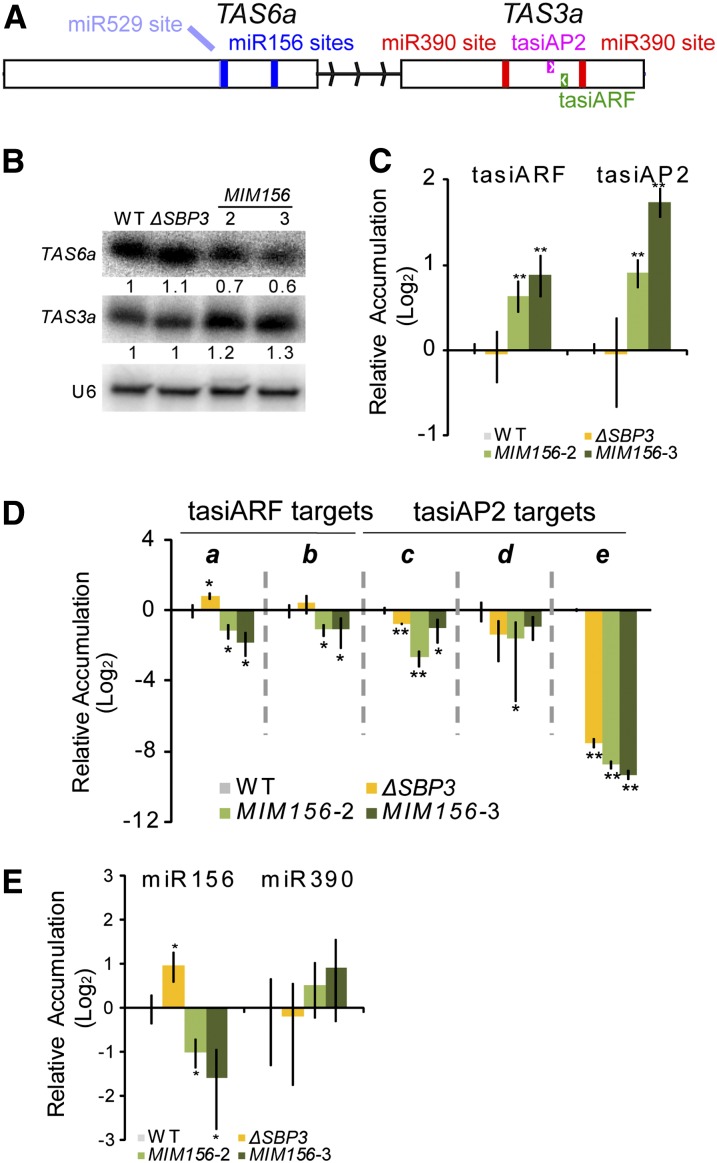
P. patens ΔRNA DEPENDENT RNA POLYMERASE (RDR6) plants also have an accelerated production of leafy gametophores (Talmor-Neiman et al., 2006). RDR6 is required for the accumulation of TRANS-ACTING SIRNA3 (TAS3) family tasiRNAs derived from non-protein coding RNA precursors processed by miR390-directed slicing (Axtell et al., 2006; Talmor-Neiman et al., 2006). Out of the large population of P. patens TAS3-derived tasiRNAs, two are known to direct slicing of target mRNAs in trans: tasiAP2, which directs slicing of mRNAs encoding an ERF-like single N-terminal AP2 domain, and tasiARF, which directs slicing of mRNAs encoding B3 and ARF domains (Talmor-Neiman et al., 2006; Axtell et al., 2007). Although there are six P. patens TAS3 loci (TAS3a-f), TAS3a contributes the majority of tasiAP2 and tasiARF production (see Supplemental Table 1 online) and dominates the overall small RNA profile from this family (Arif et al., 2012). We and our colleagues recently discovered another family of P. patens TAS loci, TAS6, characterized by miR156- and miR529-directed slicing and close genomic proximity to TAS3 loci (note that miR156 and miR529 are related in sequence) (Arif et al., 2012). Degradome analysis provided evidence that both miR156 sites are sliced, and phased siRNAs dependent upon RDR6 and DCL4 arise from the region between the two miR156 sites (Arif et al., 2012). A small RNA gel blot demonstrated that TAS6a small RNA accumulation was also dependent upon RDR6 and DCL4 (see Supplemental Figure 7 online). The opposite phenotypes of ΔRDR6 and MIM156 plants, as well as the close proximity of miR156-sliced TAS6 loci to miR390-sliced TAS3 loci, suggested that the functions of P. patens miR156 and miR390 may be directly intertwined. In support of this hypothesis, cDNA cloning revealed that TAS6a and TAS3a share a single primary transcript, with a central 256-nucleotide-long intron separating the two tasiRNA generating regions (Figure 4A). TAS6b/TAS3f and TAS6c/TAS3d also occur in tandem arrangements that suggest origins from a common primary transcript (Arif et al., 2012).
Figure 4.
miR156 Influences P. patens tasiRNA Accumulation and Function.
(A) The annotated TAS6a/TAS3a primary transcript including miRNA target sites, locations of functional tasiRNAs, exons (rectangles), and central intron.
(B) Small RNA gel blot analysis from 3-week-old ΔSBP3, MIM156-2, and MIM156-3 plants. The numbers indicate the intensities relative to U6 signals. WT, the wild type.
(C) Stem loop qRT-PCR of tasiARF and tasiAP2 siRNAs from 3-week-old plants. Error bars indicate sd from three biological replicates. Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
(D) qRT-PCR of tasiARF and tasiAP2 target mRNAs in 3-week-old plants. Error bars indicate sd from three biological replicates. a, 1s14_392V6.1; b, 1s280_72V6.1; c, 1s6_75V6.1; d, 1s5_432V6.1; e, 1s74_86V6.1. Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
(E) Stem loop qRT-PCR of miR156 and miR390 from 3-week-old plants. Error bars indicate sd from three biological replicates. Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
We hypothesized that miR156 affects tasiRNA accumulation via interaction with the TAS6a/TAS3a primary transcript. To test this hypothesis, we first examined accumulation of small RNAs in two independent MIM156 plants with a high level of MIM156 accumulation. Accumulation of small RNAs from the TAS6a region was reduced in MIM156 plants, while small RNAs from the TAS3a region were increased in MIM156 plants (Figure 4B). The specific tasiAP2 and tasiARF small RNAs responded similarly to the total TAS3a small RNA population, with an increased accumulation in MIM156 plants (Figure 4C). Consistent with the tasiRNA accumulation patterns in MIM156 plants, tasiAP2 and tasiARF targets generally had reduced accumulation levels in MIM156 plants (Figure 4D). These data are consistent with a hypothesis where miR156 influences the production of tasiARF and tasiAP2 production through an interaction with TAS6a-TAS3a precursor.
Surprisingly, we found that ΔSBP3 plants had an increased accumulation of miR156 (Figure 4E). The basis for the increased accumulation of miR156 in ΔSBP3 plants is not clear. One hypothesis consistent with this observation is that the SBP3 transcription factor normally acts a negative regulator of MIR156 transcription. TAS3a small RNA accumulation in the ΔSBP3 plants was not changed (Figures 4B and 4C), but two of the three tasiAP2 target mRNAs accumulated at lower levels relative to the wild type (Figure 4D). Indeed, the reductions in some of the tasiAP2 targets observed in ΔSBP3 plants were similar to those observed in the MIM156 lines, despite the opposite morphological phenotypes (Figure 3), opposite miR156 accumulation patterns (Figure 4E), and unrelated tasiRNA accumulation patterns (Figure 4C) observed in these three lines. These data demonstrate that tasiAP2 and tasiARF accumulation levels are not the sole contributors to the accumulation levels of their respective target mRNAs. Additionally, accumulation levels of the AP2 and ARF target mRNAs cannot be the sole determinant of bud initiation rate, as plants with opposite phenotypes (MIM156 and ΔSBP3) have the similar magnitude of down-regulation.
Because MIM156 plants are developmentally retarded compared with the wild type at 3 weeks of age, it was possible that the observed differences in small RNA and target accumulation were due to different tissue compositions, instead of direct molecular effects. Arguing against this possibility, we observed similar differences in TAS3a and TAS6a small RNA accumulation and tasiAP2 and tasiARF target accumulation in 7-week-old plants (see Supplemental Figure 8 online), a time point at which all lines are more similar morphologically. TAS6a produces at least one functional tasiRNA (tasiZNF), which targets a zinc finger domain mRNA (Arif et al., 2012). The accumulation of the tasiZNF target was not significantly affected in MIM156 plants (see Supplemental Figure 9 online), though accumulation of small RNAs from TAS6a was reduced (Figure 4B), which suggests that dysregulation of tasiZNF does not contribute to the MIM156 phenotype. Curiously, the tasiZNF target was significantly upregulated in ΔSBP3 (see Supplemental Figure 9 online). We suspect this is an indirect effect caused by the loss of the SBP3 transcription factor.
miR390 Represses Formation of Buds and Leafy Gametophores
We next examined the expression patterns of miR390, tasiARF, and tasiAP2 during gametophyte development (Figure 5A; see Supplemental Figure 10A online). After spore germination, miR390 accumulation declined slightly, reaching a minimum at the 3-week-old samples when buds and young gametophores are enriched and then gradually increased again later in development. This pattern is the opposite to that of miR156 (Figure 1). tasiARF and tasiAP2 also exhibited temporal expression patterns, although they were distinct from that of miR390 (Figure 5A; see Supplemental Figure 10A online). The targets of tasiARF and tasiAP2 were also temporally regulated, with many of them reaching peak accumulation levels 2 or 3 weeks after spore germination (Figure 5B; see Supplemental Figure 10B online). The patterns of target accumulation were largely unrelated to those of their corresponding tasiRNAs, especially at early developmental time points. Therefore, tasiAP2 and tasiARF are not the only factors that regulate the overall accumulation levels of their targets during development.
Figure 5.
Overexpression of MIR390C Represses Bud and Leafy Gametophore Formation.
(A) Expression pattern of miR390, tasiARF, and tasiAP2 small RNAs in wild-type plants at the indicated ages, measured by stem loop qRT-PCR. The relative accumulation levels to the 1-week sample were plotted. Error bars indicate sd from three biological replicates. Analyses of significant differences are shown in Supplemental Figure 10A online.
(B) Expression pattern of the target mRNAs of tasiARF and tasiAP2 in the samples as in (A), measured by qRT-PCR. The relative accumulation levels to the 1-week sample were plotted. Error bars indicate sd from three biological replicates. a, 1s14_392V6.1; b, 1s280_72V6.1; c, 1s6_75V6.1; d, 1s5_432V6.1; e, 1s74_86V6.1. Analyses of significant differences are shown in Supplemental Figure 10B online.
(C) Overexpression of MIR390C. RNAs from protonemata tissues of the wild type (WT) and MIR390c OE were analyzed by stem loop–mediated qRT-PCR. Error bars indicate sd from three biological replicates. Significant differences from wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
(D) Rates of bud and gametophore appearance. Seven-day-old protonemal tissues of the wild type and MIR390C OE were inoculated on media. The numbers of buds and subsequent gametophores in 16 colonies were counted every 2 d. Error bars represent the se values for those 16 colonies. Significant differences from wild type on each day were analyzed by t test (*P ≤ 0.05). The raw data for day 16 is in Supplemental Table 2 online.
(E) Stem loop qRT-PCR of tasiARF, tasiAP2, and tasiZNF in 3-week-old plants. Error bars indicate sd from three biological replicates. Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
(F) Accumulation of tasiARF and tasiAP2 target mRNAs in 3-week-old plants measured by qRT-PCR. Error bars indicate sd from three biological replicates. a to e are as in (B). Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
(G) Accumulation of the tasiZNF target mRNA 1s286_43V6.1, measured by qRT-PCR. Error bars indicate sd from three biological replicates. No significant differences relative to the wild type were found (t test, P > 0.05).
To test if miR390 directly affects the developmental transition, we generated plants overexpressing P. patens MIR390C under the control of the Ubiquitin promoter. In the MIR390C-overexpressing plants (OE), miR390 accumulated ∼64-fold higher relative to the wild type, with no detectable effect upon miR156 accumulation (Figure 5C). The MIR390C OE plants exhibited a slower gametophore transition, with a significant deviation from the wild type at the 16th day after inoculation (Figure 5D; see Supplemental Figure 11 and Supplemental Table 2 online). Accumulation of tasiAP2 and tasiARF was elevated in MIR390C OE plants (Figure 5E), probably due to the higher availability of RDR6 and DCL4 substrates produced by miR390-guided TAS3a cleavage. The higher accumulation of tasiRNAs was correlated with reduction of transcript accumulation from four out of five of the tasiRNA targets (Figure 5F). We hypothesize that the one exception to this trend, the tasiAP2 target 1s74_86V6.1 (Figure 5F), is due to factors counteracting the effect of tasiAP2, consistent with the notion that tasiAP2 accumulation is not the sole determinant of target accumulation levels. Interestingly, neither tasiZNF accumulation nor the accumulation of its target mRNA were significantly different in MIR390C OE plants (Figures 5E and 5G). Thus, miR156 affects tasiRNA accumulation from both TAS6a and TAS3a (Figure 4), but miR390 only affects TAS3a. Overall, these data show that miR390 represses the bud formation in P. patens and affects accumulation of TAS3a-derived tasiRNAs and their targets.
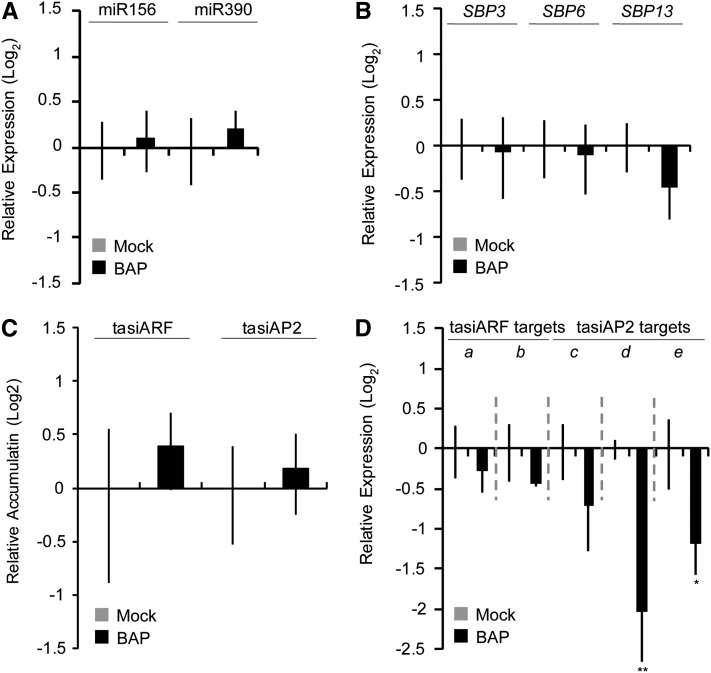
Cytokinin Application Does Not Strongly Affect Accumulation of miR156, miR390, tasiRNAs, or Their Targets
Cytokinin induces caulonemal cells to differentiate into buds in P. patens (Cove and Knight, 1993; Schumaker and Dietrich, 1997) and regulates the accumulation of miR534a (Saleh et al., 2011). Because miR156 and miR390 are involved in timing of bud and gametophore formation, we tested whether cytokinin treatment affects accumulation of these small RNAs and/or their targets. Application of benzylaminopurine (BAP; e.g., cytokinin B) to protonemata for 24 h accelerated bud formation (see Supplemental Figure 12 online), without causing significant changes in miR156 or miR390 accumulation (Figure 6A). Cytokinin also had negligible effects upon accumulation of the three SBP targets of miR156 (Figure 6B) and upon tasiARF and tasiAP2 accumulation (Figure 6C). Similarly, the accumulation of the two tasiARF target mRNAs was unaffected by BAP treatment. By contrast, tasiAP2 targets responded with a two- to fourfold reduction in mRNA accumulation after cytokinin treatment (Figure 6D). Overall, these data indicate that cytokinin treatment does not substantially affect the miR156-miR390-tasiRNA network, at least at the level of RNA accumulation. The one exception is the tasiAP2 target mRNAs, which appear to respond independent of any changes in tasiAP2 accumulation; these genes may serve as an integration point where the cytokinin-induced and miR156/miR390 pathways intersect. We conclude that the cytokinin-induced bud formation pathway in P. patens is largely independent of, or downstream from, the miR156-miR390-tasiRNA network.
Figure 6.
Cytokinin Treatment Has Mostly Negligible Effects upon RNA Accumulation Levels of Members of the miR156-miR390-tasiRNA Pathway.
(A) Stem loop qRT-PCR of the indicated mature miRNAs from wild-type P. patens treated with 5 μM BAP or with a mock control. Error bars represent sd from three biological replicates. No significant difference from the wild type was found (P > 0.05 by t test).
(B) qRT-PCR of the indicated miR156-targeted SBP mRNAs from wild-type plants treated with 5 μM BAP or with a mock control. Error bars represent sd from three biological replicates. No significant difference from the wild type was found (P > 0.05 by t test).
(C) Stem loop qRT-PCR of the indicated tasiARF and tasiAP2 siRNAs from the samples as in (A). Error bars represent sd from three biological replicates. No significant difference from the wild type was found (P > 0.05 by t test).
(D) As in (B) for tasiARF and tasiAP2 target mRNAs. a, 1s14_392V6.1; b, 1s280_72V6.1; c, 1s6_75V6.1; d, 1s5_432V6.1; e, 1s74_86V6.1. Significant differences from the wild type were analyzed by t test (*P ≤ 0.05 and **P ≤ 0.01).
A miRNA and tasiRNA Regulatory Network Controlling Developmental Timing
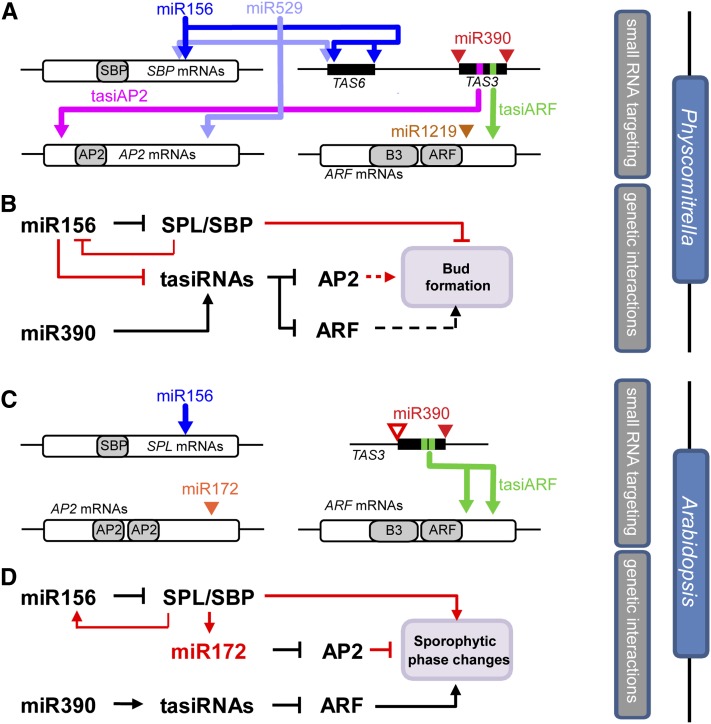
Review and synthesis of previously described miRNA and tasiRNA targets (Arazi et al., 2005; Axtell et al., 2006, 2007; Talmor-Neiman et al., 2006; Addo-Quaye et al., 2009) in light of improved P. patens gene annotations revealed a densely connected regulatory network (Figure 7A; see Supplemental Figure 13, Supplemental Table 3, and Supplemental Data Set 1 online). The conserved miRNAs miR156, miR390, and miR529 (closely related in sequence to miR156), along with the P. patens–specific miR1219 and two different TAS3-derived tasiRNAs, coordinately regulate members of three distinct transcription factor families (SBP, ERF-like AP2, and ARF). Each RNA target in this network is regulated by two distinct small RNA families. Perturbation of this network influences the gametophyte body plan by either retarding (inhibition of miR156 function in MIM156 plants, overexpression of miR390 in MIR390C OE plants) or accelerating (loss of a critical miR156 target in ΔSBP3 plants) the production of buds and leafy gametophores (Figure 7B). miR390 and miR1219 accumulation is unaffected in MIM156 and ΔSBP3 plants (Figure 4E; see Supplemental Figure 14A online), while miR529 accumulation is slightly reduced in MIM156 plants, possibly due to interaction with the miR156 target mimic RNA (see Supplemental Figure 14B online). (However, note that the similarity between miR529 and miR156 is not extensive enough to allow miR156 to also target the tasiAP2 target mRNAs: We find no evidence for this interaction from degradome sequencing data [Addo-Quaye et al., 2009], and the alignment between miR156 and the tasiAP2 target mRNAs has extensive mismatches, particularly in the critical 5′ end of the alignment [see Supplemental Figure 14C online].) Importantly, we don’t exclude the possibility of the presence of other currently unknown components in this network as well and in particular the possibility for significant feedback loops between transcription factors and the small RNAs that target them. The molecular phenotypes of the ΔSBP3 plants (increased miR156 accumulation and decreased tasiRNA target accumulation) hint at the likelihood of yet to be discovered connections in this network. In addition, it remains possible that TAS6a-derived tasiRNAs might also affect developmental progression in P. patens.
Figure 7.
Comparison of Small RNA/Target and Genetic Networks Controlling Developmental Transitions between P. patens and Arabidopsis.
(A) Small RNA–target interactions in P. patens. Colored arrows indicate small RNA targeting, open rectangles indicate open reading frames, gray regions indicate protein domains, and filled rectangles indicate regions of phased siRNA production. Not to scale.
(B) Genetic interactions controlling bud formation and leafy gametophore formation in P. patens. Interactions in red differ when compared with Arabidopsis; those in black are conserved. Dotted lines indicate hypothetical interactions.
(C) to (D) As in (A) and (B), respectively, for Arabidopsis. Empty triangle in (C) reflects the nonslicing miR390 site found in Arabidopsis TAS3 loci.
DISCUSSION
miR156 and miR390 Control tasiRNA Accumulation and Function in P. patens
In this study, we functionally characterized P. patens miR156 and miR390, two of the most ancient miRNAs in plants. MIM156 plants exhibited a delayed bud and gametophore formation and a coincident induction of SBP targets. Deletion of a single miR156 target, SBP3, resulted in a phenotype opposite to that caused by inhibition of miR156, suggesting the SBP3 is a miR156 target of major phenotypic importance. Overexpression of MIR390C also impeded the developmental transition. Accordingly, the accumulation of tasiRNAs was elevated, and target ARF and AP2 transcription factors were suppressed in MIR390C OE plants. These data indicate that miR156 and miR390 oppositely regulate developmental timing in P. patens. Previously, involvement of two miRNAs in the same developmental process was reported in Arabidopsis, in which the gradient of miR390 and miR166 defines the leaf polar axis formation (Nogueira et al., 2007). While miR166 defines abaxial fates in leaf primordia, miR390 restricts the expression domain of abaxial determinants at the adaxial region. Thus, miR166 and miR390 regulate leaf asymmetry via reciprocal expression patterns. Functional characterization of P. patens miR166 has not yet been reported. However, we have shown that P. patens miR390 and miR156 converge upon the same pathway by regulating a single TAS precursor with opposite effects on the production of key tasiRNAs and ultimately upon phenotype. We hypothesize that this convergent regulation confers a fine tuning on levels of tasiRNAs and their targets to ensure the optimal timing of the process of bud formation.
A Partially Conserved Small RNA Network Regulates Bud Formation in Mosses
The conserved miRNAs miR156, miR529, miR390, as well as the nonconserved miRNA, miR1219, and TAS3-derived tasiRNAs coordinate the regulation of three distinct transcription factor families in P. patens. While this network is not precisely replicated in angiosperms, there are several intriguing parallels (Figures 7C and 7D). The regulation of SBP domain mRNAs by miR156 (Arazi et al., 2005; Schwab et al., 2005; Wu and Poethig, 2006; Wu et al., 2009) and the regulation of ARF mRNAs by miR390-dependent, TAS3-derived tasiRNAs is shared by both lineages (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006; Axtell et al., 2007). In angiosperms, miR172 targets several mRNAs containing dual AP2 domains, including AP2 itself (Aukerman and Sakai, 2003; Chen, 2004), is directly regulated by the products of miR156-targeted SBP genes (Wu et al., 2009) and cooperates with miR156 to regulate multiple facets of developmental timing in angiosperms (Huijser and Schmid, 2011). In addition, miR156 appears to directly cooperate with miR172 to trigger-phased siRNA production from a Medicago truncatula AP2 gene (Zhai et al., 2011). miR172 is absent in P. patens; however, P. patens mRNAs containing an ERF-like single AP2 domain are targeted by tasiRNAs, which are themselves influenced by miR156. We did not find neighboring miR156 and miR390 complementary regions (“neighboring” was defined as occurring within 20 kb) in a survey of 19 seed plant genomes (one lycophyte, four monocots, and 15 eudicots), suggesting that the TAS6/TAS3 arrangement may be unique to the bryophyte lineage. Nonetheless, a potential connection between miR156 and miR390 in Arabidopsis is suggested by the unexplained influence of the Arabidopsis miR390 tasiRNA pathway upon the accumulation of the miR156 SBP domain target SPL3 (Peragine et al., 2004).
miR156, miR390, and the targets of miR156 influence vegetative developmental transitions in both mosses and flowering plants. Interestingly, the directionality of miR156’s phenotypic influence differs between flowering plants and mosses; it represses transitions in the former and promotes them in the latter. By contrast, miR390 represses developmental transitions in both lineages. The tight regulatory network found in P. patens converging upon interlocking small RNA regulation of SBP/SPL, AP2, and ARF mRNAs appears to have been partially conserved during land plant diversification. Comparing the genes directly regulated by SBP/SPL, AP2, and ARF transcription factors in different plant lineages should be a productive avenue of future research.
However, it is not yet clear if the tasiRNA-targeted ARF and AP2 genes in P. patens directly regulate bud formation or, alternatively, whether the developmental effects of MIR390 overexpression might instead be mediated by other unknown components. We favor the hypothesis that the bud formation defects seen upon overexpression of MIR390 are due to the elevated levels of AP2 and/or ARF mRNAs that are apparent, but we have not yet directly demonstrated this by analysis of the relevant P. patens ARF and AP2 mutants. In addition, accumulation levels of the relevant tasiRNAs are not strongly correlated with those of their target mRNAs during gametophore development (Figures 5A and 5B). One possible explanation for this discordance is that the AP2 and ARF mRNA levels are controlled by more factors other than just tasiRNAs. However, we cannot exclude the possibility that other, currently unknown genes, mediate the miR390-dependent phenotypes in P. patens.
Many Pathways Influence Bud Formation in P. patens
Several previous studies reported the role of small RNAs or of small RNA-related genes in the timing and rate of bud and leafy gametophore development in P. patens, including miR534a (Saleh et al., 2011), DCL3 (Cho et al., 2008), and RDR6 (Talmor-Neiman et al., 2006; Axtell et al., 2007). In this study, we demonstrated that a miR156- and miR390-regulated tasiRNA network is also involved in this process. The requirement of RDR6 for tasiRNA biogenesis integrates RDR6 into the miR156-miR390-tasiRNA network; however, to the best of our current knowledge, miR534a and DCL3 both act independently of the miR156-miR390-tasiRNA pathway to regulate bud and leafy gametophore production.
miR534a, a moss-specific miRNA targeting the BOP1 and BOP2 transcription factor mRNAs, represses bud and leafy gametophore formation (Saleh et al., 2011), which is opposite to the role of miR156 and similar to the role of miR390. Cytokinin, which is critical for bud initiation in P. patens (Cove and Knight, 1993; Schumaker and Dietrich, 1997), downregulates the MIR534a promoter and causes a correlated increase in BOP1 and BOP2 accumulation (Saleh et al., 2011). Our data show that reduction of miR156 function in MIM156 plants did not affect accumulation levels of BOP1 or BOP2, and cytokinin treatment had negligible effects on the accumulation of most of the RNAs involved in the miR156-miR390-tasiRNA pathway. Thus, the miR156-miR390-tasiRNA pathways and the cytokinin-miR534a pathways appear to independently control bud and gametophore production.
DCL3, responsible for 22- to 24-nucleotide-long siRNA production from repetitive genomic regions, negatively affects developmental timing (Cho et al., 2008; Arif et al., 2012). The accumulation of the miR156-targeted SBP mRNAs was unchanged in ΔDCL3 plants, and the accumulation of miRNAs is not affected in ΔDCL3 mutants (Cho et al., 2008). Thus, the miR156-miR390-tasiRNA pathway, the cytokinin-miR534a-BOP pathway, and the DCL3 pathway all seem to independently affect the timing of bud formation in P. patens. Nonetheless, we acknowledge that further investigation might yet uncover molecular links between these three pathways.
Why does P. patens devote so much regulatory capacity to the control of bud development? The transition from filamentous, two-dimensional growth to the production of upright leafy gametophores is a major developmental transition for mosses. Bud formation and subsequent leafy gametophore growth will only be successful if the protonematal network has become extensive enough to anchor the growing plant and can provide adequate nutrients, especially nitrogen, from the soil. Furthermore, production of buds and leafy gametophores is a prerequisite for the appearance of gametangia and subsequent attempts at sexual reproduction. Perhaps the importance of the decision to produce leafy buds and gametophores and the need to integrate multiple endogenous and environmental stimuli has led to the multiplicity of pathways that affect this trait.
METHODS
Oligonucleotides
All oligonucleotide sequences are listed in Supplemental Table 4 online.
Construction of Vectors
For construction of the MIM156 vector, the At4 gene was amplified from Arabidopsis thaliana and cloned into the pUJ3 vector. The pUJ3 vector contained the Gateway cassette under the control of the maize (Zea mays) Ubiquitin promoter and a hygromycin selection cassette, in which two DNA fragments from the 108 locus are inserted at the flanking regions of the expression and the selection cassette for targeting into the 108 locus into the moss genome (Schaefer and Zrÿd, 1997). To create a miR156 target mimic, oligos were hybridized and ligated with the HindIII digested pUJ3-At4 (Figure 2A). For the SBP3 knock out construct, two ∼1-kb regions 5′ and 3′ from the open reading frame of SBP3 were amplified and inserted into the pUQ vector (Cho et al., 2008). The vector for MIR390c overexpression was obtained by the insertion of pri-MIR390c region into the BsrGI site in the pUJ3 vector.
Transformation and DNA Gel Blot Analysis
Physcomitrella patens transformation was performed as described (Cho et al., 2008). Genomic DNAs were extracted using a Phytopure DNA extraction kit (GE Healthcare) and used for genotyping via PCR. For DNA gel blot analysis of ΔSBP3, the SphI-digested genomic DNAs were blotted onto a Hybond NX nylon membrane (GE Healthcare) and hybridized following a standard protocol (Sambrook and Russell, 2001). As a probe, hptII fragment was amplified by PCR and radiolabeled with [α-32P]dCTP using an NEblot kit (New England Biolabs).
Rapid Amplification of cDNA Ends–PCR
The full-length cDNA of TAS6a and TAS3a precursor was cloned using the GeneRacer kit (Invitrogen) following the manufacturer’s protocol.
Quantitative Real-Time RT-PCR
Total RNA for miRNA analyses were isolated using Tri-Reagent (Sigma-Aldrich) following the manufacturer’s instructions and transcribed with a Superscript III reverse transcriptase (Invitrogen) essentially as previously described (Varkonyi-Gasic et al., 2007). DNase I (Fermentas)–treated total RNAs (100 ng) were converted to cDNAs with Superscript III reverse transcriptase, primed by miRNA-specific stem loop primers using a pulsed RT reaction as described (Varkonyi-Gasic et al., 2007). As a reference, the primer for U6 was also included in the same reaction. A serial dilution was included in the same reaction plate of real-time PCR to calculate the PCR efficiency.
Total RNAs for mRNA analyses were isolated using an RNeasy plant mini kit (Qiagen) and reverse transcribed with a QuantiTect reverse transcription kit (Qiagen). EF1α was used as a reference. Real-time PCRs were performed using a QuantiTect SYBR Green PCR kit (Qiagen) with an ABI7300 teal-time PCR system (Applied Biosystems). PCR efficiencies were calculated based on serial dilution and used to determine relative accumulation levels by the using the efficiency corrected ΔΔCt (cycle threshold) method. Relative expression was normalized to the per-gene wild-type values, except Figures 1, 5A, and 5B, which were normalized to the per-gene week one value, and Figure 3A to the per-EF1α values, respectively, or as described (see Supplemental Figures 1, 3, 4, 6, and 8 to 10 online). As a statistical analysis, Student’s t tests were performed with two-tailed distribution.
Small RNA Gel Blots
Total RNAs, including small RNAs, were isolated using Tri reagent (Sigma-Aldrich) following the manufacturer’s instructions. Small RNAs were enriched as described (Blevins, 2010). Eight micrograms of small RNAs were resolved in 15% Criterion TBE-Urea gel and transferred to the Hybond-NX nylon membrane (GE Healthcare), followed by cross-linking using a 1-ethyl-3-(3-dimethylamonipropyl) carbodiimide–mediated chemical cross-linking method (Pall and Hamilton, 2008). The probes for TAS3a, TAS6a, and U6 were amplified from cDNA by PCR and radiolabeled with [α-32P]dCTP using an NEblot Kit (New England Biolabs) and hybridized to the membrane at 37°C within PerfectHyb solution (Sigma-Aldrich). Membranes were washed twice with nonstringent washing buffer (3× SSC, 5% SDS, and 25 mM NaH2PO4, pH 7.5) for 10 min and twice for 30 min at 42°C. The final wash was performed with the stringent wash buffer (1× SSC and 1% SDS) for 5 min at 42°C. The oligo probes for tasiAP2, tasiARF, miR529, miR1219, and U6 were radiolabeled with [γ-32P]dATP using T4 polynucleotide kinase (New England Biolabs) following the manufacturer’s protocol, hybridized to the nylon membrane at 42°C, and washed as in the double-stranded DNA probes at 50°C. The radioactive signals were recorded onto phosphor screens (GE Healthcare) and scanned using a STORM860 phosphor imager (Amersham Biosciences). Membranes were deprobed by washing in 1% SDS at 80°C for 30 min and hybridized with another probe. Image quantification was performed with the ImageJ program (NIH).
Small RNA Target Analyses
P. patens degradome data (National Center for Biotechnology Information Gene Expression Omnibus GSM410805; Addo-Quaye et al., 2009) were analyzed using CleaveLand 3.0.1 (http://axtell-lab-psu.weebly.com/cleaveland.html). Transcript models were the version 6.1 models, downloaded from Phytozome 7.0 in August, 2011, augmented with the genomic loci encompassing known TAS3 and TAS6 loci or, in the case of TAS6a/TAS3a, the full-length primary transcript. Strong evidence of slicing (thick lines in Supplemental Figure 13 online) reflects either prior evidence from the literature in the form of gene-specific RNA ligase–mediated 5′ rapid amplification of cDNA ends, degradome-based evidence with a P value ≤ 0.05, or both. Weak evidence for slicing (solid thin lines in Supplemental Figure 13 online) reflects degradome data with a P value > 0.05. Details are in Supplemental Table 3 and Supplemental Data Set 1 online.
Measurement of Growth Rate and BAP Treatment
For measurement of growth rate, one to two filaments of 1-week-old protonemal tissues were inoculated onto BCD media with (Figures 3E and 5D) or without (see Supplemental Figure 11 online) ammonium supplementation under standard growth conditions (25°C, 16 h day/8 h night) (Ashton and Cove, 1977). Numbers of buds and gametophores were counted every 2 d.
For BAP treatment, blended protonemata were grown on cellophane overlaid BCD media with ammonium. BAP (5 μM; Sigma-Aldrich) dissolved in 40 μM NaOH was added on top of the protonemata and incubated for 24 h. As a mock control, 40 μM NaOH treatment was performed simultaneously.
Accession Number
Sequence data for the TAS6a/TAS3a primary transcript from this article can be found in the GenBank/EMBL data libraries under accession number JN674513.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Accumulation of miR156 and Its Targets during P. patens Gametophyte Development.
Supplemental Figure 2. The Life Cycle of Physcomitrella patens
Supplemental Figure 3. Functional Knockdown of miR156 by Target Mimicry.
Supplemental Figure 4. Accumulation of SBP Transcripts during the P. patens Gametophyte Development.
Supplemental Figure 5. Targeted Knockout of SBP3.
Supplemental Figure 6. Transcript Levels of Genes Related to the Gametophore Transition in P. patens.
Supplemental Figure 7. Small RNA Gel Blots of TAS6a and TAS3a in ΔDCL4 and ΔRDR6.
Supplemental Figure 8. miR156 Influences P. patens tasiRNA Accumulation and Their Targets in Older Tissues.
Supplemental Figure 9. Transcript Level of the Target of tasiZNF in ΔSBP3 and in MIM156s in 3-Week-Old Samples.
Supplemental Figure 10. Accumulation of miR390-Regulated tasiRNAs and Their Targets
Supplemental Figure 11. Comparison of Bud and Leafy Gametophore Formation in MIM390C OE, ΔSBP3, and ΔRDR6-19 Plants.
Supplemental Figure 12. Cytokinin Treatment Accelerates Bud Formation.
Supplemental Figure 13. Detailed Small RNA–Target Interactions in P. patens.
Supplemental Figure 14. Accumulation of miR529 and miR1219 in MIM156-2 and ΔSBP3 Plants.
Supplemental Table 1. Origins of P. patens tasiAP2 and tasiARF.
Supplemental Table 2. Bud Formation in MIR390C-Overexpressing P. patens.
Supplemental Table 3. Summary of small RNA/Target Data.
Supplemental Table 4. Oligonucleotide Sequences Used in This Study.
Supplemental Data Set 1. Small RNA Target Alignments and Degradome Data.
Acknowledgments
This work was funded by a grant from the U.S. National Institutes of Health-National Institute of General Medical Sciences (R01 GM84051) to M.J.A. We thank Jo Ann Snyder for technical support, Zhaorong Ma and Wolfgang Frank for insights on the TAS6 family, Rajendran Rajeswaran for discussions and pilot experiments, and all members of the Axtell Lab for many constructive discussions during this study.
AUTHOR CONTRIBUTIONS
M.J.A. and S.H.C. designed the research. S.H.C. and C.C. performed research. M.J.A. analyzed degradome and target prediction data. M.J.A. and S.H.C. wrote and edited the article with input from C.C.
Glossary
- miRNA
microRNA
- tasiRNA
trans-acting small interfering RNA
- qRT-PCR
quantitative RT-PCR
- siRNA
small interfering RNA
- OE
overexpressing
- BAP
benzylaminopurine
References
- Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.F., Sunkar R., Axtell M.J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43: 837–848 [DOI] [PubMed] [Google Scholar]
- Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. (2012). DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton N.W., Cove D.J. (1977). The isolation and preliminary characterization of auxotropic and analogue-resistant mutants of the moss Physcomitrella patens. Mol. Gen. Genet. 154: 87–95 [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Snyder J.A., Bartel D.P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19: 1750–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T. (2010). Northern blotting techniques for small RNAs. In Plant Epigenetics, I. Kovalchuk and F.J. Zemp, eds (Totowa, NJ: Humana Press), pp. 87–107.
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.H., Addo-Quaye C., Coruh C., Arif M.A., Ma Z., Frank W., Axtell M.J. (2008). Physcomitrella patens DCL3 is required for 22-24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 4: e1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D.J., Knight C.D. (1993). The moss Physcomitrella patens, a model system with potential for the study of plant reproduction. Plant Cell 5: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Fahlgren N., Carrington J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. (2010). Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Nogueira F.T., Madi S., Chitwood D.H., Juarez M.T., Timmermans M.C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G.S., Hamilton A.J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (2009). Small RNAs and developmental timing in plants. Curr. Opin. Genet. Dev. 19: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese M., Höhmann S., Saedler H., Münster T., Huijser P. (2007). Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 401: 28–37 [DOI] [PubMed] [Google Scholar]
- Saleh O., Issman N., Seumel G.I., Stav R., Samach A., Reski R., Frank W., Arazi T. (2011). MicroRNA534a control of BLADE-ON-PETIOLE 1 and 2 mediates juvenile-to-adult gametophyte transition in Physcomitrella patens. Plant J. 65: 661–674 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schaefer D.G., Zrÿd J.P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schumaker K.S., Dietrich M.A. (1997). Programmed changes in form during moss development. Plant Cell 9: 1099–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Talmor-Neiman M., Stav R., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T. (2006). Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 48: 511–521 [DOI] [PubMed] [Google Scholar]
- Tanahashi T., Sumikawa N., Kato M., Hasebe M. (2005). Diversification of gene function: Homologs of the floral regulator FLO/LFY control the first zygotic cell division in the moss Physcomitrella patens. Development 132: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. (2007). Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]