This work shows that the chromatin remodeling ATPase BRAHMA ensures that postgermination growth arrest is triggered only upon sensing of the water stress hormone ABA. BRAHMA directly represses expression of ABA-responsive genes, including ABI5 by modulating nucleosome occupancy and position. Furthermore, brahma mutants display drought tolerance.
Abstract
The survival of plants as sessile organisms depends on their ability to cope with environmental challenges. Of key importance in this regard is the phytohormone abscisic acid (ABA). ABA not only promotes seed dormancy but also triggers growth arrest in postgermination embryos that encounter water stress. This is accompanied by increased desiccation tolerance. Postgermination ABA responses in Arabidopsis thaliana are mediated in large part by the ABA-induced basic domain/leucine zipper transcription factor ABA INSENSITIVE5 (ABI5). Here, we show that loss of function of the SWI2/SNF2 chromatin remodeling ATPase BRAHMA (BRM) causes ABA hypersensitivity during postgermination growth arrest. ABI5 expression was derepressed in brm mutants in the absence of exogenous ABA and accumulated to high levels upon ABA sensing. This effect was likely direct; chromatin immunoprecipitation revealed BRM binding to the ABI5 locus. Moreover, loss of BRM activity led to destabilization of a nucleosome likely to repress ABI5 transcription. Finally, the abi5 null mutant was epistatic to BRM in postgermination growth arrest. In addition, vegetative growth defects typical of brm mutants in the absence of ABA treatment could be partially overcome by reduction of ABA responses, and brm mutants displayed increased drought tolerance. We propose a role for BRM in the balance between growth or stress responses.
INTRODUCTION
When seed dormancy is broken by the appropriate environmental and endogenous cues, the radicle penetrates the seed coat during germination (Bewley, 1997). The newly germinated embryo next initiates a series of developmental changes prior to entering the seedling developmental program (Bewley, 1997; Lopez-Molina et al., 2001). Most notably, the germinated embryo must ensure appropriate food and water supply by switching to autotrophic growth (photosynthesis) and by elongating the root, respectively. These reprogramming events occur during the first 48 h after dormancy is broken in the postgermination embryo and culminate with seedling establishment and onset of vegetative development (Lopez-Molina et al., 2001). During postgermination, the embryo is no longer protected by the seed coat and thus is particularly vulnerable to drought stress. If plants encounter water stress during this developmental window, a growth arrest is triggered that helps protect germinated embryos against water stress–mediated cell and tissue damage (Lopez-Molina et al., 2001). The growth arrest and induction of the quiescent state involves similar signaling and response mechanisms to those that operate during seed development to induce desiccation tolerance and dormancy (Lopez-Molina et al., 2001, 2002; Bensmihen et al., 2002; Finkelstein et al., 2008).
When plants sense water stress, the levels of the stress hormone abscisic acid (ABA) rise (Nambara and Marion-Poll, 2005). ABA sensing triggers a signal transduction cascade that allows SnRK2-type kinases to phosphorylate and activate basic domain/leucine zipper (bZIP) family transcription factors, which leads to the upregulation of ABA-responsive element (ABRE)–dependent gene expression (Cutler et al., 2010; Hubbard et al., 2010; Raghavendra et al., 2010; Umezawa et al., 2010; Fujita et al., 2011). Both ABA and the bZIP transcription factor ABI5 are important for osmotic stress responses during late seed maturation and for execution of the ABA-dependent growth arrest prior to photosynthetic growth (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001; Bensmihen et al., 2002; Brocard et al., 2002). ABA INSENSITIVE5 (ABI5) is also implicated in control of radicle emergence (germination) (Lopez-Molina et al., 2001, 2002). Loss of ABI5 function causes reduced ABA sensitivity, whereas ectopic expression of ABI5 enhances ABA sensitivity and drought resistance (Lopez-Molina et al., 2001; Brocard et al., 2002). ABI5 expression is the most abundant in dry seeds and decreases during postgermination development (Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001; Finkelstein et al., 2005). Although ABI5 expression is low after seedling establishment, ABI5 is induced upon drought sensing also during vegetative development, in an ABA signaling–dependent manner (Brocard et al., 2002; Zhu et al., 2007; Nakashima et al., 2009; Mizoguchi et al., 2010; Gonzalez-Guzman et al., 2012).
Another key transcription factor important for establishment of desiccation tolerance and dormancy is the B3 domain transcription factor ABI3 (Parcy et al., 1997). ABI3 has also been linked to regulation of germination (Parcy et al., 1994; Nambara et al., 2000). Importantly, ABI3 has a key role in promoting postgermination growth arrest under osmotic stress conditions and acts upstream of ABI5 in this process (Lopez-Molina et al., 2002). ABI3 is abundant in maturing and mature seeds, but ABI3 mRNA and protein levels become undetectable upon seedling establishment (Parcy et al., 1994; Perruc et al., 2007). ABI3 cannot be induced by ABA during vegetative development (Nakashima et al., 2006).
Altered transcriptional responses to environmental stimuli, such as abiotic stress, have been linked to chromatin regulation (Chinnusamy and Zhu, 2009; Kim et al., 2010). Chromatin-mediated control of inducible gene expression is performed by two general types of activities. One mechanism involves enzymes that covalently modify histones and/or the DNA, such as histone-modifying enzymes or DNA (de)methylases (Li et al., 2007). A second general mechanism for chromatin-mediated control of inducible gene expression is noncovalent alteration of the nucleosome position, occupancy, conformation, and composition by chromatin remodeling ATPases. Among the chromatin remodeling ATPases, the SWI2/SNF2 subgroup has been studied extensively (Li et al., 2007; Clapier and Cairns, 2009; Hargreaves and Crabtree, 2011).
SWI2/SNF2 subgroup ATPases are conserved from yeast to humans and plants (Flaus et al., 2006). Plant genomes contain three types of SWI2/SNF2 subgroup chromatin remodeling ATPases, which are called BRAHMA (BRM), SPLAYED (SYD), and MINUSCULE (MINU) (Flaus et al., 2006; Jerzmanowski, 2007; Kwon and Wagner, 2007; Sang et al., 2012). SWI2/SNF2 ATPases act in large protein complexes that are required for full activity in vivo and use the energy derived from ATP hydrolysis to alter histone–DNA interactions (Clapier and Cairns, 2009; Hargreaves and Crabtree, 2011). SWI2/SNF2 complexes can increase or decrease accessibility of the genomic DNA and hence activate or repress transcription, respectively (Tang et al., 2008; Clapier and Cairns, 2009; Hargreaves and Crabtree, 2011). Selectivity of SWI2/SNF2 activity is due to recruitment to target loci by sequence-specific proteins and/or regulation of complex activity by posttranslational modifications or complex composition (Clapier and Cairns, 2009; Hargreaves and Crabtree, 2011).
Constitutive activation of ABA signaling by removing negative regulators of the ABA pathway or by enhancing transcriptional response to ABA causes ABA hypersensitivity and enhanced drought tolerance (Lopez-Molina et al., 2001; Kang et al., 2002; Fujita et al., 2005; Rubio et al., 2009). However, it also causes impaired growth under normal growth conditions (Kang et al., 2002; Fujita et al., 2005). This is because the abiotic stress responses divert resources from normal growth and development (Boyer, 1982; Grill and Ziegler, 1998; Cramer et al., 2011; Less et al., 2011). It is therefore critical that desiccation responses are repressed in nonstress conditions. Here, we describe a role for the Arabidopsis thaliana BRM SWI2/SNF2 chromatin remodeling complex components in direct transcriptional repression of ABI5 during postgermination development. BRM promotes high occupancy of a well-positioned nucleosome at the ABI5 locus, which may contribute to repression of ABI5 expression in the absence of exogenous ABA. Consistent with these findings, germinated brm mutant embryos display enhanced ABA-mediated growth arrest. This brm phenotype is dependent on ABI5 activity. Derepression of ABI5 in brm is also observed later during vegetative development and adversely affects growth of brm mutants. Our data reveal a mechanism for chromatin-mediated regulation of appropriate water stress responses prior to seedling establishment and during vegetative growth.
RESULTS
Increased ABA Sensitivity in brm Mutants
To investigate a possible link between SWI2/SNF2-dependent chromatin remodeling complexes and postgermination ABA responses, we probed the effect of mutations in each of the four Arabidopsis SWI2/SNF2 ATPases, SYD, BRM, MINU1, and MINU2 (Farrona et al., 2004; Flaus et al., 2006; Sang et al., 2012) on ABA-dependent growth arrest. Of all mutants tested, those in BRM displayed the most dramatic change in ABA sensitivity relative to the wild type (see Supplemental Figures 1A and 1B online). We therefore focused further analyses on the role of the BRM complex in ABA-mediated postgermination growth inhibition.
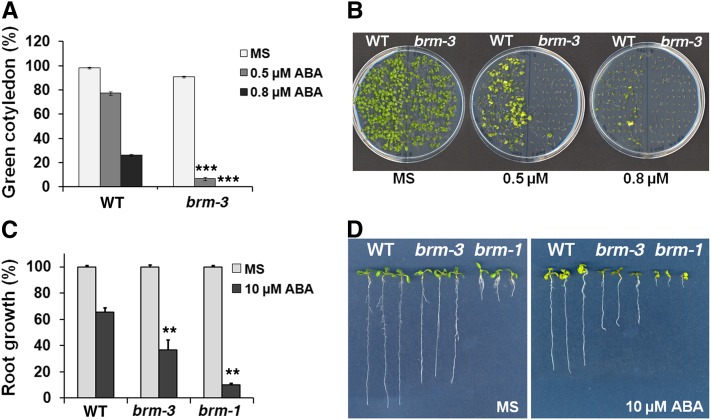
After germination of brm-3 hypomorphic (Farrona et al., 2007) mutants on agar plates containing submicromolar ABA concentrations, the mutant germinated embryos failed to develop green cotyledons and the first pair of true leaves at the lowest ABA concentration tested (Figures 1A and 1B). The wild type did not display growth arrest in this condition. Moreover, when we transferred brm-3 and brm-1 null (Hurtado et al., 2006) mutants to plates containing ABA, the growth of the primary root of was inhibited by ABA to a greater extent than wild-type roots (Figures 1C and 1D). Thus, relative to the wild type, brm mutants were hypersensitive to ABA.
Figure 1.
brm Mutants Are Hypersensitive to ABA.
(A) The percentage of germinated embryos that developed green cotyledons in the presence of 0.5 or 0.8 µM ABA in the wild type (WT) and in the hypomorph brm-3 mutant. Values are mean ± se from three independent experiments. Asterisks indicate statistical significance compared with wild-type values based on χ2 test (n = 250, P < 1E-10).
(B) Representative pictures for the data shown in (A). Photographs were taken 11 (MS) and 18 (ABA) d after stratification.
(C) Root growth inhibition of brm-1 null and brm-3 hypomorph mutants. Values are mean ± se from two independent experiments. Asterisks indicate statistical significance compared with wild-type values based on one-tailed Student’s t test (n = 10, P < 0.001).
(D) Representative pictures for data shown in (C). Photographs were taken 10 d after stratification.
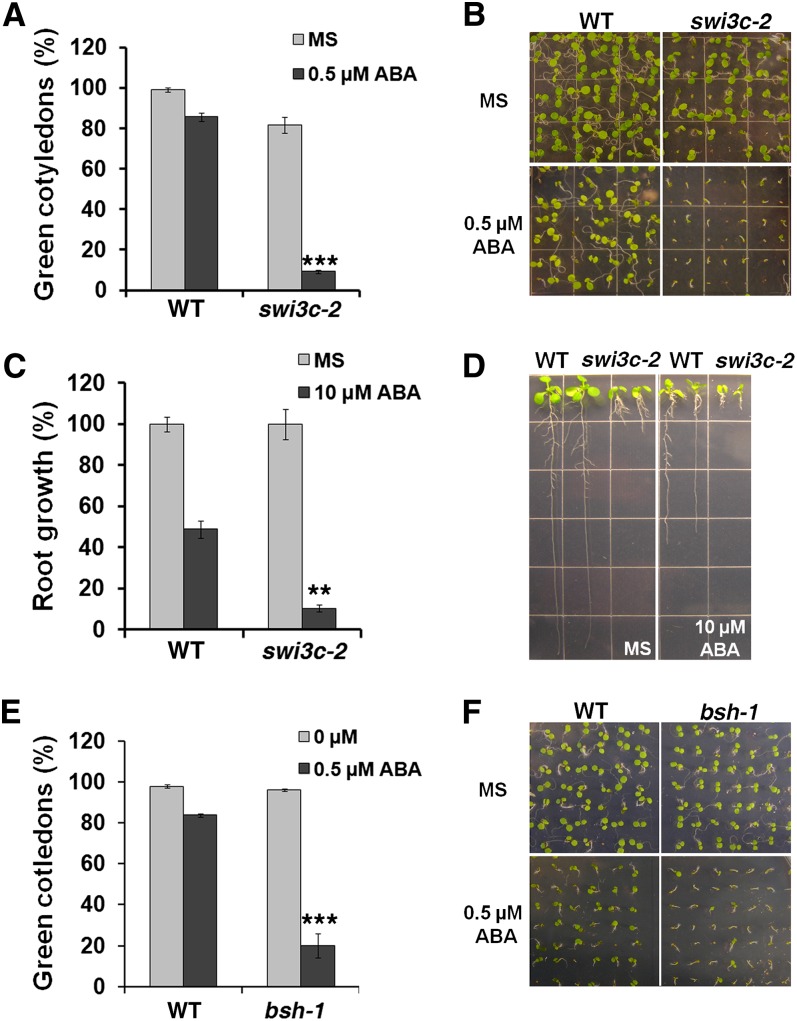
The evolutionally conserved SWI2/SNF2 core complex consists of one ATPase, two SWI3 subunits, and one SNF5 complex component (Phelan et al., 1999; Jerzmanowski, 2007; Kwon and Wagner, 2007; Hargreaves and Crabtree, 2011). The Arabidopsis genome encodes four SWI3 subunit genes (called SWI3A-D) and one SNF5 subunit gene (termed BUSHY) (Brzeski et al., 1999; Sarnowski et al., 2005). The morphological defects observed in brm null mutants are very similar to those of swi3c null mutants (Sarnowski et al., 2005; Archacki et al., 2009). Moreover, BRM and SWI3C show strong direct physical interaction (Hurtado et al., 2006), suggesting that SWI3C may be a dedicated BRM complex component. Therefore, we next examined the role of SWI3C and BUSHY (BSH) in postgermination ABA responses. Null swi3c-2 mutants (Sarnowski et al., 2005) showed an ABA-hypersensitive phenotype similar to brm mutants both with respect to cotyledon greening (Figures 2A and 2B) and growth of the primary root (Figures 2C and 2D). Likewise, the hypomorphic bsh-1 mutant (Tang et al., 2008) displayed ABA hypersensitive growth arrest (Figures 2E and 2F).
Figure 2.
swi3c-2 and bsh-1 Mutants Are Hypersensitive to ABA.
(A) The percentage of germinated embryos that developed green cotyledons in the presence of 0.5 µM ABA in the wild type (WT) and in the swi3c-2 mutant. Asterisks indicate statistical significance based on χ2 test (n = 100, P < 1E-10).
(B) Representative pictures for data shown in (A) 7 d after stratification.
(C) Root growth inhibition of the wild type and in the swi3c-2 mutant in the presence of 10 µM ABA relative to that observed on MS media. Asterisks indicate statistical significance based on Student’s t test (n = 20, P < 0.001).
(D) Representative pictures for data shown in (C) 10 d after stratification.
(E) The percentage of germinated wild-type and bsh-1 embryos that developed green cotyledons in the presence of 0.5 µM ABA. Values are mean ± se from two independent experiments. Asterisks indicate statistical significance based on χ2 test (n = 200, P < 1E-10).
(F) Representative pictures for the data shown in (E) 5 d after stratification.
We next examined seed germination (radicle emergence) in brm mutants relative to the wild type using a range of ABA concentrations. In radicle emergence assays (Müller et al., 2006), brm-3 hypomorphic mutants did not display significantly altered sensitivity to ABA (see Supplemental Figure 2A online). By contrast, brm-1 null mutants were significantly more sensitive to low ABA concentrations than the wild type with respect to germination (see Supplemental Figure 2B online). The combined data suggest that BRM affects germination and postgermination response to ABA with a very prominent role for BRM in cotyledon greening.
Derepression of ABA-Responsive Genes in the Absence of the Stress Hormone in brm Mutants
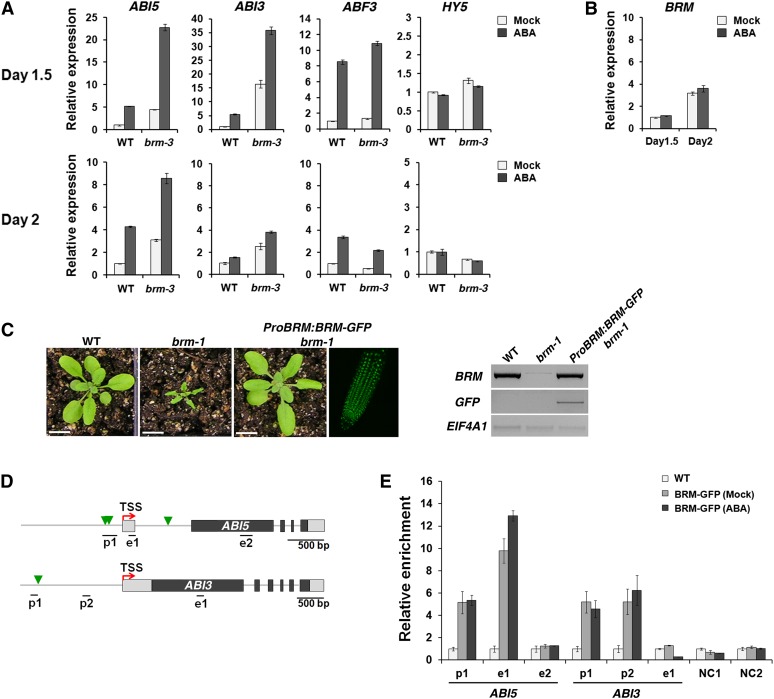
To gain insight into the molecular underpinnings of the observed brm mutant ABA hypersensitivity, we analyzed the expression of ABA-responsive genes in brm-3 mutant and wild-type embryos during postgermination development. We employed the hypomorphic brm-3 allele because, unlike the brm-1 null mutant, it is fertile and thus facilitates testing of homozygous mutant embryos. We examined expression of the bZIP transcription factor ABI5 and the B3 transcription factor ABI3, key regulators of dormancy and desiccation tolerance in germinated embryos (Giraudat et al., 1992; Parcy et al., 1997; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001, 2002). In addition, we quantified expression of the bZIP transcription factor ABF3, which has been shown to act in part in a pathway parallel to ABI5 (Kang et al., 2002; Finkelstein et al., 2005; Yoshida et al., 2010) and HY5, a component of the light signal transduction pathway and direct upstream regulator of ABI5 (Chen et al., 2008). Gene expression was examined in plants grown in continuous light at day 1.5 and day 2.0 after stratification. These time points were chosen because growth arrest is triggered by ABA only before seedling establishment, in the first 48 h after stratification (Lopez-Molina et al., 2001). We observed derepression of ABI5 expression in brm-3 relative to the wild type at both time points (4.4-fold and 3.1-fold at days 1.5 and 2, respectively; Figure 3A). The level of ABI5 mRNA was also much higher in brm-3 mutants relative to the wild type 1 h after ABA sensing; however, the rate of ABI5 induction by ABA was similar in both genotypes (5.2-fold and 5.1-fold at day 1.5 in the wild type and in brm-3, respectively; Figure 3A). ABI3 expression, by contrast, was strongly derepressed only at day 1.5 in brm-3 mutants relative to the wild type. At day 2, ABI3 derepression in brm mutants and induction by ABA was much less pronounced. Again, there was no increase in the fold induction of ABI3 expression by ABA in the brm mutant relative to the wild type. ABF3 expression was only marginally increased in brm-3 in any condition tested, while HY5 expression was not at all altered in the brm-3 mutant (Figure 3A). Thus, partial loss of BRM function led to altered expression of select ABA-responsive genes; most notably derepression of ABI5 and ABI3 expression in the absence of exogenous ABA application.
Figure 3.
BRM Represses Expression of ABI5 and ABI3 during Postgermination Development.
(A) Quantitative RT-PCR in 1.5- and 2-d-old wild-type (WT) and brm-3 mutants 1 h after mock or ABA (50 µM) treatment.
(B) Quantitative RT-PCR in 1.5- and 2-d-old wild-type plants 1 h after mock or ABA treatment. Quantitative RT-PCR expression was normalized over that of EIF4A1, and expression levels in the mock-treated wild type were set to 1. Values are mean ± se of three technical replicates from one representative experiment.
(C) Left: 3-week-old wild-type, brm-1, and brm-1 ProBRM:BRM-GFP plants. Center: GFP expression monitored by confocal microscopy in 2-d-old brm-1 ProBRM:BRM-GFP roots. Right: BRM expression (top panel), GFP expression (center panel), and EIF4A1 expression (bottom panel) tested by semiquantitative PCR. Bars = 1cm.
(D) Diagram of the loci tested. Horizontal lines below the schematic, regions amplified by qPCR; green arrowheads, ABREs; gray box, 5′ or 3′ untranslated region; black box, exon; gray line, intergenic region or intron.
(E) qPCR after anti-GFP ChIP in 1.5-d-old brm-1 ProBRM:BRM-GFP plants after mock or ABA (50 µM) treatment for 1 h. Relative enrichment is the percentage of input fold change after the percentage of input of the wild type was set to 1. Negative controls: exon regions of the retrotransposon TA3 (NC1) and of BRM (NC2). Values are mean ± se of three technical replicates from one representative experiment.
BRM Directly Represses Transcription of ABI5
Since SWI/SNF complexes can both activate and repress transcription (Kwon et al., 2005; Tang et al., 2008; Hargreaves and Crabtree, 2011), it is possible that the effect of BRM on ABI5 and ABI3 mRNA accumulation in the absence of the stress hormone is direct. The expression of BRM was consistent with a possible role in regulation of gene expression at this stage. BRM was expressed in both 1.5- and 2-d-old germinated embryos (Figure 3B). To test for binding of BRM to either the ABI5 or the ABI3 locus, we used a green fluorescent protein (GFP)–tagged biologically active version of BRM (ProBRM:BRM-GFP; Wu et al., 2012), which fully rescued the morphological defects of the brm-1 null mutant and displayed wild-type levels of BRM expression (Figure 3C). Using the brm-1 ProBRM:BRM-GFP as a substrate for chromatin immunoprecipitation (ChIP), we detected strong BRM binding to the ABI5 promoter and to the promoter proximal exon 1 of ABI5, but not to exon 2 (Figures 3D and 3E). In addition, we detected BRM association with the promoter of ABI3 (Figures 3D and 3E). To confirm these results, we generated an Hemagglutinin (HA)-tagged version of BRM, which rescued the brm-1 null mutant and displayed wild-type levels of BRM expression (see Supplemental Figure 3A and Supplemental Methods 1 online). ChIP using brm-1 ProBRM:BRM-HA yielded qualitatively similar results as brm-1 ProBRM:BRM-GFP (cf. Figure 3E and Supplemental Figure 3B online). The association of BRM with the ABI5 and ABI3 loci, in combination with the observed derepression of ABI5 and ABI3 expression in brm mutants, supports the hypothesis that BRM directly acts on ABI5 and ABI3 expression. Two of the BRM-bound regions (p1 in ABI5 and p1 in ABI3) contain ABREs (Figure 3D), cis-elements known to be involved in ABA-induced transcriptional responses (Yamaguchi-Shinozaki and Shinozaki, 2005; Gómez-Porras et al., 2007). We observed high BRM binding at the two loci both in the absence and in the presence of ABA treatment (Figure 3E). This finding was surprising, given that the main effect of loss of BRM activity is derepression of ABI5 and ABI3 expression in the absence of ABA treatment (Figure 3A). The data suggest that BRM is constitutively bound to the ABI5 and the ABI3 locus.
ABI5 Acts Downstream of BRM and Is Required for brm ABA Hypersensitivity
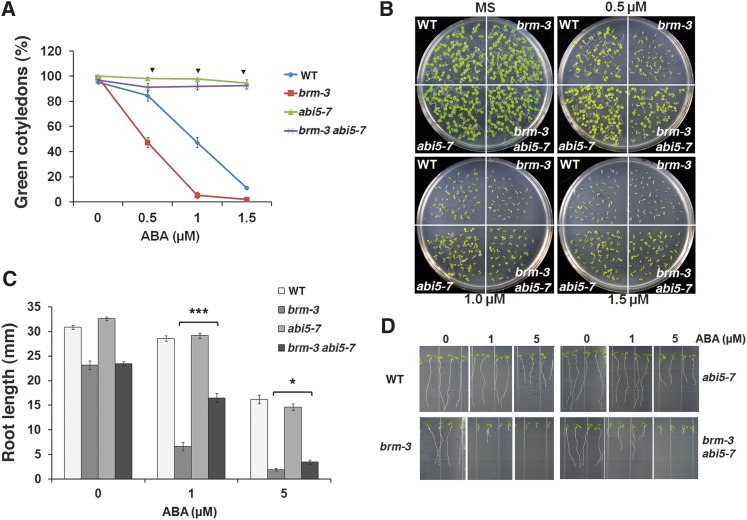
Prior molecular and genetic experiments have shown that ABI3 acts upstream of ABI5 in the ABA-mediated growth arrest of germinated embryos (Lopez-Molina et al., 2002). To elucidate the placement of BRM in this genetic pathway, we generated a double mutant between brm-3 and the abi5-7 null mutant (Yamagishi et al., 2009). The brm-3 allele was employed so we could assay the response in homozygous germinated embryos. While brm-3 was hypersensitive to ABA with respect to inhibition of cotyledon greening, abi5-7 was not responsive to any of the ABA concentrations tested (Figures 4A and 4B), consistent with previous reports (Nambara et al., 2002). Interestingly, the brm-3 abi5-7 double mutant was also not responsive to any of the ABA concentrations tested; like abi5-7, it developed green cotyledons even at the highest dose of ABA tested (Figures 4A and 4B). The data suggest that, with respect to cotyledon greening, ABI5 is epistatic to BRM. This finding, combined with the observed ABI5 derepression in brm mutants and BRM binding to the ABI5 locus, support the hypothesis that ABI5 acts downstream of BRM.
Figure 4.
The Hypersensitive brm Phenotype Is Due to Derepression of ABI5.
(A) Percentage of the germinated embryos that developed green cotyledons in the presence of 0.5, 1.0, and 1.5 µM ABA in the brm-3 abi5-7 double mutants compared with abi5-7, brm-3, and the wild type (WT) 7 d after stratification. Values are mean ± se from three independent experiments. Inverted triangles: no statistical significance compared with wild-type values (n > 100, P > 0.01).
(B) Representative pictures for the data shown in (A).
(C) Root length in the absence or presence of ABA (1 and 5 µM) in brm-3, abi5-7, and brm-3 abi5-7 double mutant plants compared with the wild type. Two-day-old plants were transferred to MS media containing ABA, and roots were measured at day 7. Asterisks: statistical significance based on one-tailed Student’s t test (n > 36, *P < 0.01, ***P < 1E-10).
(D) Representative pictures for the data shown in (C).
We also tested the ABA response of brm-3 abi5-7 double mutant with respect to inhibition of primary root growth. As previously reported (Finkelstein et al., 2005; Miura et al., 2009), the growth of abi5-7 (nulls) roots is inhibited by ABA (Figures 4C and 4D), suggesting redundant activities of other ABA-dependent transcription factors in root growth arrest. Nevertheless, brm-3 abi5-7 roots were significantly less sensitive to 1 or 5 µM ABA than those of brm-3 (Figures 4C and 4D). These data suggest that the increased ABA-dependent inhibition of root growth in the brm-3 mutants is in part attributable to the elevated ABI5 expression.
BRM Contributes to Placement and Occupancy of the Transcription Start Site Proximal Nucleosome at the ABI5 Locus
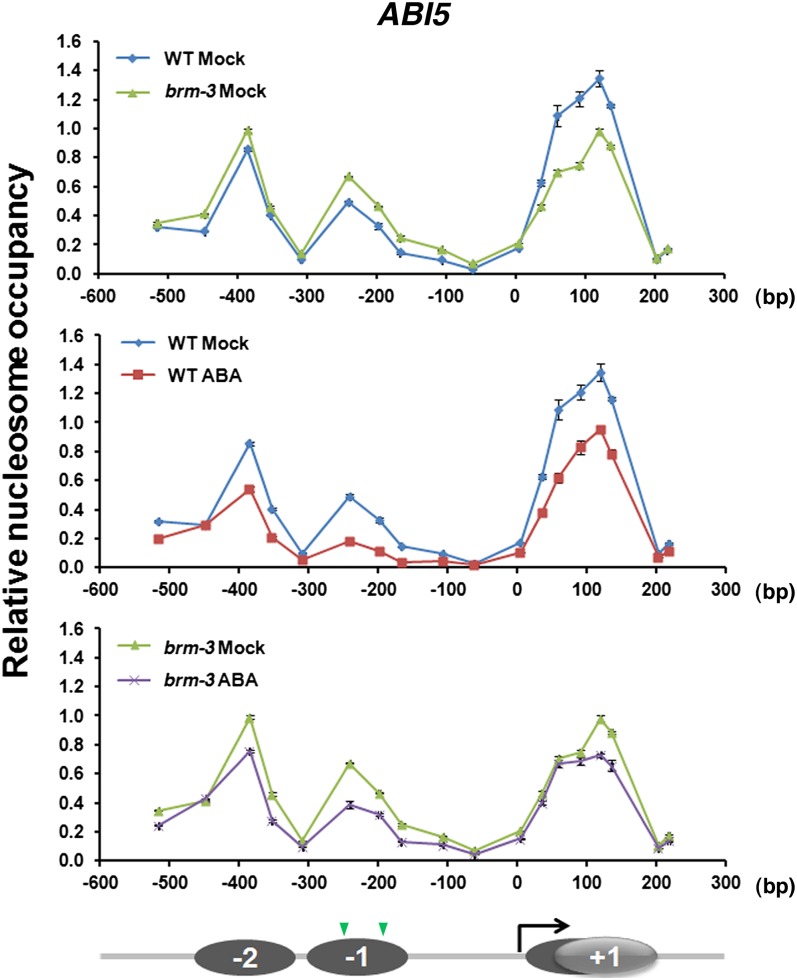
To gain insight into the mechanism by which the SWI2/SNF2 chromatin remodeling ATPase BRM might represses ABI5 expression in the absence of stress hormone treatment, we next examined nucleosome positioning and occupancy at the ABI5 promoter using high-resolution MNase mapping (Chodavarapu et al., 2010; Rafati et al., 2011). We identified two well-positioned nucleosomes in the ABI5 promoter region (−2 and −1 nucleosome) upstream of a 150-bp nucleosome-depleted region (−150 to 0 bp) (Figure 5). Nucleosome-depleted regions just upstream of the transcription start site (TSS) are common in eukaryote promoters (Yen et al., 2012). A typical nucleosome protects ∼147 bp of genomic DNA from MNase digestion (Yen et al., 2012), as was the case for the −2 and −1 nucleosomes at the ABI5 locus (Figure 5). However, the +1 ABI5 nucleosome just downstream of the TSS protected ∼200 bp of DNA, suggesting that this nucleosome may be present in two alternative positions. A nucleosome position prediction program (NuPop; Xi et al., 2010) identified nucleosome start sites around position +45, while the MNase mapping revealed start of the +1 nucleosome close to the +1 position. The data suggest that a subset of the +1 nucleosomes are positioned more TSS proximal than predicted.
Figure 5.
BRM Is Required to Maintain High Occupancy of the +1 Nucleosome at the ABI5 Locus.
MNase digestion followed by tiled primer qPCR to monitor nucleosome positioning and occupancy at the ABI5 locus. MNase qPCR was performed after a 1-h mock or ABA treatment in 2-d-old wild-type (WT) and brm-3 mutants. The fraction of undigested genomic DNA amplified for each amplicon was normalized to that of the −73 position of the negative control locus (gypsy-like retrotransposon; see Supplemental Figure 4 online). Values are mean ± se of three technical replicates from one representative experiment. The number on the x axis denotes distance (bp) from the TSS (0 bp). Below: Diagram of the positioned nucleosomes. Gray ovals, nucleosomes; black arrow, TSS; gray lines, genomic DNA; green arrowheads, ABREs.
In brm mutant plants, we observed derepression of ABI5 expression in the absence of ABA treatment (Figure 3A). Consistent with this observation, we reproducibly found a moderate (∼40%) reduction in nucleosome occupancy at the + 1 position of the ABI5 locus coupled with a shift away from the TSS in the absence of ABA treatment in brm mutant relative to wild-type germinated embryos (Figure 5). No BRM-dependent alteration in nucleosome positioning or occupancy was observed at the −2 or −1 nucleosome of the ABI5 locus (Figure 5). Likewise, no strong change in either occupancy or positioning of nucleosomes was observed at a control locus, a gypsy-like retrotransposon gene (see Supplemental Figure 4 online). Thus, BRM may be required to promote high occupancy and TSS proximity of the +1 nucleosome at the ABI5 locus.
In addition, we detected reduced occupancy of all three nucleosomes in response to ABA treatment (Figure 5). The observed ABA-dependent change in nucleosome occupancy was similar in germinated brm-3 and wild-type embryos, suggesting that this effect was likely BRM independent (Figure 5). The reduced nucleosome occupancy in response to ABA was specific to the ABI5 locus; it was not observed at the control locus (see Supplemental Figure 4 online). Finally, we noted development-dependent changes in the −1 nucleosome occupancy at the ABI5 promoter just prior to seedling establishment. The occupancy of the −1 nucleosome was very low at day 1.5 in both mock-treated wild-type and brm-3 plants but increased at day 2 (see Supplemental Figure 5 online; Figure 5). We did not detect a strong increase in the occupancy of the −2 and +1 nucleosome between days 1.5 and 2.
brm Mutant Vegetative Growth Defects Are Partly Due to Derepressed ABA Responses
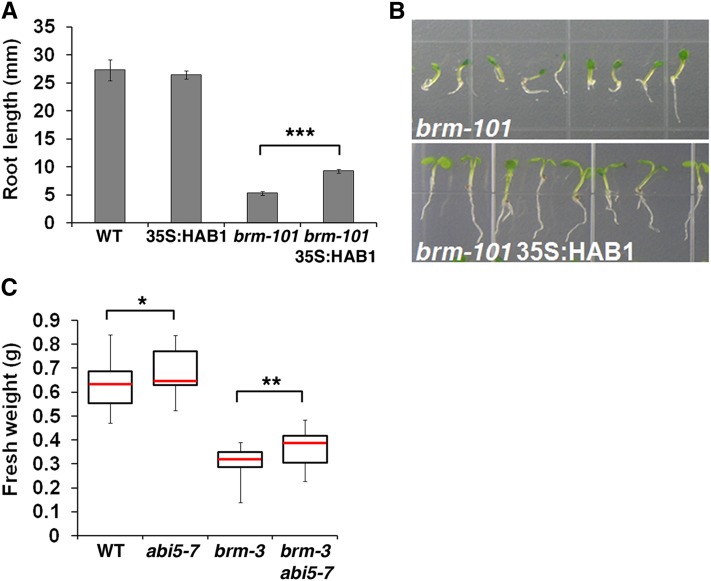
In the absence of ABA, brm plants are small with short roots (Farrona et al., 2004; Kwon et al., 2006). Given our finding that BRM represses ABA responses in the absence of the stimulus during postgermination development, we wondered whether some of the brm mutant vegetative growth defects are attributable to derepressed ABA responses. We therefore monitored root length in double mutants of brm and mutants that display reduced ABA sensitivity. Since ABI5 derepression is only partly responsible for root growth inhibition in brm mutants (Figure 4C), we employed a genetic background that displays reduced ABA sensitivity, 35S:HAB1 (Saez et al., 2004). HAB1 encodes for a PP2C phosphatase, a negative regulator of ABA signaling, that prevents phosphorylation of SnRK2-type kinases (Vlad et al., 2009) and, hence, activation of the ABA-responsive transcription factor ABI5 (Nakashima et al., 2009). 35S:HAB1 inhibits ABA responses in the absence of ABA treatment because low levels of endogenous ABA are able to partially activate ABA-responsive transcription factors in non-ABA-treated plants (Rodrigues et al., 2009).
For these assays, we used the brm-101 null nonsense allele (Kwon et al., 2006) to avoid silencing of the 35S:HAB1 transgene by the T-DNA present in brm-1 or brm-3. As previously reported, the growth of 35S:HAB1 was indistinguishable from the wild type in the absence of applied ABA (Figure 6A; Saez et al., 2004). However, 35S:HAB1 was able to partly rescue the root growth defects of brm mutants under these conditions (Figures 6A and 6B). In addition, overall growth of brm-101 35S:HAB1 was more vigorous than that of brm-101. At day 7, the cotyledons of the double mutant were fully expanded, while those of brm-101 mutants were closed and small (Figure 6B). We also measured plant fresh weight in the wild type, abi5-7 null mutants, brm-3 mutants, and brm-3 abi5-7 double mutants (Figure 6C). Removal of ABI5 activity from brm-3 mutants caused a partial but significant rescue of the brm mutant vegetative growth defect in the absence of ABA treatment (Figure 6C). In combination, the data suggest that the growth defects of brm mutants are in part due to the derepressed ABA response.
Figure 6.
The Growth Defects of the brm Mutant Are Partially Due to ABI5 Derepression and Enhanced ABA Response.
(A) Root growth of the brm mutant in an ABA-insensitive mutant background (35S:HAB1). The root length of the wild type (WT), 35S:HAB1, brm-101, and brm-101 35S:HAB1 double mutant was measured 7 d after stratification. Values are mean ± se. Sample size was as follows: the wild type (n = 28), 35S:HAB1 (n = 27), brm-101 (n = 49), and brm-101 35S:HAB1 (n = 75). Asterisks indicate statistical significance based on one-tailed Student’s t test (P < 1E-10).
(B) Representative pictures of data shown in (A).
(C) Fresh weight of 4-week-old wild type, abi5-7, brm-3, and brm-3 abi5-7 double mutants grown in soil with sufficient water. n > 22 from three independent experiments. Asterisks indicate statistical significance (*P < 0.01 and **P < 0.001).
The partial rescue of vegetative growth defects by removal of ABI5 activity suggested that ABI5 levels may also be elevated in brm mutants during vegetative development. We therefore analyzed expression of ABA-responsive genes in 3-week-old soil-grown brm-1 null mutants and the wild type. ABI3 expression is repressed after seedling establishment and remains repressed during vegetative development even upon ABA treatment (Nakashima et al., 2006; Perruc et al., 2007; Tang et al., 2008). Likewise, ABI3 mRNA was not detectable in the absence or presence of exogenous ABA in the brm-1 mutant (see Supplemental Figure 6A online). ABI5 expression, on the other hand, was derepressed (2.5-fold) in the absence of the stimulus and more strongly induced in response to ABA in the brm-1 null mutant relative to the wild type (see Supplemental Figure 6B online). In addition, we tested expression of the bZIP transcription factors, ABF3 and AREB1/ABF2; both genes are strongly induced by ABA, salt, and drought during vegetative development (Fujita et al., 2005). ABF3 expression was elevated in brm-1 mutants both in the absence and presence of exogenous ABA, while AREB1/ABF2 expression was not strongly altered. Thus, BRM is also required for repression of ABA-responsive genes during vegetative development, including that of the bZIP transcription factors ABI5 and ABF3. At least in the case of ABI5, the observed effect was direct: BRM associated with the ABI5 promoter at this stage, based on ChIP (see Supplemental Figure 6C online).
brm Mutants Display Enhanced Drought Tolerance
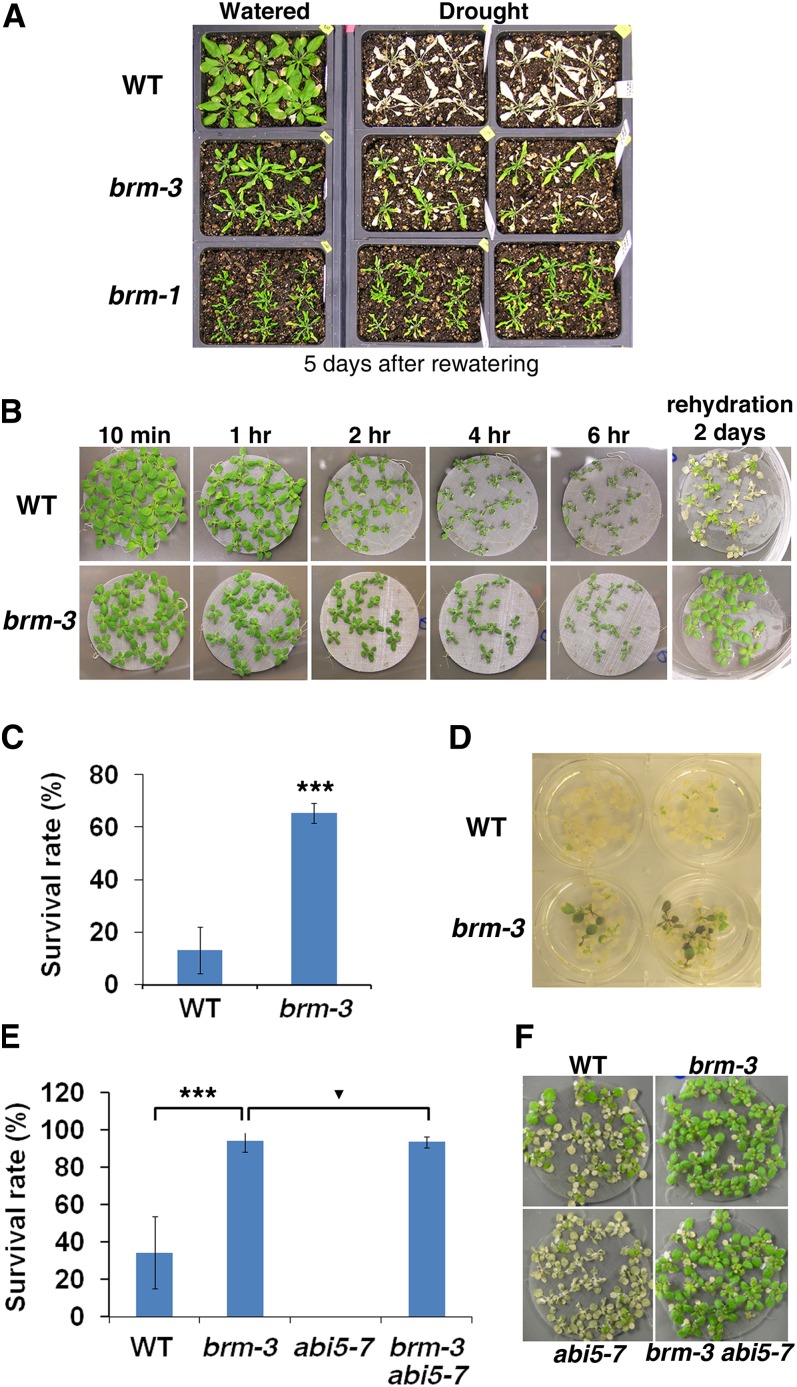
Mutants with increased sensitivity to ABA, such as pp2c mutants or plants overexpressing ABA-responsive transcription factors, display increased dehydration tolerance (Kasuga et al., 1999; Rubio et al., 2009). brm mutants were hypersensitive to ABA and showed derepression of ABA/drought-responsive gene expression; hence, we wondered whether brm mutants might display increased drought stress tolerance. To test this possibility, 3-week-old brm mutants and wild-type plants grown on soil were subjected to drought treatment. After 3 weeks of growth, water was withheld for 15 d (see Supplemental Figure 7A and Supplemental Methods 1 online). After water withholding, wild-type and brm-3 plants looked dehydrated and displayed severe tissue damage, while brm-1 plants were healthy looking and maintained greenish leaves. Upon rewatering, both brm mutants recovered quickly from the drought stress, while the wild type failed to recover (Figure 7A).
Figure 7.
brm Mutants Have Increased Dehydration Tolerance.
(A) Wild-type (WT), weak brm-3, and null brm-1 mutant plants grown in soil for 3 weeks followed by continued watering (left) or after drought treatment and rewatering (right).
(B) The effect of dehydration on 2-week-old plate-grown plants. The wild type and brm-3 mutant during and after drought treatment. The pictures farthest to the right were taken 2 d after rehydration.
(C) Survival rate (%) of 2-week-old wild-type and brm-3 seedlings after dehydration for 3 h under air flow. Values are mean ± se from four experiments (n = 42). Asterisks indicate statistical significance compared with wild-type values (P < 1E-10).
(D) Representative pictures for data shown in (C).
(E) Survival rate (%) of 2-week-old wild type, brm-3, abi5-7, and brm-3 abi5-7 double mutants after dehydration for 6 h. Values are mean ± se from two independent experiments (n > 53). Asterisks indicate statistical significance (***P < 1E-10). Inverted triangle indicates no statistical significance (P > 0.01).
(F) Representative pictures for data shown in (E).
While the drought tolerance of brm was remarkable (similar to that described for pp2c triple mutants; see Supplemental Figure 7B online), we cannot rule out that it is at least in part attributable to the different morphology of the brm mutant leaves, which are curled and smaller than those of the wild type. We therefore challenged younger (2-week-old) seedlings grown on plates with water stress. At this developmental stage, the brm-3 mutant is morphologically very similar to the wild type (Figure 7B; Farrona et al., 2007). Upon drought treatment, wild-type plants wilted faster than the brm mutants; in addition, they did not recover as well from the drought stress (Figure 7B). brm-3 plants again exhibited a significantly higher survival rate than the wild type after drought stress and rewatering (Figures 7C and 7D). The data are consistent with the hypothesis that brm mutant drought tolerance may be due to altered ABA-response gene expression.
To further test this hypothesis, we examined whether the drought tolerance of brm-3 plants was due to elevated ABI5 expression. Overexpression of ABI5 was previously shown to lead to increased drought tolerance (Lopez-Molina et al., 2001). However, brm-3 abi5-7 plants were as drought tolerant as brm-3 alone (Figures 7E and 7F). Thus, either ABI5 does not contribute to the drought tolerance of brm mutant or it does so redundantly with other ABA-responsive transcription factors, whose expression is also derepressed in brm mutants, such as ABF3 (see Supplemental Figure 6B online).
DISCUSSION
A Role for BRM in Repressing Water Stress Responses during Postgermination Development in the Absence of the Stimulus
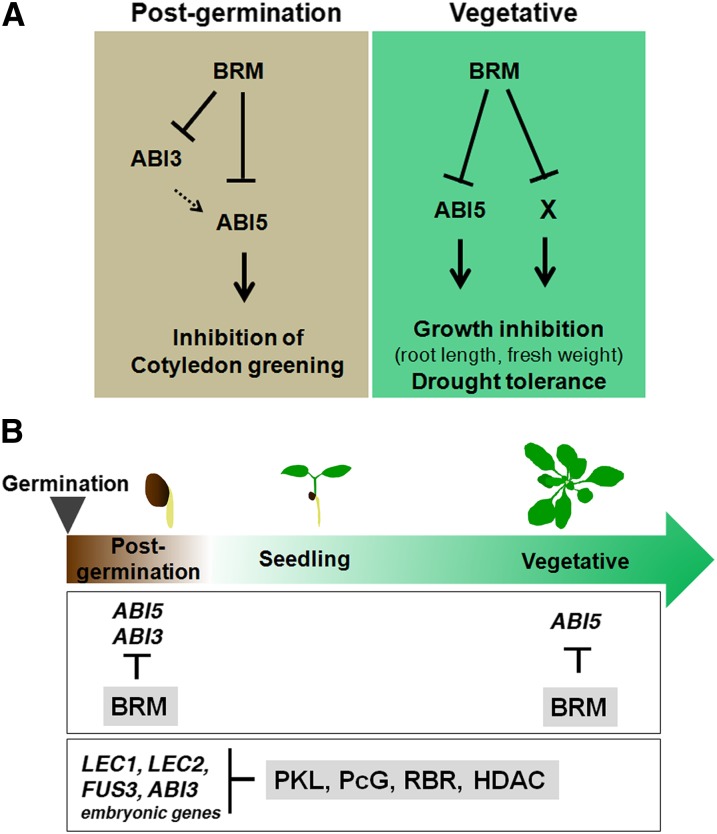
Newly germinated plant embryos are particularly vulnerable to drought stress, which triggers a growth arrest similar to that operating during seed development to induce desiccation tolerance (Lopez-Molina et al., 2001). Here, we implicate the SWI2/SNF2 ATPase BRM in ensuring that the growth arrest pathway is triggered in germinated embryos only upon drought sensing/increased endogenous ABA levels. brm mutants were hypersensitive to ABA, especially with respect to cotyledon greening and selectively derepressed expression of a subset of ABA response genes in the absence of the stimulus, among them ABI3 and ABI5. Moreover, BRM bound to the regulatory regions of both genes. ABI3 acts upstream of ABI5 during postgermination development (Lopez-Molina et al., 2002). In agreement with these combined observations, ABI5 was epistatic to BRM with respect to inhibition of cotyledon greening, indicating that derepression of ABI5 expression is the likely cause of the brm mutant’s ABA hypersensitivity during this stage of development. Thus, BRM, ABI3, and ABI5 interact in a simple genetic pathway, which corresponds to a type 2 coherent feed-forward loop (Mangan and Alon, 2003; Alon, 2007), to regulate cotyledon greening (Figure 8A). The type 2 coherent feed-forward loop displays an “off” delay (Mangan and Alon, 2003; Alon, 2007); upon ABA sensing, the upregulation of ABI3 and ABI5 would be delayed. This would ensure that growth arrest occurs only after a prolonged water stress or ABA signal has been perceived. By contrast, the “on” switch in this type of feed-forward loop is rapid (Mangan and Alon, 2003; Alon, 2007). Thus, when ABA/water stress levels fall below a certain threshold, BRM would rapidly repress ABI3 and ABI5 expression. The BRM/ABI3/ABI5 module is well suited to manage resource allocation to growth versus the stress responses.
Figure 8.
Model for Role of BRM in ABA Responses.
(A) Role of BRM in ABA response at different developmental stages. Left: Inhibition of cotyledon greening during postgermination development. BRM negatively regulates the expression of two key ABA-related transcription factors ABI5 and ABI3. ABI3 acts upstream of ABI5 (Lopez-Molina et al., 2002). Solid arrows, direct regulation; dashed arrows, direct or indirect regulation. Right: Inhibition of growth during vegetative development. Additional direct BRM targets remain unidentified that act in parallel with ABI5. ABI5 has been implicated in drought tolerance (Lopez-Molina et al., 2001), although the increase of ABI5 expression alone was not responsible for the brm mutant drought tolerance.
(B) Role of chromatin regulators in expression of ABA-responsive transcription factors during postgermination and vegetative development. BRM represses ABI5 expression during postgermination and vegetative development and ABI3 during postgermination development. Several chromatin regulators influence the developmental transition from postgermination development to seedling establishment. HDAC, histone deacetylase; PcG, Polycomb; RBR, Retinoblastoma-related protein.
Not surprisingly, since abi5 mutants show ABA-responsive root growth (Finkelstein et al., 2005; Miura et al., 2009), the ABA-triggered inhibition of root elongation in brm mutants was only partially due to ABI5 derepression. Thus, it is likely that BRM represses the expression of other transcription factors that act in parallel with ABI5 in root growth inhibition (Figure 8A). Several additional transcription factors have been shown to have a role in the inhibition of root elongation in response to ABA, including WRKY transcription factors (Chen et al., 2010), Auxin Responsive Factor 2 (Wang et al., 2011), MYB transcription factors (Zheng et al., 2012), and other bZIP transcription factors, such as ABF3 (Yoshida et al., 2010), with the latter two reported to act as least in part in parallel with ABI5.
Regulation of ABI5 Expression by BRM-Dependent and BRM-Independent Alteration of Nucleosome Positioning and Occupancy
brm mutants cause derepression of ABI5 in the absence of the ABA as well as an ∼40% reduction of the +1 nucleosome occupancy, with a preferential loss from the TSS proximal position. The +1 nucleosome is a frequent target of chromatin remodeling (Yen et al., 2012). +1 nucleosomes positioned closer to the TSS are repressive and can interfere with the assembly or activity of the transcription initiation complex (Yen et al., 2012). In addition, transcriptional activation of gene expression is associated with positioning of the +1 nucleosome away from the TSS. Thus, we hypothesize that BRM represses ABI5 transcription in the absence of water stress/ABA by promoting high occupancy of the +1 nucleosome and by directing this nucleosome from a more favorable predicted position to a position more proximal to the TSS. There is precedent for this model. Recently, derepression of HIV expression was observed upon loss of the human BAF SWI2/SNF2 subfamily complex activity, which resulted in a reduction in the occupancy of the +1 nucleosome (Rafati et al., 2011). Consistent with the idea that BRM causes increased occupancy and more TSS proximal positioning of the +1 nucleosome at the ABI5 locus, BRM very strongly associated with the region of ABI5 locus occupied by the +1 nucleosome.
We also observed stress hormone- and development-dependent alterations of the nucleosome occupancy at this locus that may explain observed gene expression changes. ABI5 expression is induced upon drought or ABA sensing (Lopez-Molina et al., 2001). Perception of the ABA stress hormone led to a destabilization (reduced occupancy) of all three nucleosomes at this locus. The most pronounced reduction in nucleosome occupancy was observed at the −1 position. This nucleosome is positioned over 2 cis-regulatory elements linked to ABA-responsive gene expression (Yamaguchi-Shinozaki and Shinozaki, 2005) and may hence modulate transcription factor access to their binding sites. The nucleosome destabilization by ABA was not BRM dependent; it was observed both in the brm-3 mutants and in the wild type. Consistent with this, the fold increase in ABI5 mRNA levels upon ABA treatment was similar in the wild type and in the brm mutant. It is possible that another chromatin regulator causes reduced nucleosome occupancy at the ABI5 locus upon stress sensing; alternatively, the nucleosome destabilization could be caused by increased transcriptional activity (Radman-Livaja and Rando, 2010).
Seed maturation is characterized by chromatin condensation, which is reversed during imbibition and germination (van Zanten et al., 2011). Consistent with an open chromatin configuration in young germinated embryos, the −1 ABI5 nucleosome was essentially absent at this stage. At the end of the postgermination phase, ABI5 expression is developmentally repressed (Lopez-Molina et al., 2001; Brocard et al., 2002). In agreement with this, the occupancy of the −1 nucleosome strongly increased at this stage. It is likely that the observed chromatin changes underlie the developmental repression of ABI5 expression at the end of postgermination development. Indeed, the ABI5 locus continues to display high occupancy of the −1 nucleosome during later stages of vegetative development (Chodavarapu et al., 2010).
The developmentally induced chromatin condensation at the ABI5 locus was observed in both the wild type and in brm mutants. In agreement with this finding, brm mutants displayed normal developmental downregulation of ABI5 expression (see Supplemental Figure 8 online). The chromatin condensation at the ABI5 locus during seedling establishment may hence be triggered by other chromatin regulators. Mutations in several different chromatin regulators, histone deacetylases, Polycomb-repressive complexes, a putative histone methyltransferase, retinoblastoma proteins, and the CHD domain chromatin remodeling ATPase PICKLE (PKL), delay the switch from the embryo to the seedling stage (Ogas et al., 1999; Tanaka et al., 2008; Aichinger et al., 2009; Bouyer et al., 2011; Gutzat et al., 2011; Kim et al., 2012; Tang et al., 2012; Zhang et al., 2012). Activity of these chromatin regulators likely contributes to the developmental downregulation of ABI5 and ABI3 (Figure 8B). Although BRM is not required for developmental repression of ABI5, it does play a role in repression of a subset of seed storage proteins (Tang et al., 2008).
Regulation of BRM Activity
In response to ABA treatment, we observed a similar upregulation of ABI5 mRNA abundance relative to mock-treated plants and a similar ABI5 promoter nucleosome destabilization in both wild-type and brm mutant germinated embryos. Thus, ABA induction of ABI5 expression is apparently BRM independent. Since BRM was still bound to the ABI5 locus upon ABA sensing, we hypothesize that BRM may be inactivated in the presence of ABA. A possible mechanism for BRM inactivation is alteration of BRM complex composition. Alternatively, BRM complex activity may be repressed via posttranslational modification(s). Both altered complex composition and posttranslational modifications can inactivate metazoan SWI2/SNF2 subgroup complexes (Clapier and Cairns, 2009). Continued BRM presence at the ABI5 locus may ensure that the growth arrest response is rapidly turned off once the desiccation stress/ABA signal has subsided. Given this model, why do brm mutants accumulate higher absolute levels of ABI5 transcript upon ABA treatment than the wild type? The higher absolute ABI5 accumulation in brm mutants upon ABA treatment is likely due to the accumulation of transcripts for ABA-dependent transcription factors in brm mutant. ABA treatment both activates and stabilizes these transcription factors (Lopez-Molina et al., 2003; Miura et al., 2009; Nakashima et al., 2009); this is expected to lead to a high absolute level of ABI5 accumulation.
Drought Tolerance and Fitness Tradeoffs
We conclude that during postgermination development BRM ensures that costly stress responses are mounted only upon perception of water stress signals to enhance fitness of the organism. The role for BRM in restricting stress response gene expression would on one hand predict that brm mutants should display defects in growth that are due to constitutive activity of the water stress response pathway; on the other hand, brm mutants would be expected to display increased drought tolerance (Boyer, 1982; Grill and Ziegler, 1998). Both expectations were confirmed in our study, supporting the conclusion that BRM prevents stress responses in the absence of the stimulus. BRM is thus positioned at the nexus of the resource allocation decision between growth and drought tolerance.
In the coming years, we will likely encounter a global deficit in food supply due to increased drought (Battisti and Naylor, 2009; Cominelli and Tonelli, 2010). To address this challenge, it is important to develop new crops that have improved water use efficiency. Efforts to engineer drought-resistant plants showed that a single gene change is often not sufficient to produce robust drought tolerance, especially in field conditions where water stress interacts with other stressors, such as heat and high light intensity (Mittler and Blumwald, 2010; Yang et al., 2010). It was proposed that manipulating expression of a master transcriptional factor that targets multiple stress response genes would be a more promising approach (Cominelli and Tonelli, 2010). An even more global change in the plant’s drought tolerance could be achieved via altered chromatin remodeling, as this mechanism can modulate gene expression in many different pathways or of several master regulators simultaneously (Kwon and Wagner, 2007). Our studies show increased drought resistance of brm mutants at multiple developmental stages. While the molecular mechanism for this enhanced drought tolerance remains to be elucidated, a key challenge for the future is to generate conditional brm loss-of-function alleles that robustly enhance water stress survival without detrimental effects on growth or yield.
METHODS
Plant Growth
The Arabidopsis thaliana genetic resources used in this study were mostly in the Columbia ecotype and have been previously described: swi3c-2 (Sarnowski et al., 2005), brm-3 (Farrona et al., 2007), brm-1 (Hurtado et al., 2006), syd-5 (Bezhani et al., 2007), bsh-1 (Tang et al., 2008), abi5-7 (Yamagishi et al., 2009), and 35S:HAB1 (Saez et al., 2004). brm-101 (Kwon et al., 2006) was in the Landsberg erecta ecotype and partly introgressed into Columbia. The strong loss-of function minu1-2 (CS413977) and minu2-1 (SALK_057856) mutants (Sang et al., 2012) and the weak syd-6 (SALK_116266) mutant were obtained from the ABRC stock center. The pBRM:BRM-GFP construct was previously described (Wu et al., 2012). Plants on plates and in soil were stratified at 4°C for 3 d. Plant growth was in inductive photoperiod (16-h-light/8-h-dark cycles) or constant light at 22°C under white fluorescent light (fluence rate: 110 µmol/m2 s for soil-grown plants; 90 µmol/m2 s for media-grown plants). Plant growth on plates was in the presence of 1% Suc unless indicated otherwise.
ABA and Drought Treatments
For germination assays, wild-type, brm-3, and brm-1/+ seeds were placed on Murashige and Skoog (MS) plates (no Suc) and supplemented with the ABA concentration indicated. Radicle emergence was scored 3 d after stratification (Müller et al., 2006). Well-ripened seeds from plants grown under the same growth condition were used. For seedling growth (green cotyledon) assays, seeds were placed on MS media supplemented with various concentration of ABA (0, 0.5, 0.8, 1.0, and 1.5 µM). Plants that had formed green cotyledons were counted 7 d after stratification unless indicated otherwise. For root growth assays, seeds were germinated on MS plates and seedlings were grown vertically for 2 or 5 d, followed by transfer to fresh media lacking or containing ABA (1, 5, or 10 µM). Plates were incubated vertically for an additional 5 d before measuring root length. For ABA treatment for gene expression, ChIP, and MNase studies, seeds were stratified for 3 d followed by growth in constant light for the time indicated. Liquid MS media with or without 50 µM ABA (Sigma-Aldrich; A1049) was added to the plates for 1 h. For studies on 3-week-old plants, 9-d-old seedlings grown on MS plates were transplanted to soil and grown for 12 more days before treatment. 100 µM ABA in 0.5 mM Tris-HCl, pH 8.0, or 0.5 mM Tris-HCl, pH 8.0, alone was applied to 3-week-old plants by spraying the leaves with an atomizer. Details on water stress treatments can be found in the Supplemental Methods 1 online.
Gene Expression Analyses
RNA purification, reverse transcription, and quantitative PCR (qPCR) were performed as described previously (Pastore et al., 2011) except amplification was monitored by EvaGreen fluorescent dye (Biotium). The sequences of primers used are listed in the Supplemental Table 1 online. Confocal imaging was performed as previously described (Winter et al., 2011).
ChIP
For the GFP-tagged BRM ChIP, 1.5-d-old brm-1 ProBRM:BRM-GFP germinated seeds (0.2 g) were used. The ChIP procedure was as previously described (Kwon et al., 2005). Five microliters of anti-GFP rabbit polyclonal antibody (Invitrogen; A6455) was employed per 0.2 g of tissue. To quantify BRM enrichment on the genomic DNA, qPCR was performed using a StepOnePlus Real-Time PCR system (Applied Biosystems) with EvaGreen fluorescent dye (Biotium). The percentage of input was calculated by determining 2−∆Ct (= 2−[Ct(ChIP)-Ct(Input)]) as per the ChampionChIP qPCR user manual (SABioscience). To facilitate comparison of different genotypes and treatments, the calculated percent input of the wild type (control) at the regions tested was set to 1. The relative enrichment represents the fold change to the wild type. The exon region of retrotransposon TA3 (Johnson et al., 2002) was used as negative control. Primer sequences are listed in the Supplemental Table 1 online.
MNase Assay
A total of 0.2 g of 1.5- or 2-d-old plants was harvested in liquid nitrogen after cross-linking in 1% formaldehyde. Nuclei and chromatin were isolated as previously described (Chodavarapu et al., 2010) with the following changes. The isolated nuclei were washed twice with HBB buffer (25 mM Tris-Cl pH 7.6, 0.44 M Suc, 10 mM MgCl2, 0.1% Triton-X, 10 mM beta-mercaptoethanol), and the isolated chromatin was digested with 0.1 units/μL −0.2 units/μL (final concentration) of Micrococcal Nuclease (Takara) for 10 min in digestion buffer at 37°C. Subsequent steps were performed as previously described (Chodavarapu et al., 2010). Mononucleosomes were excised from 1.5% agarose gels and purified using a gel purification kit (Qiagen). The purified DNA was quantified using a NanoDrop ND-1000 spectrophotometer. Two nanograms of purified DNA were used for qPCR to monitor nucleosome occupancy. The fraction of input was calculated as 2−∆Ct (2−[Ct(mono)-Ct(gDNA)]) using undigested genomic DNA (Gévry et al., 2009) followed by normalization over that of gypsy-like retrotransposon (At4g07700) −73 loci for each sample. The tiled primer sets used for real-time PCR are listed in Supplemental Table 1 online.
Statistical Analysis
For root length and fresh weight measurement, P values were calculated with the one-tailed Student’s t test. For green cotyledons, germination, and survival rate assays, χ2 analysis was performed. Two random variables (ex. the wild type and brm-3) with two types of data (survival or death) were entered in a 2 × 2 contingency table and χ2 statistic and P values were calculated using a java-based script (http://www.physics.csbsju.edu/stats/contingency_NROW_NCOLUMN_form.html). For brm-1/+ populations, we assumed Mendelian inheritance of a recessive trait. For statistical significance cutoff, we employed a P value lower than 0.01.
Accession Numbers
Sequence data for the genes in this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: BRM (AT2G46020), SWI3C (AT1G21700), SYD (AT2G28290), MINU1 (AT3G06010), MINU2 (AT5G19310), BSH (AT3G17590), ABI5 (AT2G36270), ABI3 (AT3G24650), ABF3 (AT4G34000), HY5 (AT5G11260), ABF2/AREB1 (AT1G45249), EIF4A1 (AT3G13920), TA3 (AT1G37110), and gypsy-like retrotransposon (AT4G07700). Mutants investigated in this study are listed in Methods.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ABA Responses of SWI2/SNF2 Chromatin Remodeling Mutants.
Supplemental Figure 2. Germination Assay of brm Mutants.
Supplemental Figure 3. Anti-HA ChIP in 1.5-d-Old brm-1 Plants Expressing Biologically Active ProBRM:BRM-HA.
Supplemental Figure 4. Nucleosome Occupancy at a Control Locus.
Supplemental Figure 5. Developmental Change in Nucleosome Occupancy at the ABI5 Locus.
Supplemental Figure 6. BRM Directly Represses ABI5 Expression during Vegetative Development.
Supplemental Figure 7. Measurement of Soil Water Loss during Drought Treatment and Drought Resistance of Positive Control Plants (pp2c Triple Mutants).
Supplemental Figure 8. Developmental Regulation of ABI5 and ABI3 Expression in the Wild Type and in brm Mutants.
Supplemental Table 1. List of Oligonucleotide Sequences.
Supplemental Methods 1. Chromatin Immunoprecipitation Using brm-1 ProBRM:BRM-HA and Dehydration and Drought Treatment.
Acknowledgments
We thank Kim Gallagher, Konstantinos Vlachonasios, John Wagner, and Nobutoshi Yamaguchi for critical comments on the article. We acknowledge Jose C. Reyes, Andrzej Jerzmanowski, and Eiji Nambara for providing the brm-3, swi3c-2, and abi5-7 mutant seed, respectively. The work was supported in part by National Institutes of Health Grant R01 GM64650-01 to D.W. and Ministerio de Ciencia e Innovacion grants BIO2008-00221 and BIO2011-23446 to P.L.R. Y.S. and M.-F.W. were supported by National Science Foundation IOS 0849298.
AUTHOR CONTRIBUTIONS
S.-K.H. performed the majority of the experiments. Y.S. generated and characterized the HA-tagged BRM line. A.R. and P.L.R. identified the ABA hypersensitivity of chromatin regulator mutants. The students and instructor of BIOL425F2010 optimized the MNase procedure and performed the initial nucleosome position mapping on the ABI5 promoter. M.-F.W. generated the biologically active GFP-tagged BRM line. S.-K.H. and D.W. conceived of the study and wrote the article.
Glossary
- ABA
abscisic acid
- bZIP
basic domain/leucine zipper
- ABRE
ABA-responsive element
- GFP
green fluorescent protein
- ChIP
chromatin immunoprecipitation
- HA
to be defined
- TSS
transcription start site
- MS
Murashige and Skoog
- qPCR
quantitative PCR
References
- Aichinger E., Villar C.B., Farrona S., Reyes J.C., Hennig L., Köhler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. (2007). Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Archacki R., Sarnowski T.J., Halibart-Puzio J., Brzeska K., Buszewicz D., Prymakowska-Bosak M., Koncz C., Jerzmanowski A. (2009). Genetic analysis of functional redundancy of BRM ATPase and ATSWI3C subunits of Arabidopsis SWI/SNF chromatin remodelling complexes. Planta 229: 1281–1292 [DOI] [PubMed] [Google Scholar]
- Battisti D.S., Naylor R.L. (2009). Historical warnings of future food insecurity with unprecedented seasonal heat. Science 323: 240–244 [DOI] [PubMed] [Google Scholar]
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D. (1997). Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani S., Winter C., Hershman S., Wagner J.D., Kennedy J.F., Kwon C.S., Pfluger J., Su Y., Wagner D. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.S. (1982). Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Brocard I.M., Lynch T.J., Finkelstein R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J., Podstolski W., Olczak K., Jerzmanowski A. (1999). Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 27: 2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lai Z., Shi J., Xiao Y., Chen Z., Xu X. (2010). Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang J., Neff M.M., Hong S.W., Zhang H., Deng X.W., Xiong L. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu R.K., et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Cominelli E., Tonelli C. (2010). Transgenic crops coping with water scarcity. New Biotechnol. 27: 473–477 [DOI] [PubMed] [Google Scholar]
- Cramer G.R., Urano K., Delrot S., Pezzotti M., Shinozaki K. (2011). Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975 [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Reyes J.C. (2007). A nucleosome interaction module is required for normal function of Arabidopsis thaliana BRAHMA. J. Mol. Biol. 373: 240–250 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S.S., Lynch T.J., Thomas T.L., Rock C.D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59: 253–267 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry N., Svotelis A., Larochelle M., Gaudreau L. (2009). Nucleosome mapping. Methods Mol. Biol. 543: 281–291 [DOI] [PubMed] [Google Scholar]
- Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F., Goodman H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Porras J.L., Riaño-Pachón D.M., Dreyer I., Mayer J.E., Mueller-Roeber B. (2007). Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernández M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., Rodriguez P.L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E., Ziegler H. (1998). A plant's dilemma. Science 282: 252–253 [DOI] [PubMed] [Google Scholar]
- Gutzat R., Borghi L., Fütterer J., Bischof S., Laizet Y., Hennig L., Feil R., Lunn J., Gruissem W. (2011). RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 138: 2977–2986 [DOI] [PubMed] [Google Scholar]
- Hargreaves D.C., Crabtree G.R. (2011). ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. (2010). Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L., Farrona S., Reyes J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62: 291–304 [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769: 330–345 [DOI] [PubMed] [Google Scholar]
- Johnson L., Cao X., Jacobsen S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Choi H.I., Im M.Y., Kim S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kim J.M., To T.K., Nishioka T., Seki M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Lee J., Eshed-Williams L., Zilberman D., Sung Z.R. (2012). EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 8: e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.S., Chen C., Wagner D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.S., Hibara K., Pfluger J., Bezhani S., Metha H., Aida M., Tasaka M., Wagner D. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230 [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Wagner D. (2007). Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 23: 403–412 [DOI] [PubMed] [Google Scholar]
- Less H., Angelovici R., Tzin V., Galili G. (2011). Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23: 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N.H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Mangan S., Alon U. (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2010). Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi M., Umezawa T., Nakashima K., Kidokoro S., Takasaki H., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 51: 842–847 [DOI] [PubMed] [Google Scholar]
- Müller K., Tintelnot S., Leubner-Metzger G. (2006). Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Nambara E., Hayama R., Tsuchiya Y., Nishimura M., Kawaide H., Kamiya Y., Naito S. (2000). The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 220: 412–423 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nambara E., Suzuki M., Abrams S., McCarty D.R., Kamiya Y., McCourt P. (2002). A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F., Valon C., Kohara A., Miséra S., Giraudat J. (1997). The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F., Valon C., Raynal M., Gaubier-Comella P., Delseny M., Giraudat J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore J.J., Limpuangthip A., Yamaguchi N., Wu M.F., Sang Y., Han S.K., Malaspina L., Chavdaroff N., Yamaguchi A., Wagner D. (2011). LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development 138: 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E., Kinoshita N., Lopez-Molina L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52: 927–936 [DOI] [PubMed] [Google Scholar]
- Phelan M.L., Sif S., Narlikar G.J., Kingston R.E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3: 247–253 [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M., Rando O.J. (2010). Nucleosome positioning: How is it established, and why does it matter? Dev. Biol. 339: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafati H., Parra M., Hakre S., Moshkin Y., Verdin E., Mahmoudi T. (2011). Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9: e1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rodrigues A., Santiago J., Rubio S., Saez A., Osmont K.S., Gadea J., Hardtke C.S., Rodriguez P.L. (2009). The short-rooted phenotype of the brevis radix mutant partly reflects root abscisic acid hypersensitivity. Plant Physiol. 149: 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. (2009). Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M.P., Nicolas C., Lorenzo O., Rodriguez P.L. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Sang Y., Silva-Ortega C.O., Wu S., Yamaguchi N., Wu M.F., Pfluger J., Gillmor C.S., Gallagher K.L., Wagner D. (October 13, 2012). Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J. http://dx.doi.org/10.1111/tpj.12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski T.J., Ríos G., Jásik J., Swiezewski S., Kaczanowski S., Li Y., Kwiatkowska A., Pawlikowska K., Koźbiał M., Koźbiał P., Koncz C., Jerzmanowski A. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146: 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Hou A., Babu M., Nguyen V., Hurtado L., Lu Q., Reyes J.C., Wang A., Keller W.A., Harada J.J., Tsang E.W., Cui Y. (2008). The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 147: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Lim M.H., Pelletier J., Tang M., Nguyen V., Keller W.A., Tsang E.W., Wang A., Rothstein S.J., Harada J.J., Cui Y. (2012). Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J. Exp. Bot. 63: 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Koini M.A., Geyer R., Liu Y., Brambilla V., Bartels D., Koornneef M., Fransz P., Soppe W.J. (2011). Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc. Natl. Acad. Sci. USA 108: 20219–20224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Laurière C., Merlot S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hua D., He J., Duan Y., Chen Z., Hong X., Gong Z. (2011). Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C.M., et al. (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20: 430–443 [DOI] [PubMed] [Google Scholar]
- Wu M.F., Sang Y., Bezhani S., Yamaguchi N., Han S.K., Li Z., Su Y., Slewinski T.L., Wagner D. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L., Fondufe-Mittendorf Y., Xia L., Flatow J., Widom J., Wang J.P. (2010). Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics 11: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K., Tatematsu K., Yano R., Preston J., Kitamura S., Takahashi H., McCourt P., Kamiya Y., Nambara E. (2009). CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiol. 50: 330–340 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yang S., Vanderbeld B., Wan J., Huang Y. (2010). Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol. Plant 3: 469–490 [DOI] [PubMed] [Google Scholar]
- Yen K., Vinayachandran V., Batta K., Koerber R.T., Pugh B.F. (2012). Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhang H., Bishop B., Ringenberg W., Muir W.M., Ogas J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K.S., Guo Y. (2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]