This work reveals that two non-DNA binding HLH factors (PRE1 and IBH1) and a DNA binding bHLH factor (HBI1) form an interaction cascade that controls cell elongation. BR, GA, light, and temperature signals regulate the expression level of PRE1; PRE1 binds to and inhibits IBH1; IBH1 interacts with and inhibits HBI1 binding to promoter DNA of two EXPANSIN genes.
Abstract
Environmental and endogenous signals, including light, temperature, brassinosteroid (BR), and gibberellin (GA), regulate cell elongation largely by influencing the expression of the paclobutrazol-resistant (PRE) family helix-loop-helix (HLH) factors, which promote cell elongation by interacting antagonistically with another HLH factor, IBH1. However, the molecular mechanism by which PREs and IBH1 regulate gene expression has remained unknown. Here, we show that IBH1 interacts with and inhibits a DNA binding basic helix-loop-helix (bHLH) protein, HBI1, in Arabidopsis thaliana. Overexpression of HBI1 increased hypocotyl and petiole elongation, whereas dominant inactivation of HBI1 and its homologs caused a dwarf phenotype, indicating that HBI1 is a positive regulator of cell elongation. In vitro and in vivo experiments showed that HBI1 directly bound to the promoters and activated two EXPANSIN genes encoding cell wall–loosening enzymes; HBI1’s DNA binding and transcriptional activities were inhibited by IBH1, but the inhibitory effects of IBH1 were abolished by PRE1. The results indicate that PREs activate the DNA binding bHLH factor HBI1 by sequestering its inhibitor IBH1. Altering each of the three factors affected plant sensitivities to BR, GA, temperature, and light. Our study demonstrates that PREs, IBH1, and HBI1 form a chain of antagonistic switches that regulates cell elongation downstream of multiple external and endogenous signals.
INTRODUCTION
Plant cell elongation is regulated by a wide range of environmental and hormonal signals, including light, temperature, brassinosteroid (BR), gibberellin (GA), auxin, and ethylene (Neff et al., 2006; Lau and Deng, 2010; Zhong et al., 2012). These signals act through distinct signal transduction pathways, which have been studied in detail; however, the molecular connections between these pathways are less understood, and it remains an outstanding question how different signaling pathways coordinately regulate cell elongation. Recent studies supported an emerging model that multiple signaling pathways converge on a core transcription network to control cell elongation (Bai et al., 2012; Oh et al., 2012; Gallego-Bartolomé et al., 2012; Zhong et al., 2012).
Light signals trigger seedling deetiolation/photomorphogenesis by inhibiting hypocotyl elongation and inducing cotyledon opening and chloroplast development. Light signaling mediated by photoreceptors affects the accumulation and activity of several transcription factors that directly regulate primary light-responsive genes, leading to developmental responses. Among the light-signaling transcription factors, phytochrome-interacting factors (PIFs), a small family of basic helix-loop-helix (bHLH) transcription factors, accumulate in the dark or shade to promote cell elongation but are degraded upon light activation of phytochromes (Leivar and Quail, 2011). Additional environmental and endogenous signals, including temperature, the circadian clock, and ethylene, control cell elongation at least partly by regulating the expression levels of PIF family members (Leivar and Quail, 2011; Zhong et al., 2012), whereas GA signaling activates PIF proteins by inducing ubiquitination-mediated degradation of the DELLA proteins, which directly interact with PIFs to inhibit their DNA binding (de Lucas et al., 2008; Feng et al., 2008). As such, PIFs are considered central transcription factors that mediate growth responses to multiple environmental and endogenous signals.
BR also plays an essential role in the regulation of cell elongation and photomorphogenesis. In Arabidopsis thaliana, BR-deficient or -insensitive mutants undergo constitutive photomorphogenesis in the dark and are also insensitive to GA and high temperature in hypocotyl elongation (Clouse et al., 1996; Szekeres et al., 1996; Li and Chory, 1997; Bai et al., 2012; Oh et al., 2012). BR binds to the BRASSINOSTEROID-INSENSITIVE1 (BRI1) receptor kinase at the cell surface to activate a signal transduction cascade that leads to dephosphorylation and activation of the BRASSINOZALE-RESISTANT1 (BZR1) and BZR2 (also named BES1) transcription factors, which directly regulate BR-responsive gene expression and plant development (Kim and Wang, 2010; Sun et al., 2010; Tang et al., 2011). Recent studies show that BZR1 interacts with PIF factors, and together they interdependently regulate a transcriptome that supports skotomorphogenesis and promotes cell elongation (Oh et al., 2012). In addition, BZR1 is also directly inactivated by the DELLA proteins, and GA-induced DELLA degradation releases BZR1, as well as PIFs, to promote cell elongation (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Oh et al., 2012). Therefore, DELLA, BZR1, and PIFs interact with each other to form a central module that mediates growth responses to multiple environmental and hormonal signals (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Oh et al., 2012).
Promotion of cell elongation by the BZR1-PIF4 module requires activation of members of the paclobutrazol-resistant (PRE) family of helix-loop-helix (HLH) factors. PRE1 was initially identified as a positive regulator of GA responses (Lee et al., 2006), but other studies have elucidated roles of PRE1 and its homologs in growth regulation by a wide range of signals, including BR, auxin, and light (Zhang et al., 2009; Bai et al., 2012; Oh et al., 2012; Chapman et al., 2012). Overexpression of PRE1 and its rice (Oryza sativa) homolog INCREASED LAMINA INCLINATION1 (ILI1) increased BR-induced cell elongation in both Arabidopsis and rice (Zhang et al., 2009), and overexpression of PRE3/ATBS1/TMO7 suppressed the bri1 mutant (Wang et al., 2009). PRE6/KIDARI has been implicated in light responses (Hyun and Lee, 2006), whereas PRE3/ATBS1/TMO7 is a target of an auxin response factor and required for root development (Schlereth et al., 2010). Among the six PRE members, PRE1, PRE5, and PRE6/KIDARI are direct targets of both BZR1 and PIF4 and are induced by BR, GA, and high temperature but repressed by light (Bai et al., 2012; Oh et al., 2012). Suppressing the expression of four PRE family members (PRE1, PRE2, PRE5, and PRE6/KIDARI) by artificial microRNA leads to dwarfism and hyposensitivity to BR, GA, and high temperature but hypersensitivity to light (Bai et al., 2012; Oh et al., 2012), indicating an essential role of PREs downstream of these hormonal and environmental signals.
PREs are HLH proteins similar to the human Inhibitor of DNA binding (Id) protein, which lacks the basic domain required for DNA binding but dimerizes with DNA binding bHLH factors to inhibit their DNA binding (Ruzinova and Benezra, 2003; Zhang et al., 2009). Similarly, PREs also antagonize their interacting HLH or bHLH proteins. ILI1 and PRE1 heterodimerize with and inhibit ILI1 BINDING bHLH PROTEIN1 (IBH1) in rice and Arabidopsis, whereas PRE3/ATBS1 antagonizes with four atypical HLH factors named ATBS1-interacting factors (AIF1 to AIF4) (Wang et al., 2009; Zhang et al., 2009). Overexpression of IBH1 leads to dwarfism in Arabidopsis, and this phenotype is suppressed by overexpression of PRE1, indicating that PRE1 promotes cell elongation by inactivating IBH1 (Zhang et al., 2009). However, how these HLH factors regulate gene expression remains unknown.
In this study, we performed genome expression profiling using RNA-sequencing (RNA-Seq) to identify genes downstream of PRE and IBH1. We further showed that IBH1 does not bind DNA directly but interacts with and inhibits a DNA binding bHLH factor, which we named HOMOLOG OF BEE2 INTERACTING WITH IBH1 (HBI1). Gain- and loss-of-function experiments demonstrated that HBI1 is a positive regulator of cell elongation downstream of the BR, GA, temperature, and light signaling pathways. HBI1 binds to DNA containing the G-box element, and IBH1 binds to HBI1 and inhibits HBI1’s DNA binding activity, whereas PRE1 interacts with IBH1 to prevent its inhibition of HBI1. Our results demonstrate that the balance between three antagonizing HLH/bHLH factors controls cell elongation downstream of multiple hormonal and environmental signaling pathways.
RESULTS
PRE1 and IBH1 Antagonistically Regulate Cell Elongation through Overlapping Transcriptomes
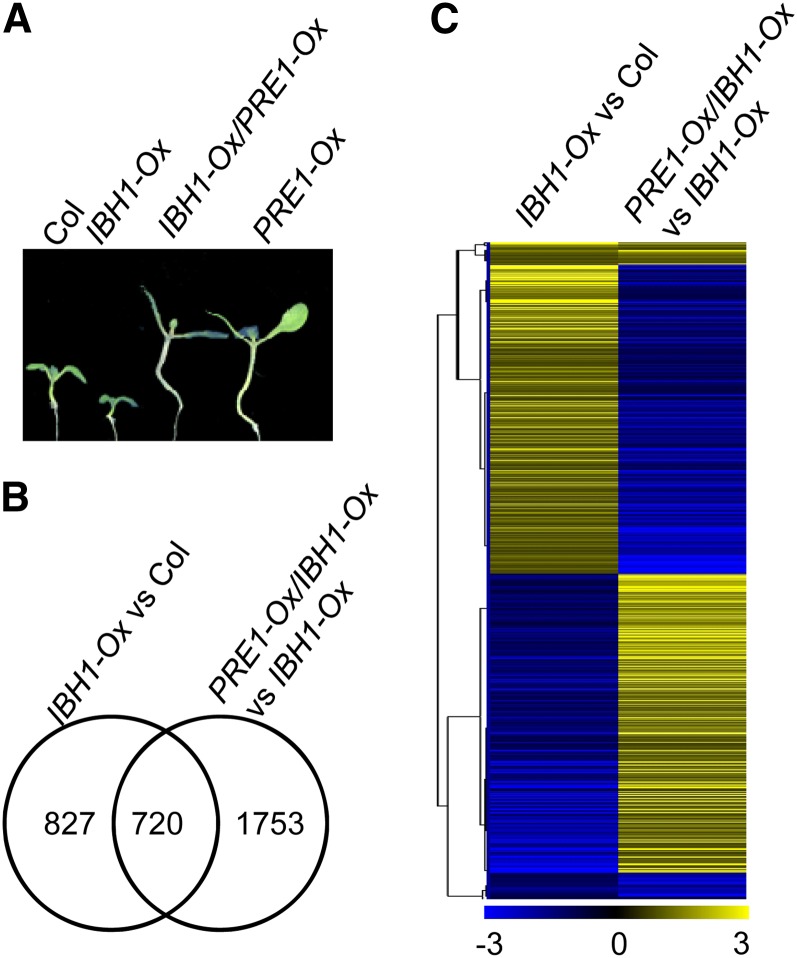
Previous studies showed that IBH1 and PRE1 interact with each other and antagonistically regulate cell elongation (Zhang et al., 2009). To further understand the function of PRE1 and IBH1 in regulating cell elongation, we performed RNA-Seq experiments to define transcriptomic changes caused by overexpression of IBH1, which results in a dwarf phenotype, and overexpression of PRE1, which suppresses the dwarf phenotype caused by IBH1 overexpression (Figure 1A). Wild-type and transgenic Arabidopsis plants overexpressing IBH1 (IBH1-Ox) or overexpressing both IBH1 and PRE1 (IBH1-Ox/PRE1-Ox) were grown on half-strength Murashige and Skoog (MS) medium with 1% Suc under constant light for 5 d. RNA-Seq analysis identified 1547 genes that increased or decreased >1.5-fold in IBH1-Ox compared with wild-type plants and 2473 genes affected in IBH1-Ox/PRE1-Ox compared with IBH1-Ox (see Supplemental Data Set 1 online). Quantitative RT-PCR analyses of nine genes confirmed the gene expression changes identified by RNA-Seq (see Supplemental Table 1 online). Of the 1547 genes affected by IBH1-Ox, 720 genes (46.5%) were also affected in IBH1-Ox/PRE1-Ox double transgenic plants compared with IBH1-Ox single transgenic plants (Figure 1B). Among these coregulated genes, 661 genes (91.8%) were affected in the opposite way by IBH1-Ox and PRE1-Ox (Figure 1C), with a correlation coefficient R = −0.76, consistent with PRE1 inhibiting IBH1.
Figure 1.
PRE1 and IBH1 Antagonistically Regulate Cell Elongation Through Overlapping Transcriptomes.
(A) PRE1 suppresses the effects of IBH1. Seedlings of Col, IBH1-Ox, PRE1-Ox, and IBH1-Ox/PRE1-Ox were grown on medium under light for 7 d.
(B) Venn diagram shows the overlap of genes regulated by IBH1-Ox and PRE1-Ox.
(C) Hierarchical cluster analysis of the genes differentially expressed in IBH1-Ox versus Col and PRE1-Ox/IBH1-Ox versus IBH1-Ox. The numerical values for the yellow-to-blue gradient bar represent log2-fold change relative to the control sample.
The Basic Domain Is Not Required for IBH1 Function
Phylogenetic analysis of IBH1 protein showed that IBH1, UPBEAT1 (UPB1), and AIFs belong to the same bHLH subfamily (see Supplemental Figure 1A online). Previous studies showed that AIFs encode atypical bHLH proteins and are speculated to have no DNA binding ability (Wang et al., 2009). However, UPB1 is reported to bind DNA and directly regulate a set of peroxidases to control the balance between cellular proliferation and differentiation (Tsukagoshi et al., 2010). Using online PRALINE multiple sequence alignment software, we analyzed the protein sequences of IBH1, UPB1, and AIFs. The results revealed that the HLH domain and AS domain were highly conserved in these proteins, whereas the basic domain was very variable and UBP1did not even contain the basic domain (see Supplemental Figure 1B online). In spite of nine basic residues at the N-terminal side of the basic domain of IBH1, IBH1 lacks the conserved residues that are necessary for binding E-box or G-box DNA (Toledo-Ortiz et al., 2003), and IBH1 belongs to a phylogenic group that contains mostly non-DNA binding HLH factors (Carretero-Paulet et al., 2010).
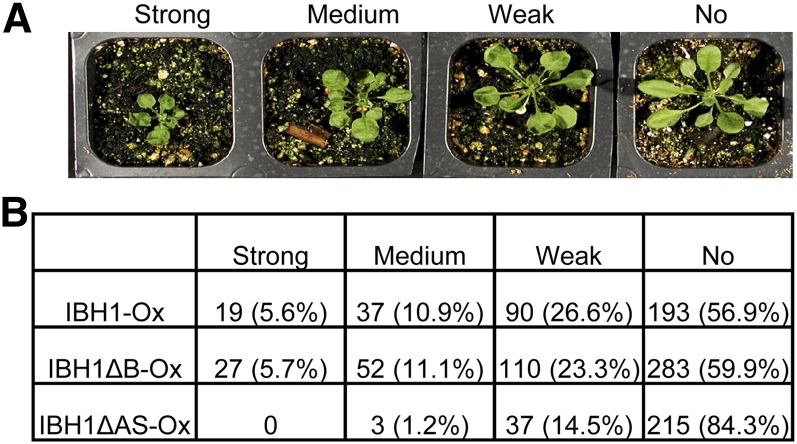
To determine the function of this basic domain and the AS domain, we deleted each of them and overexpressed the truncated proteins in transgenic plants under the control of the constitutive 35S promoter. More than 250 T1 plants for each construct were analyzed for growth phenotype. The transgenic plants overexpressing full-length wild-type IBH1 displayed a range of strong and weak dwarf phenotypes. Deletion of the basic domain had no obvious effect on the phenotype caused by overexpression, indicating that the basic domain is not necessary for IBH1’s function; thus, IBH1 is likely to function as an HLH factor without DNA binding activity. By contrast, deletion of the AS domain reduced the number of plants showing dwarf phenotypes and increased plants with no phenotype (Figures 2A and 2B), indicating that the conserved AS domain is essential for the function of IBH1 in repressing cell elongation.
Figure 2.
The Basic Domain of IBH1 Is Not Required for Its Function in Inhibition of Cell Elongation.
(A) Various degrees of dwarf phenotype observed among T1 plants overexpressing wild-type IBH1.
(B) Number and percentage of transgenic plants showing each category of phenotype severity observed among populations of transgenic plants overexpressing wild-type IBH1 or mutant IBH1 containing deletion of the basic domain (IBH1ΔB) or of the AS domain (IBH1ΔAS).
[See online article for color version of this figure.]
IBH1 Interacts with HBI1 in Vitro and in Vivo
Without a conserved basic domain to bind DNA, IBH1 is likely to heterodimerize with DNA binding transcription factors. In the public Arabidopsis interactome database, we found that IBH1 interacts with five homologous typical bHLH proteins, including BEE2, At2g18300 (named here as HBI1), BPE, CIB1, and At5g48560 (Arabidopsis Interactome Mapping Consortium, 2011). The IBH1 homolog protein AIF1 interacts with BEE1 and BEE2, and AIF2 also interacts with CIB1 and BPE (Wang et al., 2009; Arabidopsis Interactome Mapping Consortium, 2011). Interactions between multiple members of the IBH1/AIFs subfamily and members of the BEE/CIB subfamily suggest that this is a conserved mechanism important for the function of these HLH/bHLH factors (see Supplemental Figure 2 online).
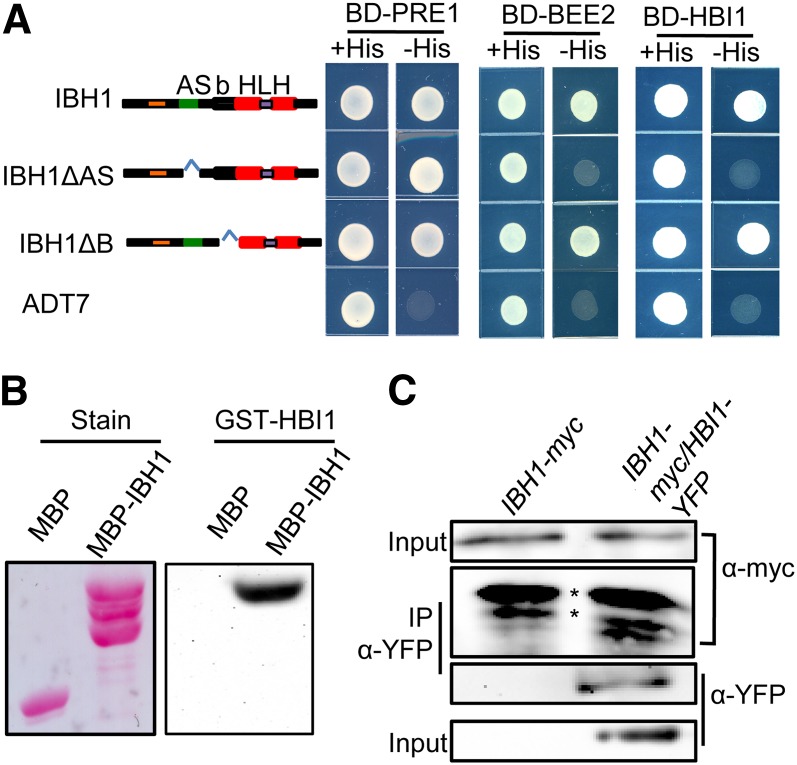
We performed yeast two-hybrid assays to confirm some of these interactions. The results showed that both HBI1 and BEE2 interact with IBH1 in yeast, and this interaction needs the IBH1 AS domain but not the basic domain (Figure 3A). By contrast, IBH1 interacts with PRE1 through its HLH domain and does not require the AS domain or basic domain (Figure 3A; see Supplemental Figure 3 online), indicating that the AS domain is specifically required for the interaction between IBH1 and HBI1. We also performed overlay assays to test the direct interaction between IBH1 and HBI1 in vitro. Incubating gel blots of MBP and MBP-IBH1 with glutathione S-transferase (GST)-HBI1 and anti-GST antibodies detected strong binding of GST-HBI1 to MBP-IBH1, but not to MBP, indicating that GST-HBI1 interacts with MBP-IBH1 in vitro (Figure 3B). Consistent with the yeast and in vitro assays, coimmunoprecipitation assays showed that IBH1 interacts with HBI1 in vivo (Figure 3C). These results demonstrated that IBH1 binds to HBI1 through the AS domain of IBH1.
Figure 3.
IBH1 Interacts with HBI1 in Vitro and in Vivo.
(A) Yeast two-hybrid assays of interactions between indicated IBH1 proteins and PRE1, BEE2, or HBI1. The AS domain is required for the IBH1 interaction with HBI1 but not for interaction with PRE1.
(B) A gel blot of MBP and MBP-IBH1 was probed with GST-HBI1 followed by horseradish peroxidase–labeled anti-GST antibody or stained with Ponceau S (Stain).
(C) Coimmunoprecipitation assays show IBH1 interacts with HBI1 in plant cells. 35S:HBI1-YFP was transformed into the protoplasts prepared from the 35S:IBH1-myc transgenic plants. Immunoprecipitation was performed using anti-YFP antibody, and immunoblots were probed with anti-myc or anti-YFP antibodies.
[See online article for color version of this figure.]
HBI1 Promotes Cell Elongation
Previous study showed that single or double mutants of BEE2 and its two homologous genes BEE1 and BEE3 did not have any obvious developmental phenotype, and only the bee1 bee2 bee3 triple knockout mutant showed a slightly shorter hypocotyl than that of wild-type seedlings, suggesting that these BEEs redundantly promote cell elongation (Friedrichsen et al., 2002). The subtle phenotype of the bee triple mutant suggests that other homologous genes may have redundant roles (Friedrichsen et al., 2002). HBI1 is the closest homolog of BEE2 in the phylogenetic tree, and microarray analysis showed that HBI1 is preferentially expressed in hypocotyl and cotyledon (see Supplemental Figure 2 online), suggesting that HBI1 may play a major role in hypocotyl elongation and cotyledon expansion.
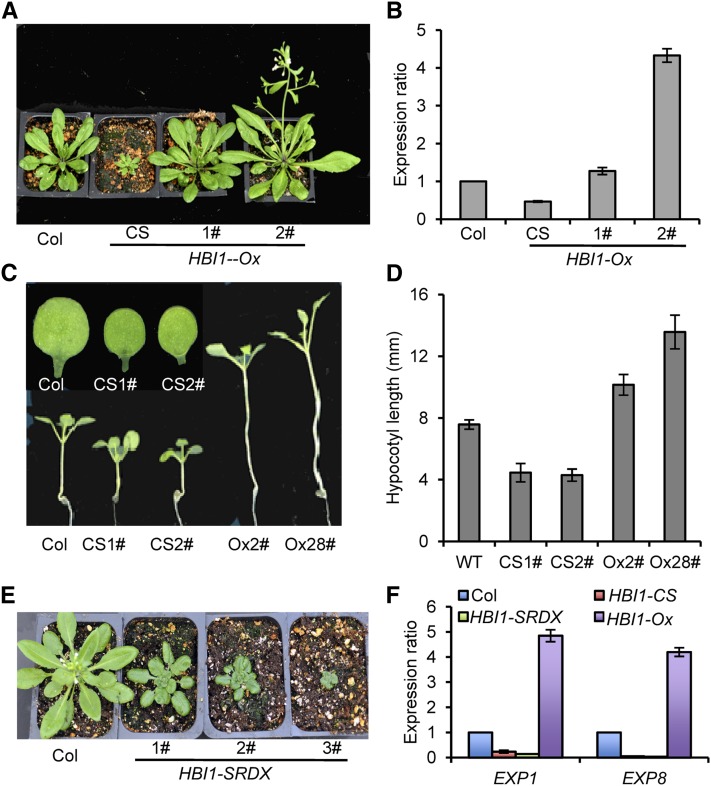
To study the function of HBI1, we transformed wild-type Arabidopsis with a construct of cauliflower mosaic virus 35S promoter driving expression of the HBI1 cDNA fused with yellow fluorescent protein (YFP). A total of 65 transgenic lines were generated, and about two-thirds of them (44 plants) showed no obvious phenotype. The other lines showed opposite phenotypes: Nine plants showed dwarf, late-flowering, and short-petiole phenotypes, and 12 plants flowered early and had long petioles (Figure 4A). To address why these 35S:HBI1-YFP transgenic plants showed opposite phenotypes, we analyzed the HBI1 expression level in these plants. The quantitative RT-PCR results showed that the HBI1 expression levels were lower in the dwarf plants but higher in the plants with long petioles than in wild-type plants, indicating the dwarf phenotype was caused by HBI1 cosuppression (HBI1-CS), and the long-petiole phenotype was caused by HBI1 overexpression (HBI1-Ox) (Figure 4B). We also analyzed the expression levels of HBI1 homologous genes in the HBI1 cosuppression lines. The results showed that in addition to HBI1, the transcript levels of BEE2, CIB1, CIB3, and CIB1L also decreased (see Supplemental Figure 4 online). The HBI1-CS plants also had short hypocotyls and small cotyledons, but the HBI1-Ox plants had long hypocotyls (Figures 4C and 4D). These results indicated HBI1 and its homologs are positive regulators of cell elongation and expansion.
Figure 4.
HBI1 Is a Positive Regulator of Cell Elongation.
(A) Representative plants of the wild type and HBI1 cosuppression (CS) and overexpression (#1 and 2#) lines were grown in soil for 4 weeks.
(B) Quantitative RT-PCR analyses of HBI1 expression in plants represented in (A) using PP2A as the internal control. Error bars indicate sd.
(C) Seedling phenotypes of HBI1-CS and HBI1-Ox transgenic plants grown on half-strength MS medium under dim light for 14 d. The top images show the smaller cotyledon of HBI1-CS lines.
(D) Quantification hypocotyl lengths of wild-type (WT) and HBI1 transgenic plants. Error bars represent sd.
(E) Phenotype of HBI1-SRDX plants grown in soil for 4 weeks.
(F) Quantitative RT-PCR analyses of expression of EXP1 and EXP8 in wild-type, HBI1-CS, HBI1-SRDX, and HBI1-Ox plants. PP2A was used as the internal control. Error bars indicate sd from three biologic repeats.
Considering the high level of functional redundancy of BEE family members, we generated a dominant repressor version of HBI1 using chimeric repressor silencing technology, in which HBI1 was fused with a 12–amino acid EAR motif repressor domain (SRDX) (Hiratsu et al., 2003). Expression of HBI1-SRDX driven by the constitutive 35S promoter resulted in dwarf transgenic plants similar to the HBI1-CS plants but opposite to the HBI1-Ox plants (Figure 4E). These results suggest that HBI1 normally functions as a transcriptional activator.
BR and GA promote cell elongation partly by increasing the expression of expansins, which loosen the cell wall (Cosgrove, 2000; Bai et al., 2012). Real-time RT-PCR analyses showed that the expression levels of EXP1 and EXP8 were reduced in the HBI1-CS and HBI1-SRDX plants but increased in the HBI1-Ox plants, suggesting that HBI1 promotes cell elongation by activating the expression of expansins (Figure 4F).
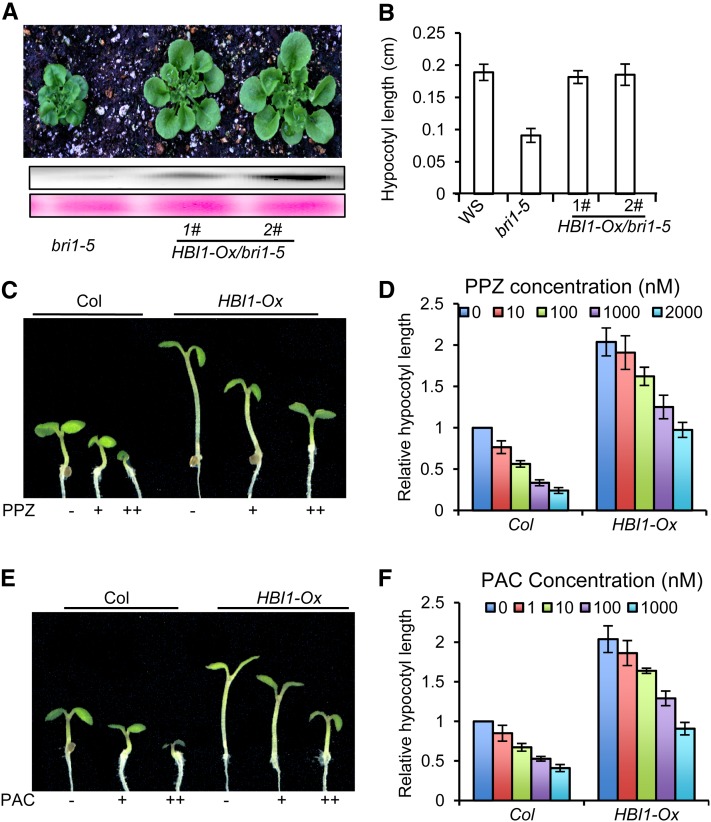
A previous study showed that BEE2 is an early BR response gene and is required for the full BR response (Friedrichsen et al., 2002). The sequence similarity between BEE2 and HBI1 suggests that HBI1 may also mediate BR response. To test this hypothesis, we overexpressed HBI1 in a weak BR-insensitive mutant, bri1-5. The transgenic plants showed longer hypocotyls and petioles than bri1-5 (Figures 5A and 5B). In addition, the HBI1-Ox plants in the wild-type background also exhibited longer hypocotyls than the wild type when grown on normal medium and on media containing the BR biosynthesis inhibitor propiconazole (PPZ) (Figures 5C and 5D) or the GA biosynthesis inhibitor paclobutrazol (PAC) (Figures 5E and 5F).
Figure 5.
HBI1 Positively Regulates BR Responses.
(A) and (B) Overexpression of HBI1 partly suppresses the bri1-5 phenotype.
(A) The bri1-5 and HBI1-Ox/bri1-5 plants were grown in soil for 4 weeks. The bottom panels show immunoblots probed with anti-YFP antibodies and Ponceau S staining for loading control.
(B) The average hypocotyl lengths of the wild type (WS), bri1-5, and HBI1-Ox/bri1-5 grown on half-strength MS medium under light for 6 d. Error bars show sd.
(C) and (D) Overexpression of HBI1 reduces sensitivity to the BR biosynthesis inhibitor PPZ. Wild-type and HBI1-Ox plants were grown on half-strength MS medium containing 100 nM (+), 2 μM (++) PPZ (C), or with indicated concentrations of PPZ (D).
(D) The hypocotyl lengths were measured from at least 15 plants. Error bars represent sd.
(E) and (F) The HBI1-Ox plants show longer hypocotyls than the wild type in the presence of the GA biosynthesis inhibitor PAC. Wild-type and HBI1-Ox plants were grown on half-strength MS medium containing 10 nM (+), 1 μM (++) PAC (E), or with indicated concentrations of PAC (F) under constant light. The average hypocotyl lengths were measured from at least 15 plants. Error bars represent sd.
IBH1 Inactivates HBI1, and PRE1 Inactivates IBH1 to Activate HBI1
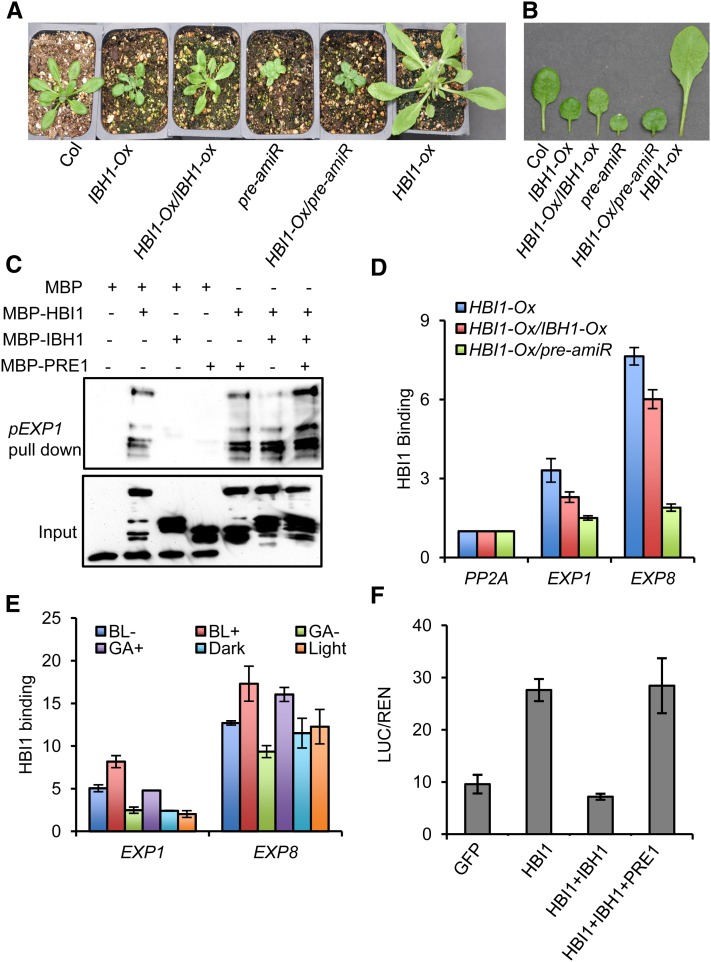
The facts that PRE1 and HBI1 promote cell elongation, IBH1 represses cell elongation, and IBH1 interacts with both PRE1 and HBI1 raised the possibility that the ratio between these positive and negative regulators determines the outcome of cell elongation. To test this possibility, we crossed the HBI1-Ox transgenic plants with IBH1-Ox and pre-amiR. The F1 plants showed phenotypes intermediate between the parental plants, consistent with IBH1 inactivating HBI1 and PREs being required to inactivate IBH1 (Figures 6A and 6B).
Figure 6.
IBH1 and PRE1 Antagonistically Regulate the Activity of HBI1.
(A) and (B) Overexpression of IBH1 or knockdown the expression of PREs suppresses the phenotype of HBI1-Ox. Plants (A) or detached leaves (B) were photographed after growth in soil for 4 weeks.
(C) In vitro DNA pull-down assays of HBI1 DNA binding activity. The indicated MBP or MBP fusion proteins purified from Escherichia coli were incubated with a biotinylated DNA fragment of the EXP1 promoter immobilized on streptavidin beads. The DNA-bound proteins were immunoblotted using anti-MBP antibody.
(D) ChIP-qPCR analysis of HBI1 binding to the EXP1 and EXP8 promoters and the effects of IBH1-Ox and pre-amiR on HBI1 DNA binding in vivo. Heterozygous transgenic 35S:HBI1-YFP/Col-0, 35S:HBI1-YFP/35S:IBH1-myc, and 35S:HBI1-YFP/35S:pre-amiR F1 plants grown in a greenhouse for 4 weeks were used for the ChIP-qPCR analysis. Error bars indicate sd of three biological repeats.
(E) ChIP-qPCR analysis of the effects of BR, GA, and light on HBI1 DNA binding in vivo. ChIP-qPCR was performed using 35S:HBI1-YFP and 35S:YFP grown in half-strength MS liquid medium with or without 2 µM PPZ or 1 µM PAC for 5 d under constant light or in the dark. The plants grown on 2 µM PPZ were treated with mock solution (BL−) or 100 nM BL (BL+) for 6 h; plants grown on 1 µM PAC were treated with mock solution (GA−) or 10 µM GA3 (GA+) for 6 h; plants grown in the dark were treated with white light (35 µM/m2/s) or kept in the dark for 6 h. Error bars indicate sd of three biological repeats.
(F) Transient assays show HBI1 activation of the pEXP1:LUC reporter gene. Arabidopsis protoplasts were transformed with the dual luciferase reporter construct containing pEXP1:LUC (luciferase) and 35S:REN (renilla luciferase) and constructs overexpressing the indicated effecters. The LUC activity was normalized to REN. Error bars indicate sd of three biological repeats.
The basic domain of HBI1 contains the Glu-13, Arg-16, and Arg-17 residues conserved in most known DNA binding bHLH proteins (see Supplemental Figure 5 online), and its homolog CIB1 has been shown to bind to the G-box (Liu et al., 2008), suggesting that HBI1 can bind the E box (5′-CANNTG-3′) or G-box (5′-CACGTG-3′) (Toledo-Ortiz et al., 2003). HBI1 activates the transcription of EXP1 (Figure 4F), and there are three G-box and 13 E-box motifs in the promoter of EXP1 (see Supplemental Figure 6 online), suggesting that HBI1 may bind to the EXP1 promoter. We thus performed DNA-protein pull-down experiments to test whether HBI1 binds DNA and whether IBH1 and PRE1 affect its DNA binding ability. We amplified a DNA fragment of EXP1 containing one G-box and four E-box motifs using biotin-labeled primers (see Supplemental Figure 6 online) and incubated it with MBP-HBI1 and/or MBP, MBP-IBH1, and MBP-PRE1. The results showed that the biotinylated EXP1 DNA fragment could pull down MBP-HBI1 but not MBP, MBP-IBH1, or MBP-PRE1 (Figure 6C), confirming the specific HBI1 binding to the EXP1 promoter and the lack of DNA binding activity for IBH1 and PRE1. Incubation of MBP-HBI1 with MBP-IBH1 dramatically reduced HBI1 binding to DNA, whereas addition of MBP-PRE1 recovered HBI1-DNA binding. These results indicate that IBH1 inhibits HBI1-DNA binding, but PRE1 blocks the IBH1’s inhibitory effect (Figure 6C).
To test whether IBH1 and PRE1 affect the DNA binding of HBI1 in vivo, we performed chromatin immunoprecipitation followed by quantitative real-time PCR (ChIP-qPCR) assays. ChIP-qPCR results showed that HBI1 binds to the promoters of EXP1 and EXP8 in vivo. Overexpression of IBH1 (IBH1-Ox) significantly reduced HBI1 binding to these promoters (Figure 6D). Suppression of PREs (pre-amiR) also reduced HBI1-DNA binding, consistent with PRE1 being required for inhibiting IBH1 and possibly IBH1 homologs too. Treatments of plants with BR and GA, which activate PRE1 expression, increased the HBI1 DNA binding in vivo, whereas light treatment showed no obvious effect (Figure 6E). Finally, we analyzed the effects of IBH1 and PRE1 on the transcription activity of HBI1 in protoplast transient assays. The expression level of an EXP1 promoter-luciferase reporter gene was increased by overexpression of HBI1. This HBI1-mediated transcription increase was abolished by co-overexpression of IBH1 but was recovered by additional co-overexpression of PRE1 (Figure 6F). Together, these results indicate that IBH1 interacts with HBI1 and inhibits HBI1 binding to DNA, but PRE1 binds to IBH1 to prevent its inhibition of HBI1, thereby activating HBI1. BR and GA act through this cascade of interacting HLH/bHLH factors to promote HBI1 activation of target gene expression and cell elongation.
The PRE-IBH1-HBI1 Regulatory Chain Controls Cell Elongation Downstream of Multiple Hormonal and Environmental Signals
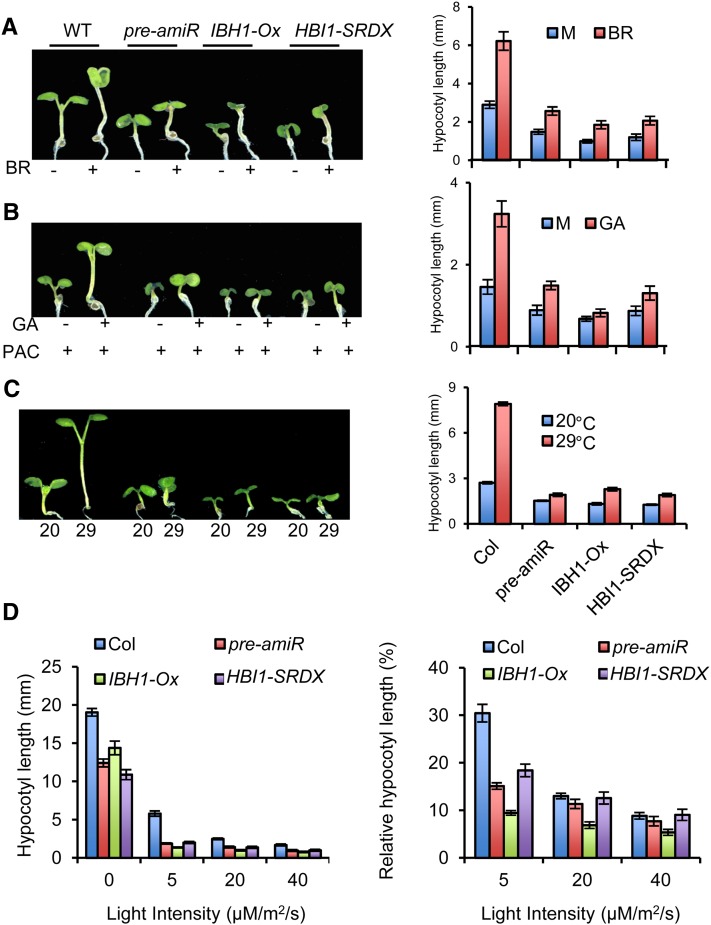
Previous studies showed that PREs play an essential role in cell elongation responses to BR, GA, temperature, and light (Lee et al., 2006; Wang et al., 2009; Zhang et al., 2009; Bai et al., 2012; Oh et al., 2012). To confirm that the PRE-IBH1-HBI1 module mediates regulation of cell elongation by these signals, we examined the hypocotyl responses to these signals in the pre-amiR, IBH1-Ox, and HBI1-SRDX plants. BR, GA, and elevated temperature dramatically promoted hypocotyl elongation in wild-type plants, whereas the hypocotyls of the pre-amiR, IBH1-Ox, and HBI1-SRDX transgenic plants were much less sensitive to these stimuli (Figures 7A to 7C). These transgenic plants also showed enhanced sensitivity to light (Figure 7D). These results support that the regulatory chain formed by PREs, IBH1, and HBI1 is a central mechanism of cell elongation regulation shared by these hormonal and environmental signaling pathways.
Figure 7.
PREs, IBH1, and HBI1 Mediate Hypocotyl Elongation Responses to BR, GA, Temperature, and Light.
(A) and (B) Wild-type (WT) and transgenic 35S:pre-amiR, 35S:IBH1-myc, and 35S:HBI1-SRDX plants were grown under constant light for 7 d in half-strength MS medium with (+) or without (−) indicated hormones: 100 nM BR (A), 1 μM GA3, and 100 nM PAC (B). Right panels: The hypocotyl lengths were measured from at least 15 plants. Error bars represent sd.
(C) Wild-type and transgenic 35S:pre-amiR, 35S:IBH1-myc, and 35S:HBI1-SRDX plants were grown at 20 or 29°C for 7 d. The hypocotyl lengths were measured from at least 15 plants. Error bars represent sd.
(D) Wild-type and transgenic 35S:pre-amiR, 35S:IBH1-myc, and 35S:HBI1-SRDX plants were grown under different light conditions for 7 d. The hypocotyl lengths and relative lengths (percentage of length in the dark) were calculated from at least 10 plants. Error bars represent sd.
DISCUSSION
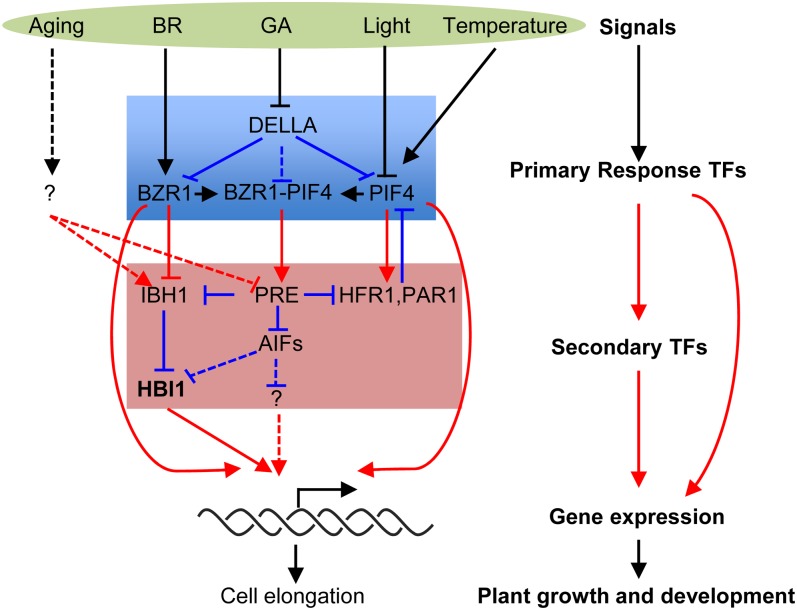
Many studies have demonstrated that the PRE family of HLH factors plays a key role in promoting cell elongation and plant growth downstream of many environmental and hormonal signals (Lee et al., 2006; Hyun and Lee, 2006; Zhang et al., 2009; Wang et al., 2009; Mara et al., 2010; Schlereth et al., 2010). Several PRE family members have been shown to interact with and inhibit atypical bHLH factors, including IBH1 and AIFs (Hyun and Lee, 2006; Zhang et al., 2009; Wang et al., 2009). How these HLH/bHLH factors regulate gene expression has been an outstanding question. Our study demonstrates that PRE1 and IBH1 interact with each other and antagonistically regulate the expression of a large number of genes through IBH1 interaction with the DNA binding bHLH factor HBI1. This study therefore fills a major gap in the transcription network that links multiple signaling pathways to the transcriptome for cell elongation. The results reveal a mechanism of activating bHLH factor by antagonistic interaction between two non-DNA binding HLH factors. Together with previous studies showing transcriptional regulation of PREs and IBH1 by the primary transcription factors of these signaling pathways (i.e., PIF4, BZR1, and ARF5) (Zhang et al., 2009; Schlereth et al., 2010; Bai et al., 2012; Oh et al., 2012), this study illustrates a second tier of interacting transcription factors that mediate coordinated regulation of plant cell elongation according to the environmental conditions and hormonal status (Figure 8).
Figure 8.
Diagrams of the Signaling Network Mediating Cell Elongation Regulation by Multiple Environmental and Hormonal Signals.
The diagram on the right side summarizes conceptually the detailed diagram on the left, where the primary and secondary transcription factors (TFs) are grouped by colored boxes, arrows show activation and bar-ended lines show inhibition, blue lines show regulation by protein–protein interactions, and red lines show transcriptional regulation. Dashed lines show unknown or speculated mechanisms. The component and mechanisms discovered in this study are marked by bold text and thicker lines, respectively.
The bHLH proteins are one of the largest families of transcription factors in eukaryotes, with ∼170 members in Arabidopsis (Carretero-Paulet et al., 2010). The bHLH factors are known to form homo- or heterodimers through the HLH domain and to bind to specific DNA sequences known as the E-box (CANNTG) or G-box (CACGTG) through the basic domains (Toledo-Ortiz et al., 2003). About 26% of plant bHLH proteins are predicted to contain only the HLH domain but no functional basic domain (Carretero-Paulet et al., 2010). Such HLH or atypical bHLH factors are known to dimerize with bHLHs to form non-DNA binding heterodimers (Toledo-Ortiz et al., 2003; Carretero-Paulet et al., 2010). One such factor in humans is known as Id, which inactivates several bHLH factors and plays important roles in cell proliferation, differentiation, and cancer (Ruzinova and Benezra, 2003). In contrast with the human Id protein, PRE1 activates bHLH factors by dimerizing with another non-DNA binding atypical HLH factor (IBH1) that inhibits DNA binding activity of the bHLH factors. This additional step of negative regulation of the inhibitor of a DNA binding protein potentially provides additional control point for integration of multiple signaling inputs. It thus appears that plants have evolved not only a larger number of HLH/bHLH factors (Riechmann et al., 2000) but also a more complex interaction network and additional regulatory mechanisms.
PRE1/ILI1 and IBH1 were identified as a pair of antagonizing HLH/bHLH factors that regulates cell elongation (Zhang et al., 2009; Bai et al., 2012). In this study, we show that PRE1 and IBH1 antagonistically regulate an overlapping transcriptome. Consistent with the phenotypic suppression of IBH1-Ox by PRE1-Ox, 91% of the commonly regulated genes were affected in opposite ways by IBH1-Ox and PRE1-Ox. However, several lines of evidence indicate that IBH1 does not bind DNA directly. While IBH1 was previously predicted to function as a DNA binding bHLH factor based on the presence of nine basic residues at the N-terminal side of the HLH domain (Zhang et al., 2009; Carretero-Paulet et al., 2010), these basic residues do not match the conserved residues required for DNA binding. In addition, the IBH1 orthologs in rice and poplar (Populus spp) were predicted to be non-DNA binding HLH factors (Carretero-Paulet et al., 2010). Furthermore, deletion of the basic domain showed no obvious effect on the IBH1 activity in vivo. Finally, in vitro DNA–protein interaction assays showed no DNA binding activity for IBH1, whereas DNA binding was detected for HBI1. Therefore, IBH1 appears to regulate gene expression through interaction with HBI1 and possibly other DNA binding bHLH factors.
HBI1 is a typical DNA binding bHLH protein, belonging to bHLH subfamily 25 together with the BR-induced bHLH factors BEE1, 2, and 3, and the cryptochrome-interacting bHLH factor CIB1 (Liu et al., 2008; Carretero-Paulet et al., 2010). CIB1 has been shown to bind to the G-box element (Liu et al., 2008). Our in vitro and in vivo assays demonstrated that HBI1 binds to the EXP1 and EXP8 promoters and activates their expression. Gain-of-function and loss-of-function studies showed that HBI1 is a major positive regulator of cell elongation. Both in vitro and in vivo experiments showed that IBH1 directly interacts with HBI1 and inhibits HBI1 binding and activation of the EXP1 and EXP8 promoters. The fact that the conserved AS domain of IBH1 is required for both HBI1 binding and growth inhibition further supports that IBH1 inhibition of cell elongation requires interaction with HBI1. Inclusion of PRE1 in the in vitro and in vivo assays abolished the effects of IBH1 and recovered HBI1’s DNA binding and activation of the EXP promoters. Our results demonstrate that PRE1 promotes cell elongation by preventing IBH1 from inhibiting HBI1, which directly activates genes encoding cell wall–loosening enzymes.
The PRE-IBH1-HBI1 cascade plays important role in regulation of cell elongation downstream of a wide range of signals, including BR, GA, light, and temperature, as perturbation of each of the three components alters plant sensitivities to these signals. Recent studies have illustrated that these signal transduction pathways primarily regulate the expression levels of PREs and IBH1. BR activation of the BRI1 receptor kinase triggers a cascade of signal transduction events, which include activation of BSK1 and CDG1 kinases, BSU1 phosphatase, inactivation of the BIN2 kinase, and PP2A-mediated dephosphorylation of the BZR1 family of transcription factors (Li and Chory, 1997; Li and Nam, 2002; Wang et al., 2002; Yin et al., 2002, 2005; Mora-García et al., 2004; He et al., 2005; Tang et al., 2008, 2011; Kim et al., 2009; Hothorn et al., 2011; Kim et al., 2011; She et al., 2011). Dephosphorylated BZR1 moves into the nucleus and regulates a large number of genes, including PRE1 and IBH1 (He et al., 2005; Yin et al., 2005; Bai et al., 2007; Gampala et al., 2007; Ryu et al., 2007; Zhang et al., 2009; Sun et al., 2010). The expression levels of PRE1 and IBH1 are increased and reduced, respectively, by BR signaling due to opposite transcriptional activities of BZR1 at these promoters (Zhang et al., 2009). Such double regulation through both direct binding by PRE1 and transcriptional repression by BZR1 is expected to effectively decrease cellular level of available IBH1. This mechanism is conserved in Arabidopsis and rice (Zhang et al., 2009). BR does not affect the transcript level of HBI1 (Friedrichsen et al., 2002); therefore, BR most likely activates HBI1 mainly by decreasing IBH1 level through BZR1-mediated transcriptional repression and PRE1-mediated posttranslational inhibition.
Interestingly, the close homologs of HBI1, namely, BEE1, 2, and 3, were identified as BR-induced genes based on BR induction of their transcript levels (Friedrichsen et al., 2002). The triple bee1 bee2 bee3 mutant showed slightly reduced hypocotyl elongation and BR sensitivity. However, overexpression of BEE1 enhanced BR response only in root but caused no obvious hypocotyl phenotype. Yeast two-hybrid assays showed that IBH1 interacts with not only HBI1 but also BEE2 and seven additional homologous bHLHs (see Supplemental Figure 2 online; Ikeda et al., 2012). BEE2 and these other bHLH factors may play redundant roles with HBI1, and BR may regulate their activities at transcriptional and/or posttranslational levels. Additional PRE1- and IBH1-related HLH factors may also contribute to BR responses. In addition to PRE1, PRE5 and PRE6 are also induced by BR and thus may play a redundant role with PRE1. An overexpressor of PRE3/ATBS1//TMO7 was identified as a bri1 suppressor, and overexpression of members of the AIF family of atypical bHLH factors caused dwarf phenotypes (Wang et al., 2009). Whether AIFs act through HBI1 or related bHLH factors remains to be investigated. These studies together suggest that BR regulation of cell elongation is mediated by balances between multiple members of the PRE and IBH1/AIF families of HLH factors and HBI1/BEE family of bHLH factors.
PRE1 was initially identified as a positive regulator of GA response, and its transcription level is increased by GA signaling through a GID receptor- and DELLA- dependent mechanism (Lee et al., 2006). Recent studies show that transcription of PRE1, PRE5, and PRE6 is directly activated by the BZR1-PIF4 heterodimer, which is inactivated by DELLA when GA levels are low. Our results show that BR and GA increase the HBI1 binding to DNA in vivo, most likely through activation of PREs, which prevent IBH1 from inhibiting HBI1. Although 6-h light treatment of dark-grown seedlings had no obvious effect on HBI1 binding to the promoters of EXP1 and EXP8, this is possibly because light’s effect on HBI1 through PREs was cancelled by its effect on PIFs, which likely compete with HBI1 for binding to the same G-box or E-box motifs of the EXP1 and EXP8 promoter. The BZR1-PIF4-DELLA module, which integrates BR, GA, light, and temperature signals (Bai et al., 2012; Oh et al., 2012), directly regulates PRE expression to control IBH1 and HBI1 activities. Therefore, the PRE-IBH1-HBI1 module forms a second tier of interacting transcription factors downstream of the primary transcriptional regulators controlled posttranslationally by the signal transduction pathways of BR, GA, phytochrome, and temperature (Figure 8).
Antagonistic interactions between HLH and bHLH factors are likely to play important roles in a broad range of plant developmental processes. High expression levels of PRE1 and IBH1 correlate with growing young floral organs and growth-arrested mature ones, respectively (Zhang et al., 2009). Additional members of PRE and IBH/AIF families have been studied in a number of developmental contexts. The Arabidopsis PRE family includes six members (PRE1/BNQ1, PRE2/BNQ3, PRE3/ATBS1/TMO7, PRE4/BNQ3, PRE5, and PRE6/KDR), which display distinct but also overlapping functions in plant growth and development (Hyun and Lee, 2006; Lee et al., 2006; Wang et al., 2009; Zhang et al., 2009; Mara et al., 2010; Schlereth et al., 2010). For example, PRE1/BNQ1, PRE2/BNQ2, and PRE4/BNQ3 are direct target genes negatively regulated by AP3/PI in the petals and therefore play a role in floral development (Mara et al., 2010). The pre4/bnq3 mutant has decreased chlorophyll levels causing pale-white sepals and carpels (Mara et al., 2010). PRE3/ATBS1/TMO7 was identified as target gene of MONOPTEROS/ARF5 that is required for embryonic root development. Knockdown of the expression of PRE3/ATBS1/TMO7 using RNA interference or artificial microRNA causes aberrant division of the hypophysis and a rootless seedling phenotype (Schlereth et al., 2010), whereas its overexpression reduces lateral root initiation (Castelain et al., 2012). Among the Arabidopsis IBH1 homologs, UPB1 also plays a key role in root development. The T-DNA insertional mutant upb1-1 exhibits increased root growth. UPB1 was shown to repress several peroxidase genes and thus plays a role in the distribution of reactive oxygen species and the balance between cell division and differentiation in the root tip region (Tsukagoshi et al., 2010).
PREs also interact with additional HLH proteins distantly related to IBH1. Long HYPOCOTYL IN FAR-RED1 (HFR1), PHYTOCHROME RAPIDLY REGULATED1 (PAR1), and PAR2 are PIF4 targets genes and are transcriptionally induced by shade through a PIF-dependent mechanism (Leivar et al., 2012; Oh et al., 2012). Like IBH1, HFR1 and PAR1/PAR2 encode atypical bHLH proteins that lack DNA binding ability (Galstyan et al., 2011). HFR1 and PAR1/PAR2 interact with PIF4 and inhibit PIF4 DNA binding (Hornitschek et al., 2009; Hao et al., 2012). PRE6/KDR was shown to bind to HFR1, and PRE1 was shown to bind to PAR1; thus, PRE6/KDR and PRE1 sequester HFR1 and PAR1 away from PIF4 (Hyun and Lee, 2006; Hao et al., 2012). Therefore, PREs, HFR1/PAR1/PAR2, and PIF4 form another HLH/bHLH cascade, which provides a positive feedback loop that potentially enhances plant responses to light, BR, and GA (Figure 8).
The HLH/bHLH transcription networks apparently play central roles in plant developmental regulation. The regulatory networks are shaped by complex interactions between antagonistic and synergistic partners, which accept input from a wide range of signal transduction and developmental pathways and form not only linear regulatory cascades but also feedback and crosstalk regulatory loops. Further dissection of the protein–protein and protein–DNA interactions in these networks, as well as quantitative analysis of the network dynamics in cell-type and developmental contexts, will be important for understanding plant growth regulation.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used as the wild-type control for phenotype comparison and for generating the transgenic plants. Seeds were surface sterilized for 15 min with 75% ethanol and plated on half-strength MS basal salt medium (Phyto-Technology Laboratories) supplemented with 0.7% phytoagar with or without indicated hormones or inhibitors. After 2 d of incubation at 4°C to promote germination, seedlings were grown in the dark or under light for 6 d. For hypocotyl length measurement, seedlings were photographed and their hypocotyl lengths were measured using ImageJ software (http://rsbweb.nih.gov/ij/) (Schneider et al., 2012).
Vector Construction
Full-length cDNAs of IBH1 and HBI1 without stop codon were amplified by PCR from Arabidopsis cDNA and cloned into pENTRY/SD/D-TOPO vectors (Invitrogen) or pENTRY-SRDX and then recombined into destination vector pEarlygate 101 (35S:C-YFP), p1390-Myc (35S:C-Myc), pDEST15 (N-GST), pMAL2CGW (N-MBP), pCY86 (N-GAL4BD), and pGAL4ADGW (N-GAL4AD). The truncated IBH1 constructs, including IBH1∆AS (deleting amino acids 48 to 72), IBH1∆B (deleting amino acids 87 to 103), IBH1N (amino acids 1 to 86), IBH1C1 (amino acids 87 to 156), and IBH1C2 (amino acids 104 to 156), were created by site-directed mutagenesis or PCR, cloned into pENTRY/SD/D-TOPO vectors (Invitrogen), and then recombined into destination vector pGAL4ADGW (N-GAL4AD) and/or pEarlygate 101 (35S:C-YFP). The EXP1 promoter was amplified from Arabidopsis genomic DNA and cloned into pENTRY/SD/D-TOPO vectors, then recombined into the destination vector pGREEN-GW (promoter LUC). Oligo primers used for PCR/cloning are listed in Supplemental Table 2 online.
Transient Gene Expression Assays
The protoplast transient assays were performed following the procedure described previously (Yoo et al., 2007; Wu et al., 2009). Plasmid DNAs were extracted using the Qiagen Plasmid Maxi Kit according to the manufacturer’s instructions. Aliquots of 5 × 104 isolated mesophyll protoplasts were transfected with a mixture of 20 µg of DNA using the polyethylene glycol method and incubated overnight. Protoplasts were harvested by centrifugation and lysed in 50 μL of passive lysis buffer (Promega). Firefly and Renilla (as internal standard) luciferase activities were measured using a dual-luciferase reporter kit (Promega).
Overlay Protein Gel Blot
The gel blot containing MBP and MBP-IBH1 proteins was stained with Ponceau S for loading control or incubated with 10 µg recombinant GST-HBI1 protein, washed, and then probed with anti-GST antibody (Santa Cruz Biotechnology, 1:3000 dilution).
Coimmunoprecipitation Assays
Mesophyll protoplasts (5 × 106 cells/reaction) prepared from the 35S:IBH1-myc transgenic plants were transfected with 20 µg of 35S:HBI1-YFP DNA using the polyethylene glycol method and incubated overnight. The protoplast cells were harvested by centrifugation and lysed in 200 μL of NEBT lysis buffer (20 mM HEPES-KOH, pH 7.5, 40 mM KCl, 1 mM EDTA, 0.5% Triton X-100, and 1× protease inhibitors; Roche). The lysates were incubated with anti-YFP antibody and Protein A agarose beads for 1 h, and the beads were washed four times with wash buffer and then eluted by boiling in 2× SDS loading buffer for 5 min. Samples were analyzed by SDS-PAGE and immunoblotted with anti-YFP (homemade, 1:3000 dilution; He et al., 2005) and anti-Myc antibodies (Cell Signaling; 1:5000 dilution).
DNA-Protein Pull-Down Assays
The recombinant proteins MBP-HBI1, MBP-IBH1, and MBP-PRE1 were expressed and affinity purified from Escherichia coli using amylose resin (NEB). The EXP1 promoter fragments were amplified by PCR using the biotin-labeled EXP1-biotinF and the EXP1-promoterR primers (see Supplemental Table 2 online). The DNA and proteins were incubated, and then DNA binding proteins were pulled down using streptavidin-agarose beads and analyzed by immunoblotting, as described previously (Bai et al., 2012).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays were performed as described previously (Bai et al., 2012) using 35S:YFP, 35S:HBI1-YFP, 35S:HBI1-YFP/35S:IBH1-myc, and 35S:HBI1-YFP/35S:pre-amiR transgenic plants. An affinity-purified anti-YFP polyclonal antibody was used for immunoprecipitation. The chromatin immunoprecipitation products were analyzed by quantitative real-time PCR (primer sequences are listed in Supplemental Table 2 online), and enrichment was calculated as the ratio between the transgenic samples expressing HBI1-YFP and the 35S:YFP control sample. The ChIP experiments were performed with three biological replicates, from which the means and standard deviations were calculated.
Quantitative RT-PCR Analysis
Total RNA was extracted from 5-d-old Arabidopsis seedlings using the Spectrum Plant Total RNA kit (Sigma-Aldrich). The first-strand cDNA was synthesized using RevertAid reverse transcriptase (Fermentas) and used as RT-PCR templates. Quantitative PCR analyses were performed on a plate-based LightCycler 480 (Roche) using a SYBR Green reagent (Bio-Rad) with gene-specific primers (see Supplemental Table 2 online). The conditions for PCR amplification were as follows: 98°C for 10 min; 45 cycles of 98°C for 30 s; 65°C for 45 s and 72°C for 30 s; and one cycle 72°C for 10 min. PP2A was used to as an internal reference gene. The relative expression was calculated as ratio between the transgenic plant and wild type and then normalized by PP2A.
RNA-Seq Analysis
Wild-type Arabidopsis, 35S:IBH1-myc, and 35S:IBH1-myc/35S:PRE1-YFP plants were grown on half-strength MS medium for 5 d under constant light. Total RNA was extracted with Spectrum Plant Total RNA kit (Sigma-Aldrich), and mRNA sequencing libraries were constructed with barcodes using the TruSeq RNA Sample Preparation Kit (Illumina). Six barcoded libraries were pooled together and sequenced by Illumina HiSeq2000. The RNA-Seq experiments were performed with two biological repeats. Total reads were mapped to the Arabidopsis genome (TAIR9; www.Arabidopsis.org) using TopHat software (Trapnell et al., 2009). Read counts for every gene were generated using HTSeq with union mode. Differential expressed genes between samples were defined by DESeq using two separate models (Anders and Huber, 2010), based on fold change>1.5 and false discovery rate–adjusted P value < 0.05. The accession number for the RNA-Seq data in the Gene Expression Omnibus database is GSE41766.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: PRE1, At5g39860; PRE2, At5g15160; PRE3, At1g74500; PRE4, At3g47710; PRE5, At3g28857; PRE6, At1g26945; OsIBH1, Os04g0660100; AtIBH1, At2g43060; AIF1, At3g05800; AIF2, At3g06590; AIF3, At3g17100; AIF4, At1g09250; UPB1, At2g47270; HBI1, At2g18300; BEE1, At1g18400; BEE2, At4g36540; BEE3, At1g73830; BPE, At1g59640; CIB1, At4g34530; EXP1, At1g69530; and EXP8, At2g40610.
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. IBH1, UPB1, and AIFs Belong to the Same HLH Family.
Supplemental Figure 2. IBH1 Interacts with Five Homologous Proteins in Yeast.
Supplemental Figure 3. Yeast Two-Hybrid Assays Showed That IBH1 Interacts with PRE1 through Its HLH Domain.
Supplemental Figure 4. Quantitative RT-PCR Analyses of the Expression of HBI1 and Its Homologous Genes in Wild-Type and HBI1 Cosuppression Plants.
Supplemental Figure 5. HBI1 and Its Homologs Contain Conserved Basic Domain to Bind DNA.
Supplemental Figure 6. The Gene Structure Diagram of EXP1 Showing Promoter Regions Analyzed for HBI1 Binding.
Supplemental Table 1. Quantitative RT-PCR Validation of the RNA-Seq Data.
Supplemental Table 2. Oligo Sequences.
Supplemental Data Set 1. PRE1 and IBH1 Antagonistically Regulate Gene Expression.
Acknowledgments
We thank Yang. Bai for help with RNA-Seq data analysis and Jian-Xiu. Shang for experimental assistance. This work was supported by a grant from the National Institutes of Health (R01GM066258).
AUTHOR CONTRIBUTIONS
M.-Y.B. and Z.-Y.W. designed the experiments, analyzed the data, and wrote the article. M.-Y.B. performed RNA-Seq, statistical analysis of plant growth, yeast two-hybrid, overlay, coimmunoprecipitation, transient expression assay, and ChIP-qPCR. M.F. analyzed the function of different domains of IBH1. E.O. generated HBI1-Ox transgenic plants. M.-Y.B. performed all other experiments.
Glossary
- BR
brassinosteroid
- GA
gibberellin
- PIF
phytochrome-interacting factor
- bHLH
basic helix-loop-helix
- HLH
helix-loop-helix
- PRE
paclobutrazol-resistant
- RNA-Seq
RNA-sequencing
- MS
Murashige and Skoog
- GST
glutathione S-transferase
- YFP
yellow fluorescent protein
- PPZ
propiconazole
- PAC
paclobutrazol
- ChIP-qPCR
chromatin immunoprecipitation followed by quantitative real-time PCR
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelain M., Le Hir R., Bellini C. (2012). The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant. 145: 450–460 [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Greenham K., Castillejo C., Sartor R., Bialy A., Sun T.P., Estelle M. (2012). Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (2000). Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Nemhauser J., Muramitsu T., Maloof J.N., Alonso J., Ecker J.R., Furuya M., Chory J. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A., Cifuentes-Esquivel N., Bou-Torrent J., Martinez-Garcia J.F. (2011). The shade avoidance syndrome in Arabidopsis: A fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 66: 258–267 [DOI] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Fujiwara S., Mitsuda N., Ohme-Takagi M. (2012). A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24: 4483–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee S., Yang K.Y., Kim Y.M., Park S.Y., Kim S.Y., Soh M.S. (2006). Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. (2012). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Mara C.D., Huang T., Irish V.F. (2010). The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22: 690–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Street I.H., Turk E.M., Ward J.M. (2006). Interaction of light and hormone signaling to mediate photomorphogenesis. In Photomorphogenesis in Plants and Bacteria, E. Schäfer and F. Nagy, eds (Springer Verlag: New York), pp. 439–473.
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ruzinova M.B., Benezra R. (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13: 410–418 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E.H., Schmid M., Jürgens G., Weijers D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to Image: 25 years of image analysis. Nature Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J., Han Z., Kim T.W., Wang J., Cheng W., Chang J., Shi S., Wang J., Yang M., Wang Z.Y., Chai J. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X.Y., Cao D.M., Tang W., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S., Oh E., Patil S., Kim T.W., Ji H., Wong W.H., Rhee S.Y., Wang Z.Y. (2012). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Németh K., Koncz-Kálmán Z., Mathur J., Kauschmann A., Altmann T., Rédei G.P., Nagy F., Schell J., Koncz C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P.H., Deng X.W., Guo H. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]