Coordinated spatiotemporal-specific cell proliferation is critical to plant growth and development. This study demonstrates the function of a mitogen-activated protein kinase cascade downstream of the ERECTA receptor-like kinase in regulating localized cell proliferation, which determines the inflorescence architecture and size in Arabidopsis thaliana.
Abstract
Spatiotemporal-specific cell proliferation and cell differentiation are critical to the formation of normal tissues, organs, and organisms. The highly coordinated cell differentiation and proliferation events illustrate the importance of cell–cell communication during growth and development. In Arabidopsis thaliana, ERECTA (ER), a receptor-like protein kinase, plays important roles in promoting localized cell proliferation, which determines inflorescence architecture, organ shape, and size. However, the downstream signaling components remain unidentified. Here, we report a mitogen-activated protein kinase (MAPK; or MPK) cascade that functions downstream of ER in regulating localized cell proliferation. Similar to an er mutant, loss of function of MPK3/MPK6 or their upstream MAPK kinases (MAPKKs; or MKKs), MKK4/MKK5, resulted in shortened pedicels and clustered inflorescences. Epistasis analysis demonstrated that the gain of function of MKK4 and MKK5 transgenes could rescue the loss-of-function er mutant phenotype at both morphological and cellular levels, suggesting that the MPK3/MPK6 cascade functions downstream of the ER receptor. Furthermore, YODA (YDA), a MAPKK kinase, was shown to be upstream of MKK4/MKK5 and downstream of ER in regulating inflorescence architecture based on both gain- and loss-of-function data. Taken together, these results suggest that the YDA-MKK4/MKK5-MPK3/MPK6 cascade functions downstream of the ER receptor in regulating localized cell proliferation, which further shapes the morphology of plant organs.
INTRODUCTION
Plant growth and development require constant communication between different cells, tissues, and organs. In eukaryotes, cell–cell communication often involves cell surface receptors/sensors, which trigger protein phosphorylation, a universal signaling mechanism involved in almost all fundamental cellular processes (Cohen, 2000; Manning et al., 2002). Plant receptor-like protein kinases (RLKs) form the largest group of protein kinases in the protein kinase gene family. For instance, Arabidopsis thaliana has more than 600 different RLKs (Shiu and Bleecker, 2001). The large number of plant RLKs and the fact that a single RLK can sometimes recognize multiple stimuli allow plants to use RLKs to sense a diverse array of extracellular signals (reviewed in Yin et al., 2002; Morris and Walker, 2003; Diévart and Clark, 2004; Morillo and Tax, 2006; De Smet et al., 2009). At present, we have functional information on only a few RLKs. They are involved in sensing/recognizing plant hormones, peptide ligands, and pathogen-derived molecules (Becraft, 2002; Diévart and Clark, 2004; Vert et al., 2005; Matsubayashi and Sakagami, 2006; De Smet et al., 2009; Uchida et al., 2012).
Arabidopsis ERECTA (ER) regulates several growth/developmental processes including inflorescence architecture, stomatal formation and patterning, and ovule development (Shpak et al., 2003; van Zanten et al., 2009; Pillitteri and Torii, 2012). ER has two closely related homologs, ER-LIKE 1 (ERL1) and ERL2, which are partially redundant with ER (Shpak et al., 2004; Lee et al., 2012). The role of the ER family receptors in the stomatal development pathway has been well studied (Shpak et al., 2005; Pillitteri and Torii, 2012). After perceiving the secreted peptide ligands, EPF1 and EPF2, the ER family receptors act sequentially to specify stomatal patterning (Hara et al., 2007; Hunt and Gray, 2009; Lee et al., 2012). ER also promotes inflorescence growth by regulating localized cell proliferation. Loss of function of the ER gene results in shortened pedicels and clustered inflorescences due to reduced cell proliferation in pedicels (Shpak et al., 2003, 2004; Woodward et al., 2005). Recently, two secreted peptides, EPFL4 and EPFL6, were identified as redundant upstream components of ER-mediated inflorescence growth, possibly as the ligands in the signaling process (Abrash et al., 2011; Uchida et al., 2012). However, the signaling pathways downstream of ER in regulating inflorescence architecture remain unknown at this stage.
Mitogen-activated protein kinase (MAPK) cascades are highly conserved signaling modules in eukaryotes. They function downstream of sensors/receptors and convert signals generated at the sensors/receptors into cellular responses (Widmann et al., 1999; Chang and Karin, 2001; Schwartz and Madhani, 2004). Each MAPK cascade is composed of three kinases with MAPK at the bottom tier. Phosphorylation activation of MAPKs is performed by MAPK kinases (MAPKKs), which are in turn activated by MAPKK kinases (MAPKKKs) through phosphorylation. Plant MAPK cascades play important roles in regulating plant growth, development, and responses to stress/defense stimuli (reviewed in Pedley and Martin, 2005; Zhang, 2008; Pitzschke et al., 2009; Rodriguez et al., 2010; Tena et al., 2011). In Arabidopsis, there are 20 MAPKs, 10 MAPKKs, and ∼60 MAPKKKs (Ichimura et al., 2002; Hamel et al., 2006; Andreasson and Ellis, 2010). Our research has been focused on MPK3 and MPK6, two Arabidopsis stress-responsive MAPKs, and their tobacco (Nicotiana tabacum) orthologs wounding-induced protein kinase (WIPK), salicylic acid-induced protein kinase (SIPK), and Ntf4 (Zhang and Klessig, 1997, 1998; Ren et al., 2002; Liu and Zhang, 2004; Ren et al., 2006, 2008). In the process of acquiring loss-of-function evidence to support the functions of MPK3 and MPK6 in plant defense/stress signaling, we found that these two MAPKs also play essential roles in several developmental processes, including stomatal patterning, floral organ abscission, and ovule development (Wang et al., 2007; Cho et al., 2008; Wang et al., 2008).
In this report, we provide gain- and loss-of-function genetic evidence to support a MAPK cascade, composed of YODA (YDA), MKK4/MKK5, and MPK3/MPK6, as a key downstream component of the ER receptor in regulating inflorescence architecture. Loss-of-function mutants of MPK3/MPK6 and MKK4/MKK5 phenocopy the er mutant, whereas the gain-of-function MKK4 and MKK5 transgenes can rescue the er mutant at both morphological and cellular levels, demonstrating that the MKK4/MKK5-MPK3/MPK6 module functions downstream of the ER receptor. Furthermore, YDA was shown to be upstream of MKK4/MKK5 and downstream of ER in this signaling pathway. Our previous study demonstrated a key role of the YDA-MKK4/MKK5-MPK3/MPK6 cascade in coordinated cell differentiation leading to proper stomatal formation and patterning (Wang et al., 2007). This study reveals another function of this MAPK cascade downstream of the ER receptor in regulating coordinated local cell proliferation, which shapes the morphology of plant organs.
RESULTS
Loss-of-Function mpk3 and mpk6 Mutant Plants Phenocopy er Mutants
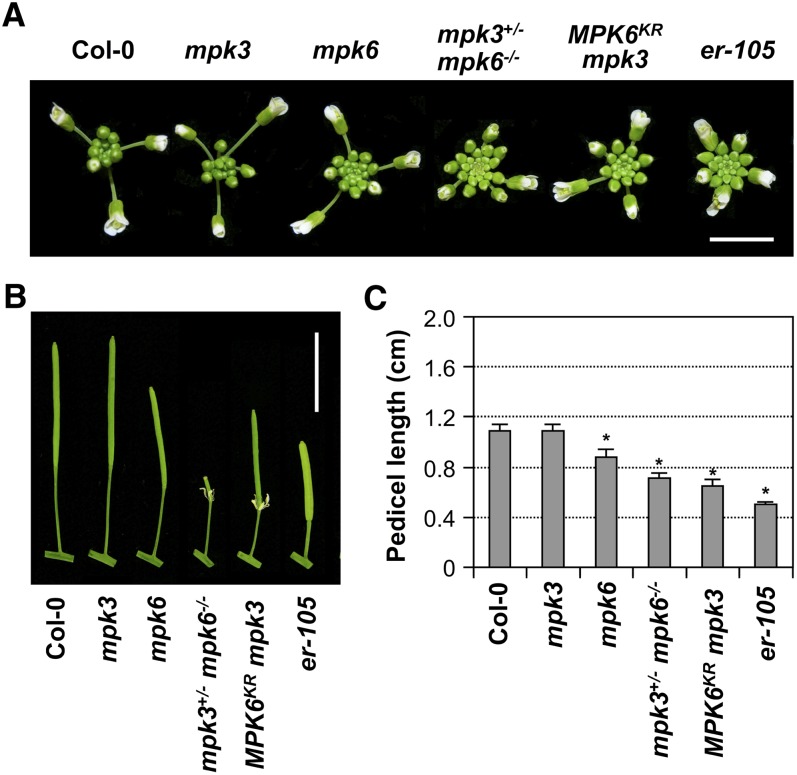
In the process of generating loss-of-function mutants to explore the roles of MPK3 and MPK6 in plant defense signaling, we found that mpk6 single mutant plants showed a weak er phenotype with a more clustered inflorescence in comparison to wild-type plants (Columbia-0 [Col-0]) (Figure 1A), whereas mpk3 single mutant plants did not display this phenotype. As with er-105, an ER null mutant allele in the Col-0 background (Shpak et al., 2003), the clustered inflorescence of the mpk6 single mutant was associated with shortened pedicels (Figures 1B and 1C). Consistent with the gene redundancy and dosage effects of MPK3 and MPK6 (Wang et al., 2007, 2008), mpk3+/− mpk6−/− plants exhibited a more compact inflorescence, which was closer to that of the er-105 mutant. Because the mpk3 mpk6 double mutant is embryo lethal, we examined another loss-of-function MPK3/MPK6 system, in which a dominant-negative MPK6KR mutant protein was expressed in the mpk3 mutant background (Cho et al., 2008). As shown in Figure 1, MPK6KR mpk3 plants also displayed a clustered inflorescence with shortened pedicels, whereas MPK6KR plants did not show alterations in inflorescence morphology. These results indicate that MPK3 and MPK6 have overlapping functions in regulating inflorescence architecture. In this process, MPK6 plays a more important role than MPK3 because mutations in MPK6, but not MPK3, conferred a more clustered inflorescence.
Figure 1.
Loss-of-Function MPK3 and MPK6 Mutant Plants Phenocopy the er Mutant in Inflorescence Architecture and Pedicel Length.
(A) Top view of Col-0, mpk3, mpk6, mpk3+/− mpk6−/−, MPK6KR mpk3, and er-105 inflorescence. Bar = 0.5 cm.
(B) Pedicels of mature siliques of Col-0, mpk3, mpk6, mpk3+/− mpk6−/−, MPK6KR mpk3, and er-105 plants. Bar = 1 cm.
(C) Lengths of mature pedicels on the main stems of 7-week-old plants. Bars represent average values + sd (n = 30 to 40 pedicels per genotype). Asterisks above the columns indicate significant differences compared with Col-0 (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
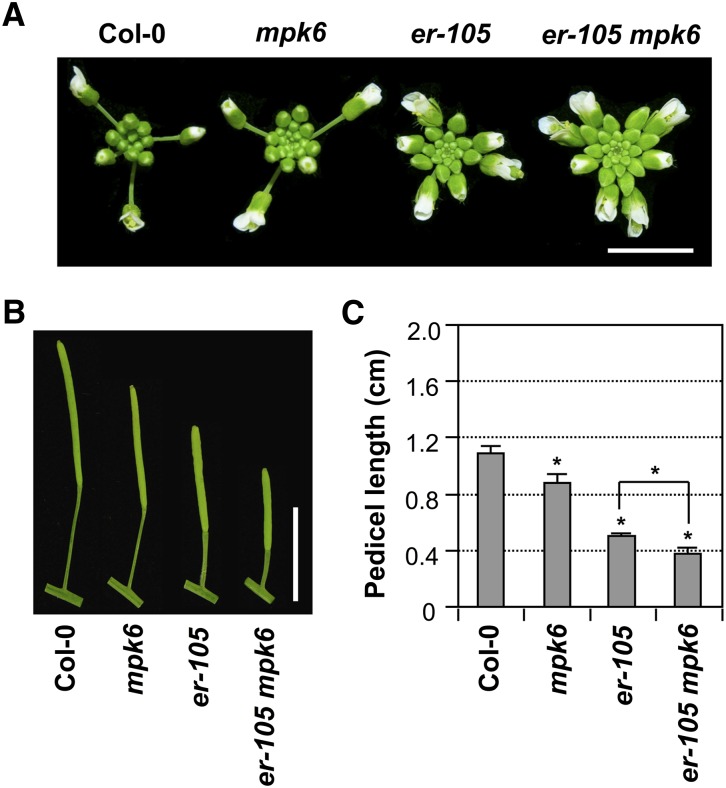
We also observed a more severely clustered flower phenotype in the er-105 mpk6 double mutant (Figure 2), an indication that these two components might function in the same pathway in regulating the inflorescence architecture. ER has two additional paralogs, ERL1 and ERL2, and the higher order mutants showed more severe phenotypes (Shpak et al., 2004). In the sensitized single er mutant background, mutation of additional components such as MPK6 in the same pathway would result in a more severe phenotype.
Figure 2.
Additive Effect of MPK6 and ER in Regulating Inflorescence Architecture and Pedicel Elongation.
(A) Inflorescence stem apices of Col-0, mpk6, er-105, and er-105 mpk6 plants. Bar = 0.5 cm.
(B) Mature siliques and attached pedicels of respective genotypes. Bar = 1 cm.
(C) Lengths of mature pedicels on the main stems of ∼7-week-old plants. Bars represent average values + sd (n = 30 to 40 pedicels per genotype). Asterisks above the columns indicate significant differences compared with Col-0 or in the marked comparisons (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
Gain of Function of GVG-Nt-MEK2DD Can Rescue the er Mutant Phenotype
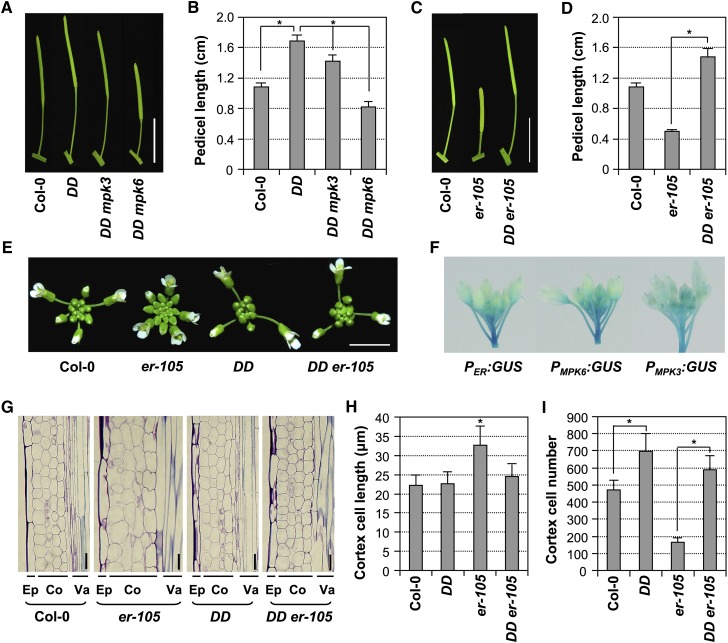
Previously, we successfully used GVG-Nt-MEK2DD transgenic plants (hereafter abbreviated as DD) as a conditional gain-of-function MPK3/MPK6 system (Ren et al., 2002, 2008; Wang et al., 2007). Nt-MEK2 is the tobacco ortholog of Arabidopsis MKK4 and MKK5. In DD plants, a constitutively active Nt-MEK2 variant (Nt-MEK2DD) driven by a steroid-inducible promoter can activate the endogenous MPK3/MPK6 after dexamethasone treatment (Ren et al., 2002, 2008). We observed that DD plants had much longer pedicels than wild-type plants (Figures 3A and 3B), which is opposite to that of the loss-of-function MPK3/MPK6 plants (Figure 1). This gain-of-function phenotype in DD plants is a result of basal level expression of DD transgene since steroid application was not required for the phenotype. In the mpk6 mutant background, the DD-mediated gain-of-function phenotype was completely reversed, whereas mutation of MPK3 had much less effect (Figures 3A and 3B), suggesting again that MPK6 plays a predominant role in this process. These results indicate that gain-of-function activation of MPK3/MPK6 could promote pedicel elongation and affect inflorescence architecture.
Figure 3.
Expression of Constitutively Active Nt-MEK2DD (DD) Rescued the Developmental Defects of the er-105 Inflorescence by Promoting Cell Proliferation.
(A) Elongated pedicel length of DD plants is dependent on downstream MPK3 and MPK6. Mature siliques and attached pedicels of Col-0, DD, DD mpk3, and DD mpk6 plants. Bar = 1 cm.
(B) Lengths of mature pedicels on the main stem of ∼7-week-old plants (n = 30 to 40 pedicels per genotype).
(C) Mature siliques and attached pedicels of Col-0, er-105, and DD er-105 plants. Bar = 1 cm.
(D) Lengths of mature pedicels on the main stem of ∼7-week-old Col-0, er-105, and DD er-105 plants (n = 30 to 40 pedicels per genotype).
(E) Inflorescence stem apices of Col-0, er-105, DD and DD er-105 plants. Bar = 0.5 cm.
(F) PER:GUS, PMPK3:GUS, and PMPK6:GUS expression patterns in inflorescences.
(G) Longitudinal sections of mature pedicels from Col-0, er-105, DD, and DD er-105 plants. Co, cortex; Ep, epidermis; Va, vasculature. Bar = 25 μm.
(H) Quantitative analysis of cortex cell length (n = 15 to 20 pedicels per genotype).
(I) Cell numbers in the longitudinal cortex file of a mature pedicel (n = 15 to 20 pedicels per genotype). Bars represent average values + sd. Asterisks above the columns indicate significant differences in the marked comparisons or compared with Col-0 (P < 0.0001, Student’s t test).
To determine if MPK3/MPK6 function downstream of ER in regulating inflorescence structure, we analyzed potential epistatic interactions by crossing the DD transgene into the er-105 background. As shown in Figures 3C to 3E, the shortened pedicel and clustered inflorescence phenotypes of er-105 were completely reversed in DD er-105 plants, suggesting that the activation of MPK3/MPK6 by DD is sufficient to rescue the loss of ER function. Previous studies showed that ER regulates inflorescence growth by promoting cell proliferation. The defect of pedicel elongation in er-105 is due to reduced cell proliferation, notably in the cortex, accompanied by compensatory cell expansion (Shpak et al., 2004; Woodward et al., 2005; Uchida et al., 2012). We observed the same cell proliferation phenotype in the er-105 mutant (Figures 3G to 3I). Interestingly, the cell size of the pedicel cortex of DD plants was similar to that of wild-type plants (Figures 3G and 3H), and the elongated pedicel of DD plants was associated with an increase in the cell number (Figure 3I). In DD er-105 plants, cell numbers in a longitudinal cortical file were significantly increased in comparison with er-105, with a concomitant increase in the final pedicel length. These results demonstrate that the activation of MPK3/MPK6 cascade promotes the cell proliferation in pedicels, which results in pedicel elongation and a less clustered inflorescence. More importantly, the rescue of the er mutant by a gain-of-function DD transgene suggests that the MPK3/MPK6 cascade functions downstream of ER in controlling the same biological process (i.e., localized cell proliferation), which in turn determines inflorescence architecture. Consistent with this conclusion, promoter–β-glucuronidase (GUS) reporter analyses indicated that ER, MPK3, and MPK6 are coexpressed in growing inflorescence stems and pedicels (Figure 3F).
Constitutively Active MKK4 and MKK5, but Not Other MAPKKs, Rescued the er Mutant Phenotype
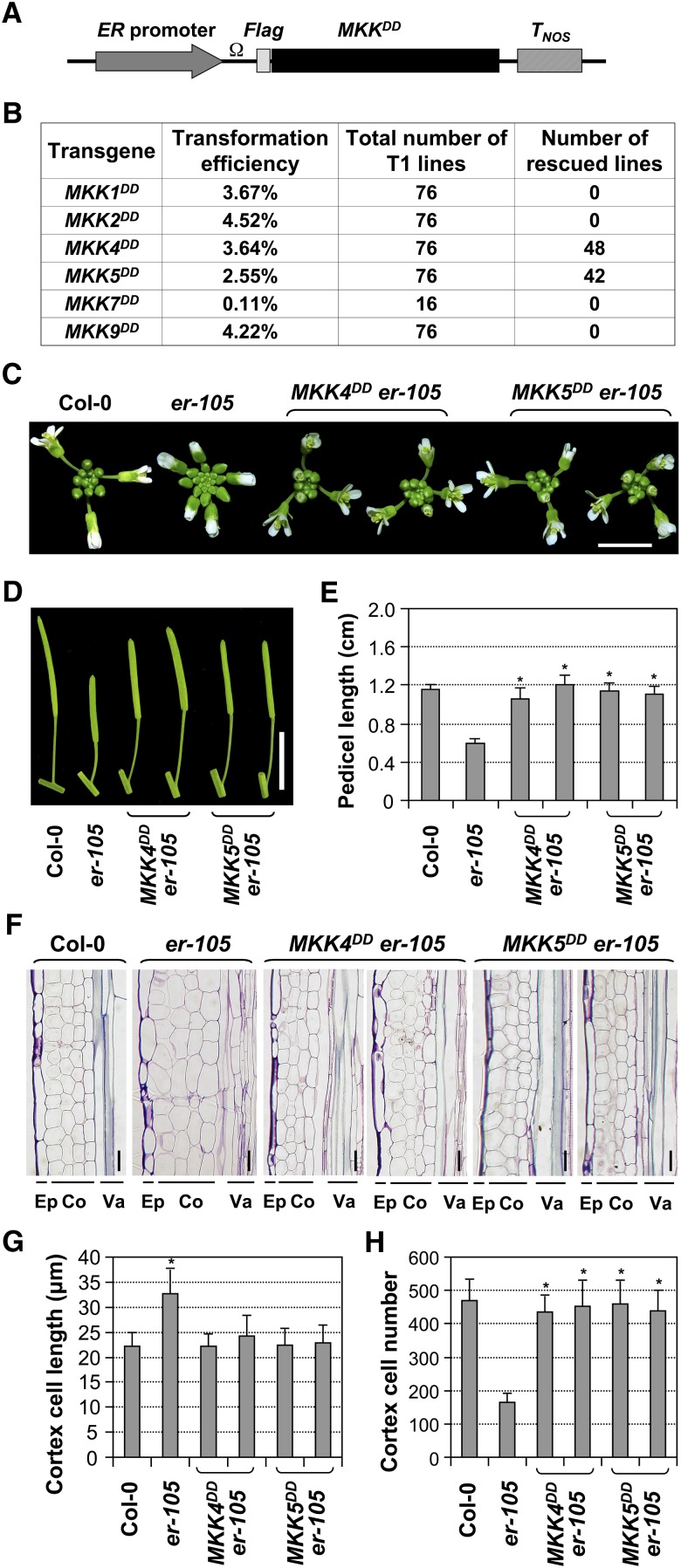
Arabidopsis MKK4 and MKK5 were shown to be functionally interchangeable with tobacco Nt-MEK2 in several biological pathways (Ren et al., 2002, 2008; Wang et al., 2007; Han et al., 2010). Several other MAPKKs, including MKK1, MKK2, MKK7, and MKK9, were also reported as upstream kinases of MPK3/MPK6 (Teige et al., 2004; Mészáros et al., 2006; Xing et al., 2008; Xu et al., 2008; Lampard et al., 2009; Popescu et al., 2009). To determine the endogenous MAPKKs downstream of ER in regulating inflorescence architecture in the ER signaling pathway, we conducted experiments by expressing constitutively active MAPKK variants (MAPKKDD) under the control of the ER promoter (Figure 4A) in the er-105 background. Besides MKK4 and MKK5, we also tested MKK1, MKK2, MKK7, and MKK9. The use of the ER promoter is intended to provide spatiotemporal activation of MPK3/MPK6 related to ER functions.
Figure 4.
Rescue of the er Inflorescence Phenotype by ER Promoter–Driven Expression of Constitutively Active MKK4 and MKK5, but Not MKK1, MKK2, MKK7, and MKK9.
(A) Schematic diagram of ER promoter–driven expression constructs for constitutively active MKKs. Ω, TMV omega translation enhancer; MKKDD, constitutively active MKK mutants; TNOS, NOS terminator.
(B) Transformation efficiencies of different MKKDD constructs in the er-105 background and frequencies of rescued T1 lines.
(C) Inflorescence stem apices of Col-0, er-105, and two representative lines each of T2 PER:MKK4DD and PER:MKK5DD transgenic plants in the er-105 background. Bar = 0.5 cm.
(D) Mature siliques and attached pedicels of the indicated genotypes. Bar = 1 cm.
(E) Lengths of mature pedicels on the main stem of ∼7-week-old plants (n = 30 to 40 pedicels per genotype).
(F) Longitudinal sections of mature pedicels from Col-0, er-105, and two representative lines of T2 PER:MKK4DD and PER:MKK5DD transgenic plants in the er-105 background. Co, cortex; Ep, epidermis; Va, vasculature. Bars = 25 μm.
(G) Quantitative analysis of cortex cell length (n = 15 to 20 pedicels per genotype).
(H) Number of cells in the longitudinal cortex file of a mature pedicel of the indicated genotypes (n = 15 to 20 pedicels per genotype). Bars represent average values + sd. Asterisks above the columns indicate significant differences compared with Col-0 (G) or er-105 ([E] and [H]) (P < 0.0001, Student’s t test).
T1 transgenic plants in er-105 were screened for the rescue of er mutant phenotypes. As summarized in Figure 4B, the typical er phenotypes were reversed in a large percentage (40 to 50%) of PER:MKK4DD and PER:MKK5DD T1 plants, which have longer pedicels and less clustered inflorescences. By contrast, none of the PER:MKK1DD and PER:MKK2DD T1 plants rescued the er phenotypes. A large percentage of PER:MKK9DD T1 plants showed defective growth and development associated with localized cell death (see Supplemental Figure 1 online), and no rescued lines were obtained. A very low transformation rate was obtained with the PER:MKK7DD construct, and none of the transgenic lines showed rescue of er phenotypes. The low transformation rate might be a result of lethality since even basal level expression of MKK7DD in the steroid-inducible pTA7002-MKK7DD transgenic plants can cause an HR-like lethal phenotype (see Supplemental Figure 2 online). Taken together, our data indicate that MKK4 and MKK5, but not MKK1, MKK2, MKK7, or MKK9, function downstream of ER in regulating inflorescence architecture. We further performed more detailed characterization of selected PER:MKK4DD and PER:MKK5DD lines in the T2 generation. As shown in Figure 4C, expression of the constitutively active MKK4 and MKK5 rescued the clustered inflorescence phenotype of er-105, a result of elongated pedicels (Figure 4D). Quantitative measurement showed that both MKK4DD and MKK5DD completely rescued the shortened pedicel phenotype of er-105 (Figure 4E).
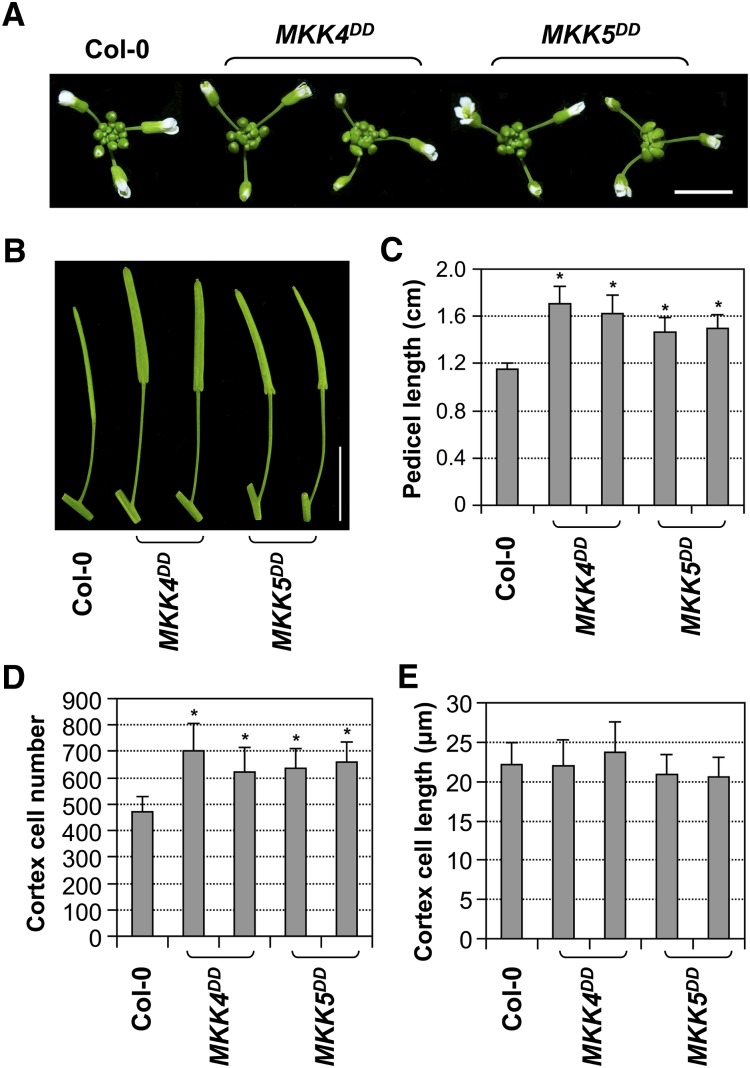
Longitudinal sectioning of pedicel tissues revealed that the cortex cells of PER:MKK4DD and PER:MKK5DD transgenic lines resembled those of wild-type plants, in contrast with the large and expanded cortex cells in er-105 (Figures 4F and 4G). In addition, PER:MKK4DD and PER:MKK5DD transgenic plants had comparable cell numbers in the cortical cell files as wild-type plants, whereas the cortex cell number was dramatically reduced in er-105 (Figure 4H). Our data suggest that gain-of-function MKK4/MKK5 could rescue the inflorescence developmental defects of er-105 at both morphological and cellular levels. In addition to the rescue of er phenotypes, ER promoter–driven expression of MKK4DD or MKK5DD in Col-0 also gave a gain-of-function phenotype with much longer pedicels than wild-type plants (Figures 5A to 5C). The longer pedicels of the gain-of-function transgenic lines were correlated with increased cortex cell numbers (Figure 5D), whereas cortex cell sizes were not significantly different between the transgenic lines and wild-type plants (Figure 5E). These results provide both genetic and cellular evidence that MKK4/MKK5 are downstream of ER in regulating pedicel elongation and inflorescence architecture.
Figure 5.
ER Promoter–Driven Expression of Constitutively Active MKK4 or MKK5 Resulted in a Gain-of-Function Phenotype with Elongated Pedicels and Enhanced Cell Proliferation.
(A) Inflorescence stem apices of Col-0 and two representative T2 lines each of PER:MKK4DD and PER:MKK5DD transgenic plants in Col-0 background. Bar = 0.5 cm.
(B) Mature siliques and attached pedicels of the indicated genotypes. Bar = 1 cm.
(C) Lengths of mature pedicels on the main stem of ∼7-week-old plants (n = 30 to 40 pedicels per genotype).
(D) Quantitative analysis of cortex cell length (n = 15 to 20 pedicels per genotype).
(E) Number of cells in the longitudinal cortex file of a mature pedicel of the indicated genotypes (n = 15 to 20 pedicels per genotype).
Bars represent average values + sd. Asterisks above the columns indicate significant differences compared with Col-0 (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
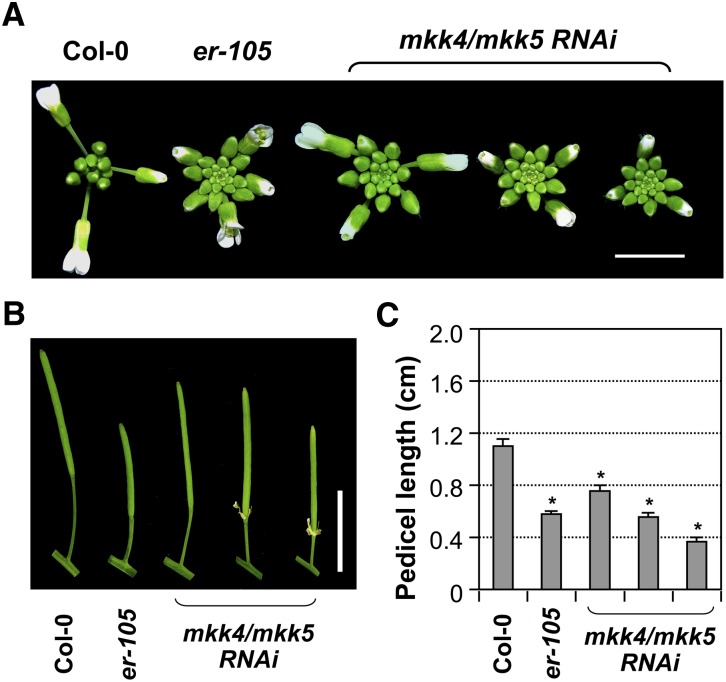
Loss-of-Function MKK4/MKK5 Plants Phenocopy the er Mutant
Gain-of-function data suggest a positive role for MKK4/MKK5 in regulating inflorescence architecture downstream of ER. To provide loss-of-function evidence, we used a tandem mkk4/mkk5 RNA interference (RNAi) construct to silence both genes because loss-of-function alleles of MKK4 and MKK5 are not available in public depositories. Previously, we used this RNAi strategy to demonstrate the function of MKK4/MKK5 as the upstream kinase of MPK3/MPK6 in regulating the stomatal development and floral organ abscission (Wang et al., 2007; Cho et al., 2008). The majority of T1 transgenic plants had severe stomata developmental defects (Wang et al., 2007) and were unable to survive beyond the seedling stage. Among the transgenic plants that survived, we found that the lines with abscission defects also displayed a clustered inflorescence phenotype nearly identical to that of er-105 (Figures 6A and 6B). There were also lines that had a weak er phenotype, but without abscission defects (Figure 6). The pedicels of these mkk4/mkk5 RNAi lines were much shorter than those of wild-type plants, reminiscent of er-105 and opposite of the gain-of-function MKK4DD and MKK5DD transgenic plants (Figures 6B and 6C). These loss-of-function data further support that MKK4/MKK5 function downstream of ER in regulating pedicel elongation and inflorescence architecture. Based on data from loss-of-function and epistasis analyses, we conclude that MKK4/MKK5 are downstream of ER and upstream of MPK3/MPK6 in regulating localized cell proliferation, which further influences pedicel elongation and inflorescence architecture. MKK4 and MKK5 have been previously reported to be upstream of MPK3/MPK6 in both defense- and development-related processes (Ren et al., 2002, 2008; Wang et al., 2007; Han et al., 2010).
Figure 6.
Tandem RNAi Suppression of MKK4 and MKK5 Resulted in a Clustered Inflorescence and Shortened Pedicels, Phenocopying the er-105 Mutant.
(A) Inflorescence stem apices of Col-0, er-105, and three representative T1 mkk4/mkk5 RNAi transgenic plants. Bar = 0.5 cm.
(B) Mature siliques and attached pedicels of the indicated genotypes. Bar = 1 cm.
(C) Lengths of mature pedicels on the main stem of ∼7-week-old plants (n = 10 to 15 pedicels per genotype). Bars represent average values + sd. Asterisks above the columns indicate significant differences compared with Col-0 (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
YDA Is the Upstream MAPKKK of MKK4/MKK5 in the ER Signaling Pathway
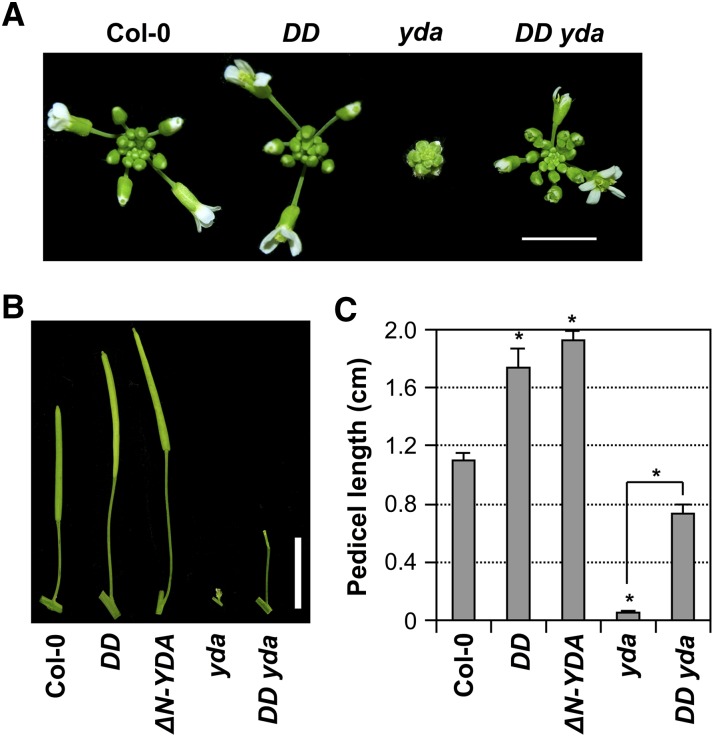
We previously showed that YDA, a MEKK1 type of Arabidopsis MAPKKK, is upstream of MKK4/MKK5 in regulating stomatal development and patterning (Wang et al., 2007). To determine whether YDA is also upstream of the MKK4/MKK5-MPK3/MPK6 module in regulating inflorescence architecture, we characterized transgenic plants expressing the constitutively active YDA mutant (ΔN-YDA) driven by its endogenous promoter (Lukowitz et al., 2004). Similar to the gain-of-function DD, MKK4DD, and MKK5DD transgenic plants, ΔN-YDA plants also displayed elongated pedicels (Figures 7B and 7C). Opposite to the gain-of-function ΔN-YDA plants, the loss-of-function yda mutant had extremely short pedicels, resulting in a very tightly compact inflorescence, along with other phenotypes such as defective floral organs and sterile plants (Figure 7). We then crossed the gain-of-function DD transgene into the yda mutant for epistatic interaction analysis. As shown in Figure 7, DD partially rescued the yda mutant phenotype. Compared with the tightly compact inflorescence of the yda mutant with almost no pedicels, DD yda plants showed significantly extended inflorescences with much longer pedicels. However, DD failed to rescue the sterile phenotype of yda (Figure 7B). This epistasis analysis suggests that YDA is upstream of MKK4/MKK5 in regulating pedicel elongation and inflorescence architecture.
Figure 7.
YDA Is Upstream of MKK4/MKK5-MPK3/MPK6 in Regulating Inflorescence Architecture.
(A) Inflorescence stem apices of Col-0, DD, yda, and DD yda plants illustrating the extremely clustered inflorescence of yda and the partial rescue of this mutant phenotype in DD yda plants. Bar = 0.5 cm.
(B) Mature siliques and attached pedicels of Col-0, DD, ΔN-YDA, yda, and DD yda plants, showing the very long pedicel of the ΔN-YDA plant, the extremely short pedicel of the yda mutant, and the partially rescued pedicel of the DD yda plant. Bar = 1 cm.
(C) Lengths of mature pedicels on the main stem of ∼7-week-old plants (n = 30 to 40 pedicels per genotype). Bars represent average values + sd. Asterisks above the columns indicate significant differences compared with Col-0 or in the marked comparison (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
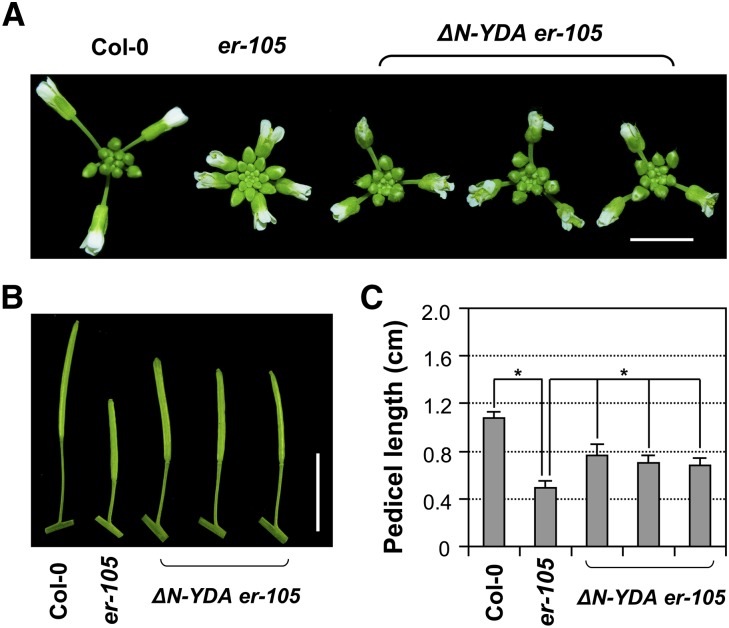
To further clarify the genetic relationship between YDA and ER, we transformed the constitutively active ΔN-YDA transgene driven by the native promoter into er-105. As shown in Figure 8, expression of ΔN-YDA partially rescued the er mutant phenotypes and conferred significantly elongated inflorescences and pedicels in comparison to those of er-105, suggesting that YDA is a downstream component of ER signaling. The incomplete rescue of the er-105 mutant phenotype by ΔN-YDA suggests the presence of a YDA-independent pathway(s), possibly mediated by other MAPKKKs, in regulating inflorescence architecture. Alternatively, the incomplete rescue of the er-105 mutant phenotype by ΔN-YDA could be a consequence of the lack of N terminus of the gain-of-function YDA transgene. Deletion of the large N-terminal domain makes ΔN-YDA constitutively active, yet in the meantime, may make it less efficient in performing other functions, such as interacting with other components in the signaling pathway, rendering it less effective in rescuing the er-105 phenotypes. By contrast, gain-of-function MAPKKs were generated by making Ser/Thr-to-Asp point mutations in their activation loop (Yang et al., 2001), which should not alter the functionalities of other domains. Nonetheless, the ΔN-YDA transgene is sufficient to confer a gain-of-function phenotype in the wild type (Figures 7B and 7C). In summary, our loss- and gain-of-function genetic data suggest that YDA, MKK4/MKK5, and MPK3/MPK6 function together in a MAPK cascade downstream of the ER receptor in regulating cell proliferation during pedicel growth/development, which determines the inflorescence architecture in Arabidopsis.
Figure 8.
Expression of Constitutively Active ΔN-YDA Partially Rescued the Inflorescence Developmental Defect of er-105.
(A) Inflorescence stem apices of Col-0, er-105, and three representative T1 lines of ΔN-YDA transgenic plants in the er-105 background. Bar = 0.5 cm.
(B) Mature siliques and attached pedicels of the indicated genotypes. Bar = 1 cm.
(C) Lengths of mature pedicels on the main stem of ∼7-week-old plants (n = 10 to 15 pedicels per genotype). Bars represent average values + sd. Asterisks above the brackets indicate significant differences between the sample group(s) and er-105 (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
Constitutively Active MKK4 and MKK5 Also Rescue the Short Petiole and Dwarf Phenotypes of the er Mutant
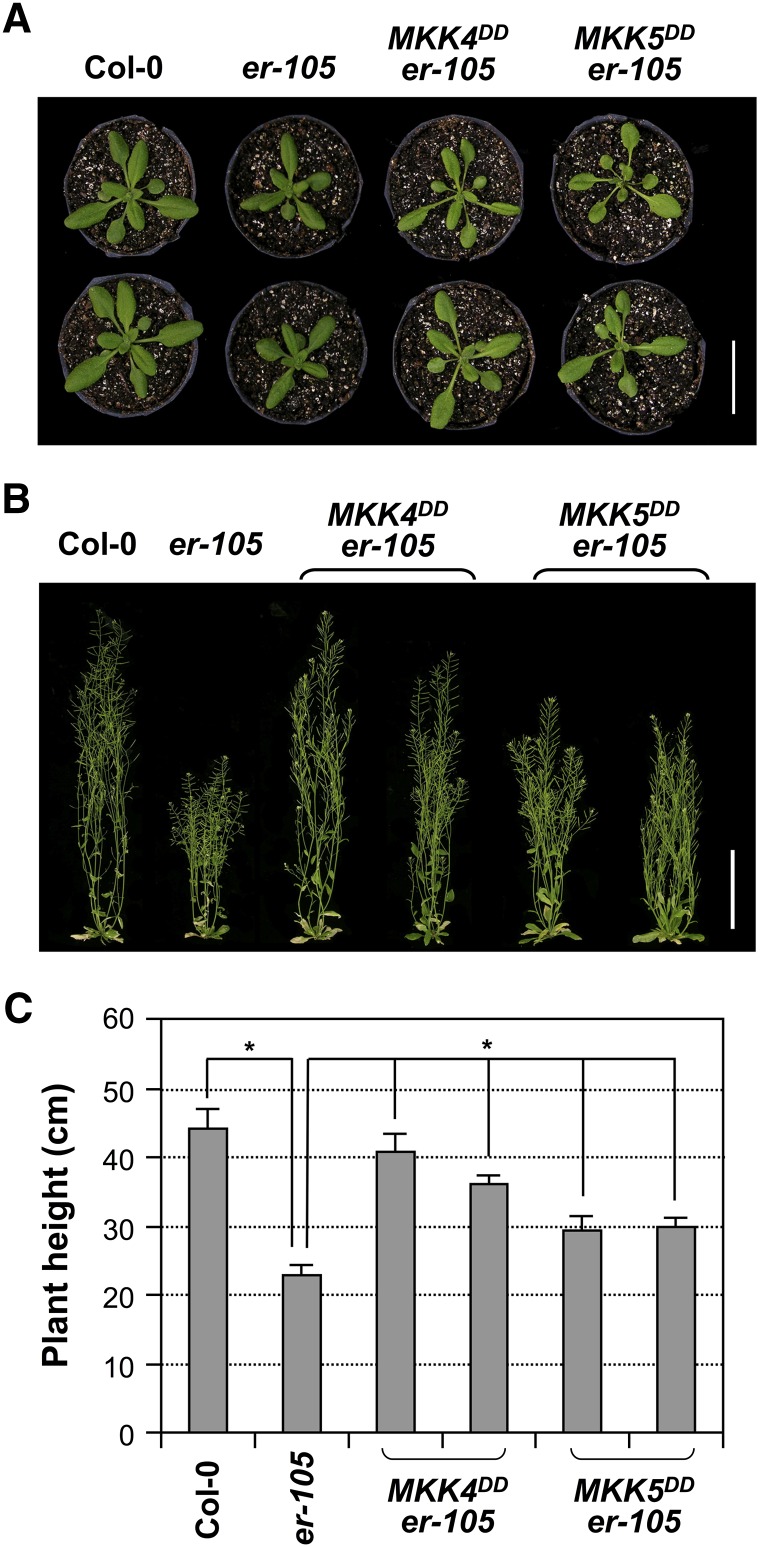
In addition to short pedicels and the resulting clustered inflorescence phenotype, the er mutant also has short leaf petioles, which causes clustered rosettes. Moreover, er displays a significant reduction in overall height (i.e., a dwarf phenotype). These organ growth defects of er were shown to be a result of reduced cell proliferation as well (Shpak et al., 2004; Woodward et al., 2005; Uchida et al., 2012). In characterizing MKK4DD and MKK5DD transgenic plants, we found that ER promoter–driven expression of MKK4DD or MKK5DD also rescued other growth defects of er-105, in addition to the shortened pedicels. As shown in Figure 9A, the short petiole phenotype of er-105 was reversed by both MKK4DD and MKK5DD. In addition, the dwarf phenotype of er-105 was almost completely rescued by MKK4DD, whereas MKK5DD partially rescued this mutant phenotype (Figures 9B and 9C). These results suggest that the YDA-MKK4/MKK5-MPK3/MPK6 cascade may function downstream of ER in regulating developmental processes in multiple organs through promoting cell proliferation.
Figure 9.
Rescue of the Short Petiole and Dwarf Phenotypes of er-105 by ER Promoter–Driven Expression of Constitutively Active MKK4 and MKK5.
(A) Four-week-old plants of Col-0, er-105, and two representative T2 lines of PER:MKK4DD and PER:MKK5DD transgenic plants in the er-105 background. Bar = 3 cm.
(B) Eight-week-old plants of the genotypes shown in (A). Bar = 10 cm.
(C) Plant heights of 8-week-old plants of the genotypes shown in (A). Bars represent average values + sd (n = 10 to 15 plants per genotype). Asterisks above the brackets indicate significant differences between the sample group(s) and er-105 (P < 0.0001, Student’s t test).
[See online article for color version of this figure.]
DISCUSSION
Cell differentiation and spatiotemporal-specific cell proliferation are critical to plant growth and development. ER is among one of the first few RLKs implicated in the regulation of plant growth and development (Torii et al., 1996; van Zanten et al., 2009). Loss of function of ER results in a clustered inflorescence and rosette, a result of defective localized cell proliferation (Shpak et al., 2003, 2004; Woodward et al., 2005). Analysis of high-order mutants of ER, ERL1, and ERL2 also revealed that they play a critical role in cell differentiation, such as stomatal differentiation (Shpak et al., 2005). Several transcription factors have been shown to be downstream of ER and ERLs in regulating stomatal formation and patterning (Pillitteri and Torii, 2012). However, the signaling process downstream of ER in regulating cell proliferation is largely unknown. This research reveals that a MAPK cascade functions downstream of ER in regulating coordinated cell proliferation, which plays a critical role in shaping organ size and structure during plant growth and development.
A Complete MAPK Cascade Downstream of ER in Regulating Inflorescence Architecture by Promoting Cell Proliferation
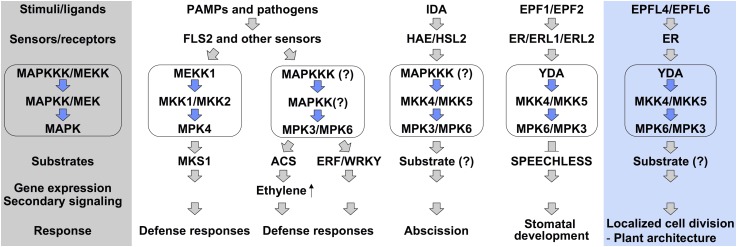
Loss-of-function mutations of MPK3/MPK6 or their upstream MKK4/MKK5 resulted in an er-like inflorescence phenotype (Figures 1 and 6), while expression of constitutively active MKK4, MKK5, or their tobacco ortholog, Nt-MEK2, led to the rescue of the clustered inflorescence phenotype of er mutants and the gain-of-function inflorescence phenotype in wild-type plants by promoting cell proliferation (Figures 3 to 5). These data suggest that the MKK4/MKK5-MPK3/MPK6 module functions downstream of ER in regulating inflorescence architecture. Moreover, gain- and loss-of-function genetic data support YDA as the upstream MAPKKK of MKK4/MKK5 in the ER signaling pathway (Figures 7 and 8). Together with the recently identified ER ligands EPFL4 and EPFL6 (Abrash et al., 2011; Uchida et al., 2012), we proposed a signaling pathway that controls inflorescence architecture in Arabidopsis from ligands, to receptors, to downstream effectors (Figure 10). After perceiving the peptide ligands, EPFL4 and EPFL6, which are specifically expressed in the stem endodermis, ER activates the downstream YDA-MKK4/MKK5-MPK3/MPK6 cascade, which transduces the signal(s) to components further downstream through the phosphorylation of unidentified MAPK substrates. These events eventually lead to spatiotemporal-specific cell proliferation within the pedicel cortex and thereby determine inflorescence architecture. In this MAPK cascade, MPK6 is likely to play a more important role than MPK3 downstream of ER because mutation of MPK6 in either wild-type or DD plants resulted in more severe effects on pedicel elongation and inflorescence architecture (Figures 1 and 3). In addition, the additive effect of er and mpk6 mutations in the er-105 mpk6 double mutant (Figure 2) suggests that MPK6 might function downstream of multiple ER family receptors in regulating inflorescence architecture.
Figure 10.
A Model Depicts the Functions of the YDA-MKK4/MKK5-MPK3/MPK6 Cascade Downstream of ER in Regulating Cell Proliferation and Plant Organ Development.
After sensing tissue/cell-specific signaling molecules (i.e., peptide ligands EPFL4 and EPFL6), ER receptors activate the YDA-MKK4/MKK5-MPK3/MPK6 cascade, either directly or through an additional mediator(s). Activated MPK3 and MPK6 phosphorylate their substrate(s), which eventually leads to tissue-specific cell proliferation and thereby regulates plant growth including pedicel, petiole, and inflorescence elongation. The shadowed area on the left highlights a typical MAPK-mediated signaling pathway downstream of receptors/sensors in eukaryotes. The placement of the YDA-MKK4/MKK5-MPK3/MPK6 cascade downstream of ER/ERLs in stomatal developmental regulation is solely based on correlative evidence. Activation of MPK3/MPK6 in response to pathogen-derived stimuli, such as the sensing of flg22 by FLS2, occurs in most cell/tissue types. By contrast, their activation after sensing of peptide ligands by HAE/HSL2 and ER/ERLs receptors is limited to specific cell types and during specific growth/developmental stages. The functions and signal specificity of MPK3/MPK6 are mostly dependent on the cell/tissue-specific activation of upstream receptors and spatiotemporal expression of downstream MAPK substrates.
[See online article for color version of this figure.]
MPK3/MPK6 Cascade Functions Downstream of ER Receptors in Regulating Multiple Biological Processes
MPK3/MPK6 and ER/ERL1/ERL2 have been implicated in regulating similar biological functions in Arabidopsis. Previously, mpk3+/− mpk6−/− and er erl1 erl2+/− plants were shown to have an ovule development defect due to reduced cell proliferation in the integuments, suggesting that the MPK3/MPK6 cascade might function downstream of ER family receptors in regulating ovule integument development (Pillitteri et al., 2007b; Wang et al., 2008). In this report, we demonstrate that the MPK3/MPK6 cascade functions downstream of ER in regulating inflorescence architecture by promoting cell proliferation in pedicels. We also found that gain-of-function MKK4DD and MKK5DD could rescue other organ growth defects of the er mutant, including short leaf petioles and reduced plant height, all of which are largely due to reduced cell proliferation (Shpak et al., 2004; Woodward et al., 2005; Uchida et al., 2012). Taken together, the MPK3/MPK6 cascade plays a pivotal role downstream of ER receptors in regulating localized cell proliferation during plant growth and development, which shapes the morphology of plant organs.
In addition to their roles in cell proliferation, ER, ERL1, and ERL2 RLKs also regulate cell differentiation by repressing asymmetric cell divisions during guard cell differentiation and anther development (Shpak et al., 2005; Hord et al., 2008). These functions are also shared with the MPK3/MPK6 cascade (Wang et al., 2007; Hord et al., 2008). Loss of function of YDA, MPK3/MPK6, or MKK4/MKK5 disrupts the coordinated cell fate specification of stomata versus pavement cells, resulting in the formation of clustered stomata, similar to that of er erl1 erl2 triple mutant plants (Bergmann et al., 2004; Shpak et al., 2005; Wang et al., 2007). Although there is no direct evidence linking the two components in the literature, several studies have suggested that the MPK3/MPK6 cascade may function downstream of ER/ERL1/ERL2 receptors in regulating stomatal development and patterning (Wang et al., 2007; Kim et al., 2012; Pillitteri and Torii, 2012).
Signaling Strength and Signaling Specificity of the ER Signaling Pathways
ER/ERL1/ERL2 and MPK3/MPK6 bear similarity in their gene dosage–dependent cellular function. Mutation of ERL1 and/or ERL2 genes further reduces the pedicel length of the er mutant, resulting in a much more clustered inflorescence (Shpak et al., 2004). Similarly, mpk3+/− mpk6−/− plants showed a more severely clustered inflorescence than the mpk6 single mutant. In addition, ERL2 is haplo-insufficient in maintaining normal cell proliferation of the integuments when ER and ERL1 are mutated (Pillitteri et al., 2007b). Similarly, MPK3 is haplo-insufficient in the mpk6 mutant background, resulting in a sterile phenotype almost identical to that of er erl1 erl2+/− plants (Wang et al., 2008). The observed gene dosage effect is likely a result of a reduction in signaling strength in the mutants. Direct measurement of the kinase activity of ER and ERLs is not possible at this stage. Because of the covalent modification of MAPKs and the fact that the active state can be preserved during protein extraction, in vivo MAPK activity can be determined. Previously, we demonstrated that wounding-induced MPK3/MPK6 activation is gene dosage dependent (Wang et al., 2008). Although mpk3+/− mpk6−/− plants are sterile and show clustered inflorescence and shortened pedicels, they have normal stomatal development. Only the mpk3 mpk6 double mutant has abnormal stomatal development and patterning (Wang et al., 2007). These findings suggest that signaling strength is very important and that different processes have different signal thresholds.
How Does MPK3/MPK6 Cascade Achieve Signaling Specificity in Different Biological Processes?
The multifunctional aspect of the MPK3/MPK6 cascade downstream of ER receptors opens up an interesting question of how the signaling specificity of MPK3/MPK6 is maintained in different growth and developmental pathways. Tissue- or cell-specific expression of input signaling molecules, especially the small peptide ligands, may play a critical role in maintaining the signaling specificity of different ER signaling pathways. Recently, several secretory peptides of the EPFL family were shown to be ligands of ER. Of these ER ligands, EPF1 and EPF2, which are expressed specifically within subsets of stomatal lineage cells, play critical roles in stomatal development (Hara et al., 2007; Hunt and Gray, 2009; Lee et al., 2012). EPFL4 and EPFL6, two redundant upstream components of ER-mediated inflorescence growth, showed the highest expression level in inflorescence stems (Abrash et al., 2011; Uchida et al., 2012). Signaling specificity can also be maintained by substrate specificity; the same MAPK can activate different substrates that are differentially expressed. For instance, MPK3/MPK6 was shown to regulate stomatal development through phosphorylation and inactivation of the basic helix-loop-helix transcription factor SPEECHLESS, which is expressed in stomatal lineage cells (Lampard et al., 2008). As a result, the MPK3/MPK6 cascade functions more like a molecular switch. Depending on the upstream and downstream components, it can affect different biological processes.
MAPK Cascades as Key Signaling Modules Downstream of Plant RLKs
RLKs, the largest kinase subfamily with more than 600 members in the model plant Arabidopsis, play diverse functions from growth/development to response to environmental stimuli in plants (Diévart and Clark, 2004; Morillo and Tax, 2006; De Smet et al., 2009). It is likely that they employ various signaling modules to transduce extracellular signals, generated upon perception of ligands, into intracellular responses. One of the best-studied examples is the brassinosteroid (BR)-BRI1 signaling pathway. BR binding to BRI1 triggers the formation of the BRI1-BAK1 receptor complex, which leads to BRI1 activation and phosphorylation of cytoplasmic BR signal kinases, ultimately culminating in activation of the transcription factors BZR1/2 and BR-responsive gene expression (Clouse, 2011). By contrast, signaling events/components downstream of other RLKs are largely unknown. It is interesting that MPK3 and MPK6, two closely related MAPKs, were recently shown to be downstream of several RLKs (Figure 10), from FLS2, a RLK involved in plant sensing of bacterial flagellin (Asai et al., 2002), to HEA/HSL2, two RLKs playing redundant function in Arabidopsis floral organ abscission (Cho et al., 2008), to CLV1/CLV2, two RLKs involved in plant stem cell population maintenance (Betsuyaku et al., 2011). In this report, we demonstrate that the ER RLK also employs MPK3/MPK6 to transduce the signal for localized cell proliferation.
In Arabidopsis, there are 18 additional MAPKs besides MPK3 and MPK6. The functions of many of them are unknown. It is likely that more MAPKs besides MPK3/MPK6 are also downstream of RLKs and are involved in converting other signals generated by ligand-activated RLK receptors into cellular responses. At this moment, how MAPK cascades carry out their functions downstream of RLKs is an actively researched area. FLS2-mediated MPK3/MPK6 activation mounts a global response that occurs in many, if not all, tissue/cell types. As a result, this process can be easily monitored biochemically using, for example, in-gel kinase activity assays and anti-phospho-specific antibodies. More importantly, several MPK3/MPK6 substrates or putative substrates, including ACS2/ACS6, ERF104, WRKY33, PHOS32, VIP1, and NIA2, have been identified (Liu and Zhang, 2004; Djamei et al., 2007; Merkouropoulos et al., 2008; Bethke et al., 2009; Wang et al., 2010; Mao et al., 2011). These substrates are involved in regulating biotic or abiotic stress responses in Arabidopsis. By contrast, the identification of MPK3/MPK6 substrates involved in regulating plant growth and development is much more difficult. The biochemical approach is not as effective since the activation of these two MAPKs occurs only in a few cells at a certain time. SPEECHLESS was identified in a genetic screen for mutants with stomatal developmental defects (MacAlister et al., 2007; Pillitteri et al., 2007a) and was later shown to be a substrate of MPK3/MPK6 (Lampard et al., 2008). Yet, it is challenging to demonstrate in vivo phosphorylation of SPEECHLESS by MPK3/MPK6 using biochemical assays because the expression of SPEECHLESS is limited to stomatal lineage cells at a specific stage of stomatal differentiation.
Using molecular genetic approaches, we demonstrated that the YDA-MKK4/MKK5-MPK3/MPK6 cascade is a key downstream signaling component in ER-mediated cell proliferation in this study. Identifying components further downstream of this MAPK cascade especially the MPK3/MPK6 substrate(s) in the pathway will ultimately reveal the molecular mechanisms underlying the regulation of localized cell proliferation during plant growth/development by this ER-MAPK signaling pathway.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Col-0 ecotype was used as the wild type. Mutant alleles of mpk3-1 (SALK_151594), mpk6-2 (Salk_073907), and er-105 were reported previously (Torii et al., 1996; Liu and Zhang, 2004). The T-DNA insertional mutant of YDA was obtained from the ABRC (SALK_105078) (Alonso et al., 2003). The transgenic lines of DD, mkk4/mkk5 RNAi, PMPK3:GUS, PMPK6:GUS, and PER:GUS were generated previously (Ren et al., 2002; Shpak et al., 2004; Wang et al., 2007, 2008). Generation of mpk3+/− mpk6−/−, MPK6KR mpk3, DD mpk3, DD mpk6, and DD yda double mutant plants was described previously (Wang et al., 2007, 2008; Cho et al., 2008; Ren et al., 2008). Homozygous DD er-105 and er-105 mpk6 F3 plants were generated by genetic crosses. For these crosses, er and mpk6 mutations were followed by PCR genotyping as described previously (Liu and Zhang, 2004; Shpak et al., 2004), and the DD transgene was followed by hygromycin resistance (Ren et al., 2002).
Seeds were plated on half-strength Murashige and Skoog medium with 0.7% phytagar and appropriate antibiotics for selection after surface sterilization and imbibing at 4°C for 3 to 5 d. Plates were incubated in a tissue culture chamber at 22°C under continuous light (70 µE/m−2 s−1) for 7 d. Seedlings were then transplanted into soil and grow in a greenhouse with a 16-h-light/ 8-h-dark cycle.
Transgenic Plant Generation
To make ER promoter–driven constitutively active MAPKK constructs, we amplified the ER promoter region using primer pair forward, 5′-GCAGAAGAGAAACCCAAGAAGA-3′, and reverse, 5′-ATTCAAAAATCCCTCTCCAACA-3′. After the addition of EcoRI and SacI sites by PCR, the product was cloned into pCAMBIA3300 digested with the same enzymes, resulting in the pCambia3300-PER intermediate construct. MKK1, MKK2, MKK4, MKK5, MKK7, and MKK9 containing constitutively active Ser-to-Asp mutations were PCR amplified from pTA7002-MKK1DD, -MKK2DD, -MKK4DD, -MKK5DD, -MKK7DD, and -MKK9DD constructs using primers that flank the omega enhancer-Flag-MKKDD-terminator cassette (forward, 5′-CTCGAGGTATTTTTACAACAATTACCAACAAC-3′, and reverse, 5′-CAGTCACGACGTTGTAAAACGAC-3′). The pTA7002-MKKDD constructs were prepared as previously described for pTA7002-NtMEK2DD (Yang et al., 2001). The PCR products were then inserted into pCAMBIA3300-PER digested with SmaI enzyme. Constructs with correct orientation were selected by PCR and then sequenced to ensure that no error was introduced by PCR.
The construct expressing constitutively active ΔN-YDA under its native promoter was kindly provided by Wolfgang Lukowitz (Lukowitz et al., 2004). Both the MAPKKDD and ΔN-YDA expression constructs were electroporated into Agrobacterium tumefaciens strain GV3101, which were used to transform wild-type and er-105 plants by the floral dipping method (Clough and Bent, 1998). Basta-resistant T1 transgenic plants in the er-105 background were screened for complementation of er mutant phenotypes. T1 plants in the wild-type background were examined for gain-of-function phenotypes (i.e., increased pedicel length). The typical lines in er-105 and wild-type backgrounds were characterized in more detail in T2 generation.
GUS Staining
Inflorescences were prefixed in prechilled 90% acetone for 20 min and washed with distilled water. After brief vacuum infiltration, the inflorescences were incubated in GUS staining buffer (10 mM EDTA, 0.1% Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 100 µg mL−1 chloramphenicol, and 1 mg mL−1 X-Gluc in 50 mM sodium phosphate buffer, pH 7.0) overnight at 37°C. After being cleared in 20% lactic acid/20% glycerol solution, the inflorescences were pictured using a digital camera.
Histological Analysis, Cell Length, and Number Measurement
Pedicel tissue samples were fixed overnight in formalin-acetic acid-alcohol solution at 4°C, dehydrated with a graded series of ethanol, and infiltrated with Technovit 7100 resin (Heraeus Kulzer) followed by embedding and polymerization. Two-micrometer sections were prepared using a Leica Reichart Ultracut S microtome (Wetzlar). The tissue sections were stained with 0.1% toluidine blue and observed using light microscopy. Three regions of each sectioned pedicel were taken. The number of cells in a middle longitudinal cortex row was determined. This number was used to calculate the total number and average length of cells in the cortex row of each pedicel. The number of cortex cells was counted in 15 to 20 sectioned pedicels for each genotype, and the average cell number and length was determined.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ER (At2g26330), MPK3 (At3g45640), MPK6 (At2g43790), MKK1 (At4g26070), MKK2 (At4g29810), MKK4 (At1g51660), MKK5 (At3g21220), MKK7 (At1g18350), MKK9 (At1g73500), and YDA (At1g62700).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Growth and Developmental Defects of PER:MKK9DD Transgenic Plants.
Supplemental Figure 2. Cell Death Phenotype Associated with the Basal-Level Expression of MKK7DD Transgene in the Steroid-Inducible pTA7002-MKK7DD Transgenic Plants.
Acknowledgments
We thank Joe Abbott for his involvement in generating the intermediate vector with ER promoter. This research was supported by a National Science Foundation grant (MCB-0950519) to S.Z. K.U.T. is a Howard Hughes Medical Institute–Gordon and Betty Moore Foundation investigator and is endowed by the University of Washington, College of Arts and Sciences.
AUTHOR CONTRIBUTIONS
X.M., H.W., and S.Z. designed research. X.M., H.W., Y.H., and Y.L. performed research. K.U.T. and J.C.W. contributed new tools. X.M., H.W., K.U.T., J.C.W., and S.Z. analyzed data. X.M. and S.Z. wrote the article.
Glossary
- RLK
receptor-like protein kinase
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- MAPKKK
MAPKK kinase
- Col-0
Columbia-0
- GUS
β-glucuronidase
- RNAi
RNA interference
- BR
brassinosteroid
References
- Abrash E.B., Davies K.A., Bergmann D.C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andreasson E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Becraft P.W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18: 163–192 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S., Takahashi F., Kinoshita A., Miwa H., Shinozaki K., Fukuda H., Sawa S. (2011). Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52: 14–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Karin M. (2001). Mammalian MAP kinase signalling cascades. Nature 410: 37–40 [DOI] [PubMed] [Google Scholar]
- Cho S.K., Larue C.T., Chevalier D., Wang H., Jinn T.L., Zhang S., Walker J.C. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2011). Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. (2000). The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 25: 596–601 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Diévart A., Clark S.E. (2004). LRR-containing receptors regulating plant development and defense. Development 131: 251–261 [DOI] [PubMed] [Google Scholar]
- Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. (2007). Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318: 453–456 [DOI] [PubMed] [Google Scholar]
- Hamel L.P., et al. (2006). Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Han L., Li G.J., Yang K.Y., Mao G., Wang R., Liu Y., Zhang S. (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord C.L., Sun Y.J., Pillitteri L.J., Torii K.U., Wang H., Zhang S., Ma H. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Ichimura K., et al. ; MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Lukowitz W., Ellis B.E., Bergmann D.C. (2009). Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Manning G., Plowman G.D., Hunter T., Sudarsanam S. (2002). Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27: 514–520 [DOI] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (2006). Peptide hormones in plants. Annu. Rev. Plant Biol. 57: 649–674 [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G., Andreasson E., Hess D., Boller T., Peck S.C. (2008). An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J. Biol. Chem. 283: 10493–10499 [DOI] [PubMed] [Google Scholar]
- Mészáros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardóczy V., Teige M., Koncz C., Peck S., Bögre L. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Morillo S.A., Tax F.E. (2006). Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 9: 460–469 [DOI] [PubMed] [Google Scholar]
- Morris E.R., Walker J.C. (2003). Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 6: 339–342 [DOI] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Bemis S.M., Shpak E.D., Torii K.U. (2007b). Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134: 3099–3109 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007a). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63: 591–614 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Ren D., Yang K.Y., Li G.J., Liu Y., Zhang S. (2006). Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 141: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Schwartz M.A., Madhani H.D. (2004). Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38: 725–748 [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., Berthiaume C.T., Hill E.J., Torii K.U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak E.D., Lakeman M.B., Torii K.U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., Whittier R.F., Komeda Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Lee J.S., Horst R.J., Lai H.H., Kajita R., Kakimoto T., Tasaka M., Torii K.U. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109: 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Snoek L.B., Proveniers M.C., Peeters A.J. (2009). The many functions of ERECTA. Trends Plant Sci. 14: 214–218 [DOI] [PubMed] [Google Scholar]
- Vert G., Nemhauser J.L., Geldner N., Hong F., Chory J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu Y., Bruffett K., Lee J., Hause G., Walker J.C., Zhang S. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Li Y., Ren D., Song C.P. (2010). Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C., Gibson S., Jarpe M.B., Johnson G.L. (1999). Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 79: 143–180 [DOI] [PubMed] [Google Scholar]
- Woodward C., Bemis S.M., Hill E.J., Sawa S., Koshiba T., Torii K.U. (2005). Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 139: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54: 440–451 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Yang K.Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wu D., Chory J. (2002). Plant receptor kinases: Systemin receptor identified. Proc. Natl. Acad. Sci. USA 99: 9090–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. (2008). Mitogen-activated protein kinase cascades in plant intracellular signaling. In Annual Plant Reviews, Vol. 33: Intracellular Signaling in Plants, Z. Yang, ed (Oxford, UK: Wiley-Blackwell), pp. 100–136.
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1998). Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 95: 7433–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]