The length and position of the spindle apparatus change significantly during the meiosis-to-mitosis transition. This study reports on the role of the germline-specific E3 ligase MATH-BTB adaptor protein MAB1 that is required for nuclei separation during meiosis II and asymmetric nuclei positioning during subsequent mega- and microspore division, respectively.
Abstract
Germline and early embryo development constitute ideal model systems to study the establishment of polarity, cell identity, and asymmetric cell divisions (ACDs) in plants. We describe here the function of the MATH-BTB domain protein MAB1 that is exclusively expressed in the germ lineages and the zygote of maize (Zea mays). mab1 (RNA interference [RNAi]) mutant plants display chromosome segregation defects and short spindles during meiosis that cause insufficient separation and migration of nuclei. After the meiosis-to-mitosis transition, two attached nuclei of similar identity are formed in mab1 (RNAi) mutants leading to an arrest of further germline development. Transient expression studies of MAB1 in tobacco (Nicotiana tabacum) Bright Yellow-2 cells revealed a cell cycle–dependent nuclear localization pattern but no direct colocalization with the spindle apparatus. MAB1 is able to form homodimers and interacts with the E3 ubiquitin ligase component Cullin 3a (CUL3a) in the cytoplasm, likely as a substrate-specific adapter protein. The microtubule-severing subunit p60 of katanin was identified as a candidate substrate for MAB1, suggesting that MAB1 resembles the animal key ACD regulator Maternal Effect Lethal 26 (MEL-26). In summary, our findings provide further evidence for the importance of posttranslational regulation for asymmetric divisions and germline progression in plants and identified an unstable key protein that seems to be involved in regulating the stability of a spindle apparatus regulator(s).
INTRODUCTION
During the development of a multicellular organism, cell polarity and asymmetric cell division (ACD) play decisive roles in cell specialization, differentiation, and fate determination and rely on the asymmetric organization of cellular components and structures. The establishment and maintenance of cell polarity and ACD are especially important in the reproductive stage of the flowering plant life cycle in both male and female germline development as well as during patterning in early embryogenesis (Scheres and Benfey, 1999; Ranganath, 2005; Paciorek and Bergmann, 2010). After completion of meiosis in male spores, ACD leads to the generation of a small germ cell and a large vegetative cell that will form the tube cell during pollen germination (Twell, 2011). The apparent polarity of the female germline is first established during primordial germ cell (megaspore mother cell) differentiation along its micropylar-chalazal axis. After meiosis, the three micropylar-most spores generally degenerate and the chalazal-most spore continues to develop and undergoes three stereotypic cycles of asymmetric free-nuclear mitotic divisions. A highly polarized eight-nucleate coenocyte is formed containing two female gametes (egg and central cell) after cellularization (Yang et al., 2010; Sprunck and Gross-Hardt, 2011). ACD is also the major characteristic of zygote division leading to daughter cells of different fates: The small cytoplasmic apical cell develops into the embryo proper and the large vacuolated basal cell forms the suspensor (Ueda et al., 2011; Zhang and Laux, 2011).
The molecular mechanisms that underlie cellular asymmetries and cell specification during germline and zygote development in plants are still poorly understood. The polar positioning of nuclei and of associated cell fate determinants appears to be achieved by the precise regulation of meiotic and mitotic divisions, alignment and elongation of mitotic spindles, vacuolization, and nuclear migration (Huang and Sheridan, 1994; Christensen et al., 1997; Sprunck and Gross-Hardt, 2011). In animals and yeast, cytoskeletal elements (F-actin and microtubules) provide the structural basis for cell polarization (Li and Gundersen, 2008). Radial arrays of microtubules (MTs) involved in nuclei positioning emanate from a discrete microtubular organizing center (MTOC) called the centrosome in animal cells and the spindle pole body in fungi. The nucleation of cytoplasmic and spindle MTs occurs from proteinaceous pericentriolar material, where γ-tubulin ring complexes act as nucleation cores (Lüders and Stearns, 2007). Proper spindle positioning and orientation is essential for ACD as it determines not only the asymmetry of cell division but also the relative location of daughter nuclei and the distribution and inheritance of future cell fate determinants (McCarthy and Goldstein, 2006; Fabritius et al., 2011). It has remained unclear how asymmetric spindle positioning and nuclear migration are controlled during ACD in plants. One of the major reasons is linked to the lack of discrete MTOCs in higher plants (Wasteneys, 2002; Ehrhardt, 2007), although a number of proteins with homology to animal MTOC components have meanwhile been identified (reviewed in Lloyd and Chan, 2004; Ehrhardt and Shaw, 2006; Zhang and Dawe, 2011). Nevertheless, plants possess highly organized MT arrays essential for cell division and expansion: cortical interphase MTs, the preprophase band (PPB), which labels the future cell division plane, mitotic and meiotic MT spindles required for chromosome segregation, and the phragmoplast, which appears during cytokinesis as a result of the reorganization of MTs. In interphase, MTs at the cell cortex are arranged parallel to one another and transverse to the direction of cell expansion. Several cell types, such as meiotic cells, lack a PPB but nonetheless are capable of forming highly oriented cell plates (Baskin and Cande, 1990; Canaday et al., 2000; Ambrose and Cyr, 2008).
Thus, the major question arises how these structural elements associated with ACD are regulated in space and time. One of the mechanisms would be through selective degradation of these components. In animals, there is increasing evidence that the temporally and spatially controlled targeting of key regulators of the cytoskeleton to the ubiquitin/26S proteasome pathway is not only required for cell cycle transition and progression but also for cell fate determination after ACD (McCarthy Campbell et al., 2009). The oocyte-to-embryo transition in nematodes represents a particular case, as it requires rapid degradation of meiotic ACD proteins to allow the switch to mitotic divisions. The first mitotic spindle differs from its meiotic counterpart in size, morphology, and intracellular location and likely requires the labeling of meiosis-specific proteins for degradation to prevent their interference with subsequent mitotic divisions (Lu and Mains, 2007). Recent studies in mammalian cells and in Caenorhabditis elegans have shown that multisubunit Cullin 3 (CUL3)–based E3 ligases are involved in regulating mitotic progression, cytokinesis, and the proper regulation of MT dynamics and spindle assembly during the meiosis-to-mitosis transition (Bowerman and Kurz, 2006; Sawin and Tran, 2006; Sumara et al., 2008). BTB (for Bric-à-Brac/Tramtrack/Broad complex) domain proteins appear to function in this complex as substrate-specific adaptors. BTB domains were reported to interact with CUL3, while secondary domains are thought to be responsible for substrate specificity (Geyer et al., 2003; Figueroa et al., 2005; Gingerich et al., 2005; Sumara et al., 2007). The female germline-specific CUL3 substrate adaptor Maternal Effect Lethal 26 (MEL-26) of C. elegans is responsible for the spatial and temporal targeting of MEI-1 (for Defective in Meiosis1), which together with another AAA-ATPase MEI-2 is a part of the MT severing katanin complex. MEI-1 was reported to be degraded at the meiosis-to-mitosis transition to allow the formation of long MTs for proper anchoring of the spindle apparatus and chromosome segregation (Pintard et al., 2003; Xu et al., 2003). Additionally, MEL-26 targets the MT-interacting protein FIGL-1 (for fidgetin-like 1 AAA-ATPase) for degradation in mitosis (Luke-Glaser et al., 2007). MEL-26 consists of a MATH (for Meprin-Associated Traf Homology) and a BTB domain on a single polypeptide chain, and this protein family is present in all multicellular eukaryotes (Stogios et al., 2005). The function and possible substrates of MATH-BTB proteins in plants still need to be elucidated. A small family of six ubiquitously expressed MATH-BTB encoding genes is present in the genome of Arabidopsis thaliana (Dieterle et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005; Thomann et al., 2005; Weber et al., 2005), while this gene family is largely expanded in grasses. For example, 68 members are found in the genome of rice (Oryza sativa; Gingerich et al., 2007). However, so far there are only two reports showing that Arabidopsis MATH-BTB proteins target the ABA transcriptional response regulator ATHB6 for degradation (Lechner et al., 2011) and interact with members of the ERF/AP2 transcription factor family (Weber and Hellmann, 2009).
Here, we characterize the maize (Zea mays) MATH-BTB protein MATH-BTB1 (MAB1). The MAB1-encoding gene is specifically expressed during male and female germline development as well as during ACD in the maize zygote. We generated mab1 (RNA interference [RNAi]) mutant plants and report the functional role of MAB1 in organizing microtubular spindles as well as nuclei positioning and identity during meiosis II and the first mitotic division in both plant germlines. We further analyzed its subcellular localization during the cell cycle in tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cells. Finally, we show that MAB1 is capable of forming homodimers and interacts with CUL3a, indicating that it is a component of a dimeric/oligomeric CUL3-based E3 ligase complex and may interact with AAA ATPase substrates, such as p60 of the MT-severing katanin complex.
RESULTS
Expansion of the MATH-BTB Protein Family in Grasses
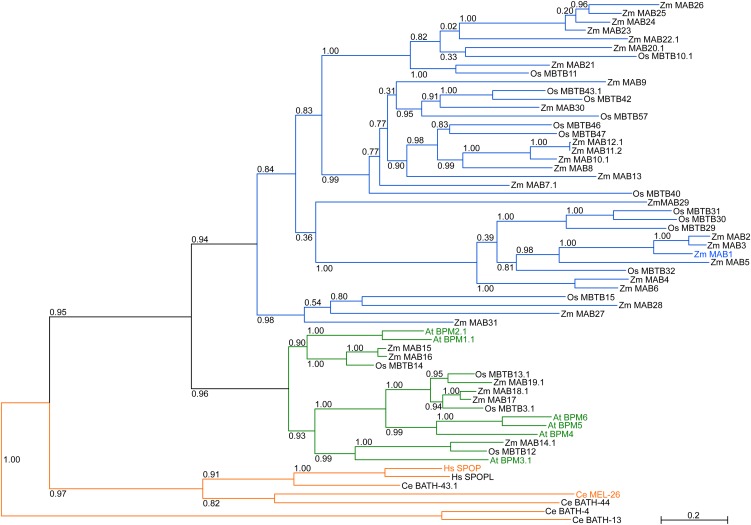
The human genome encodes only two strongly related MATH-BTB proteins, whose number is slightly increased to six in Arabidopsis but largely expanded in grasses, with 68 family members in the genome of rice (Gingerich et al., 2007). It is unknown whether the expanded grass group contains genes with a grass-specific function or whether the few genes in human and Arabidopsis are stronger regulated and modified at the posttranscriptional and posttranslational level and thus are capable of recognizing a spectrum of similar target proteins. Using maize as a grass model, we scanned the sequenced B73 genome for genes encoding MATH-BTB domain–containing proteins. Thirty-one genes were detected and named Zm MAB1-31 (for Z. mays MATH-BTB domain protein 1-31; see Supplemental Table 1 online). Phylogenetic analysis of all identified maize, Arabidopsis, and human MATH-BTB proteins as well as selected proteins from rice and C. elegans showed that animal MATH-BTB proteins form their own clade, while plant MATH-BTB genes separate into two major clades (Figure 1; see Supplemental Data Set 1 online). The core clade contains all six Arabidopsis proteins in addition to six homologous proteins from maize (Zm MAB14-19; see Supplemental Figure 1 online). The expanded group contains 25 MATH-BTB proteins from maize (Zm MAB1-13 and Zm MAB20-31) separated into four subclades and one unusual MATH-BTB protein (Zm MAB29) containing two MATH and two BTB domains.
Figure 1.
The Phylogenetic Tree of Maize, Rice, Arabidopsis, Human, and C. elegans MATH-BTB Homologs.
Entire protein sequences of 31 identified maize, six Arabidopsis, 17 selected rice, two human, and five selected nematode MATH-BTB genes were analyzed. Plant MATH-BTB proteins separate into two major groups classified by Gingerich et al. (2007) as core and expanded groups. The animal group forms its own clade. Individual members of the plant core group are represented by green branches; Arabidopsis MATH-BTB proteins are highlighted in green. The expanded group containing only grass proteins is indicated in blue and the most studied members are highlighted in orange. Zm MAB1 is highlighted in blue. The numbers on each node are the Shimodaira-Hasegawa–like test indices of statistical support provided by PhyML. Bar = 0.2 is a branch length that represents nucleotide substitutions per site. For sequence identifiers, see Supplemental Table 1 online.
To study the role of MATH-BTB proteins from the grass-specific expanded clade and to investigate the importance of the ubiquitin/26S proteasome pathway for ACD in plants, we selected MAB1 as a candidate as it is expressed in maize egg cells and was found to be strongly upregulated in zygotes before ACD (see below).
RNAi Silencing of Zm MAB1 Impairs the Positioning and Identity of Nuclei after the Meiosis-to-Mitosis Transition in the Female Germline
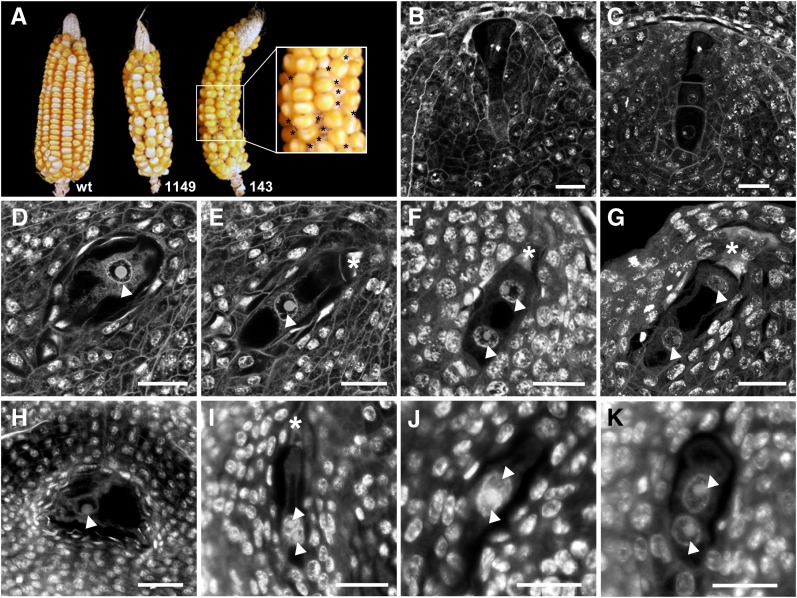
To analyze the function of MAB1, we applied RNAi to downregulate MAB1 expression in maize, as knockout lines are not available. The maize Ubi1 promoter was used in the RNAi constructs as it has been shown previously to be capable of driving marker gene expression throughout female germline development (Srilunchang et al., 2010). Thirteen independent transgenic lines were generated and the integration pattern and transgene expression were analyzed. Based on the strength of RNAi transgene expression, we selected two independent mab1 (RNAi) mutant lines (nos. 1149 and 143) for detailed functional analysis. The silencing effect was studied by real-time RT-PCR in a number of mab1 (RNAi) progeny plants of both lines. These serve as examples, as whole plants were destroyed during the procedure to isolate reproductive tissues. As shown in Supplemental Figure 2 online, downregulation of MAB1 in tissues showing the highest expression in wild-type plants (see below) varied among RNAi plants. By contrast, plants showing weak RNAi expression did not show a significant silencing effect (plants 1149_1 and 1149_8), especially progeny plants of line 143, which showed silencing of 9.5 to 33% in male and female spores, respectively, compared with messenger levels in wild-type plants. Cobs of self-pollinated T0 plants and their following T1 generations of both lines showed a significantly reduced seed set compared with self-pollinated wild-type cobs (Figure 2A). Closer examination (asterisks in Figure 2A) revealed that development was arrested in ∼40% of the ovules. To investigate the effect of MAB1 RNAi silencing in more detail, we analyzed fixed and sectioned ovules from both transgenic offspring and wild-type plants by confocal laser scanning microscopy. According to Huang and Sheridan (1994), the silk length of immature maize cobs was used as an external morphological feature to determine the corresponding stages of developing ovules and female germline cells. To compare mutant phenotypes with wild-type ovule development, we first analyzed the genotype A188 that was also used to generate transgenic plants. As shown in Figure 2B, a single hypodermal cell of the L2 layer of an ovule primordium enlarges into the primordial germ or archesporial cell that differentiates further into a highly polar meiocyte called the megaspore mother cell (Figure 2B). During meiosis, it forms a linear tetrad of haploid megaspores (Figure 2C). While the three micropylar-most megaspores degenerate, the chalazal-most megaspore develops into the functional megaspore, progressing the germline toward two female gametes. Being initially surrounded by a number of small vacuoles (Figure 2D), the megaspore nucleus becomes localized between a smaller vacuole at the chalazal pole and a large vacuole at the micropylar pole (Figure 2E). The first mitotic division separates one daughter nucleus toward the micropylar pole of the developing female gametophyte, while the second nucleus remains approximately at the position of the mother nucleus separated from the chalazal pole by a small vacuole (Figures 2F and G). By contrast, mab1 (RNAi) mutant ovules were arrested either at the one (Figure 2H) or two nucleate stage, containing both nuclei attached to each other (Figures 2I and 2J) or two nuclei that are not properly separated from each other (Figure 2K). A total of 36% (line 143; n = 20) and 41% (line 1149; n = 33) of the analyzed female spores showed either the phenotypes described or were collapsed (Table 1). One (chalazal) nucleus was observed in ∼4% (line 143) and 10% (line 1149) of the ovules, while two attached or adjacent nuclei were found in 11% (line 143) and 25% (line 1149) of the ovules, respectively. In contrast with the wild type, where 96% of all analyzed female gametophytes were fully developed, a ratio of ∼48% affected embryo sacs would be expected in the case of a full knockout in heterozygous offspring of mab1 (RNAi) lines. Although RNAi approaches are rarely 100% effective in plants (Smith et al., 2000), the high number of arrested and collapsed female spores is close to the expected number of a full knockout. Due to these early arrested phenotypes, we could not elucidate the role of MAB1 during zygotic ACD. However, the observed phenotypes indicate that MAB1 function could also be required for the proper assembly and positioning of the first mitotic spindle apparatus in the zygote, in analogy to C. elegans MEL-26 function during the meiosis-to-mitosis transition.
Figure 2.
Polar Nuclei Separation and Migration during the First Mitotic Division of Female Germline Cells Is Arrested in mab1 (RNAi) Mutants of Maize.
(A) Compared with the wild type (wt), two independent mab1 (RNAi) maize lines (no.1149 and no.143) display ∼40% undeveloped ovules (asterisks, inset).
(B) to (G) Confocal laser scanning microscopy sections of developing wild-type ovules.
(B) Primordial germ cell or female meiocyte at metaphase I during meiosis I.
(C) After completion of meiosis, the micropylar-most megaspores of the linear tetrad degenerate.
(D) The functional megaspore becomes vacuolated.
(E) At stage FG1, the nucleus of the functional megaspore is localized polar toward the chalazal pole.
(F) At early stage FG2, both nuclei become separated.
(G) At late stage FG2, one nucleus moves toward the micropylar pole.
(H) to (K) Germline phenotypes found in mab1 (RNAi) maize lines.
(H) A functional megaspore containing large vacuoles is arrested at stage FG1.
(I) and (J) Two examples showing two unseparated nuclei at the chalazal pole at stage FG2.
(K) Example showing two nuclei separated but in close contact to each other at the chalazal pole.
Arrowheads point toward nucleus of the functional megaspore or toward its daughter nuclei. Asterisks mark degenerated megaspores. Bars = 20 μm.
Table 1. Female Gametophyte Phenotypes of Heterozygous mab1 (RNAi) Silencing Lines.
| Genotype | na | One (Chalazal) Nucleus | Two Attached Nuclei | Two Adjacent Nuclei | Σ Mutant FG1-FG2 Phenotypes | Collapsed FGs | Fully Developed FGs (FG7) |
|---|---|---|---|---|---|---|---|
| RNAi | 80 (100%) | 8 (10%) | 6 (7.5%) | 14 (17.5%) | 35% | 5 (6.3%) | 47 (58.7%) |
| (line 1149) | |||||||
| RNAi | 55 (100%) | 2 (3.6%) | 2 (3.6%) | 4 (7.2%) | 14.4% | 12 (21.8%) | 35 (63.8%) |
| (line 143) | |||||||
| Wild-type | 63 (100%) | 0 | 0 | 0 | 0% | 3 (4.8%) | 60 (95.2%) |
| (A188xH99) |
Ovules were manually sectioned and examined by confocal laser scanning microscopy after period acid-Schiff staining. Two independent mab1 (RNAi) and a wild-type line of the same genetic background were compared. Note that ovules were randomly selected and studied at a silk length of 3 to 8 mm when wild-type ovaries contained fully differentiated mature female gametophytes. FG, female gametophyte.
Total no. of dissected and scanned ovules; only ovule sections generating a complete three-dimensional female gametophyte stack were counted.
Germline Progression and Identity Are Affected in mab1 (RNAi) Mutant Pollen, Generating Two Vegetative-Like Nuclei
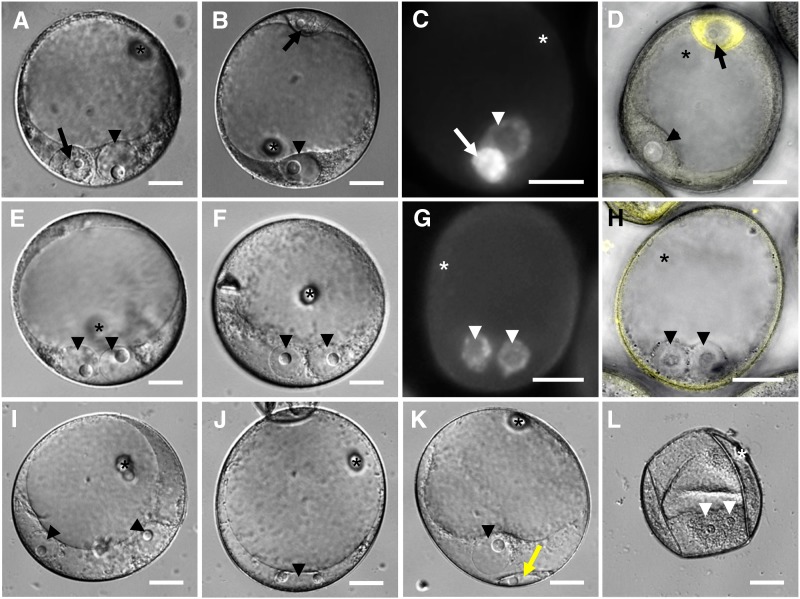
To investigate whether MAB1 activity is also required during the meiosis-to-mitosis transition during male germline development, we first studied ACD occurring during pollen mitosis I (PMI) in wild-type pollen of maize. As shown in Figure 3A, PMI generates a small germ cell containing a condensed nucleus with a small nucleolus adjacent to the large nucleus of the vegetative cell. Immediately after ACD, the germ cell migrates to the pole opposite of the vegetative nucleus (Figure 3B). The 4′,6-diamidino-2-phenylindole (DAPI) staining shows that the DNA of the germ cell nucleus is more strongly condensed compared with the vegetative nucleus (Figure 3C). The yellow fluorescent protein (YFP)-α-tubulin fusion protein, expressed under the α-tubulin promoter (Kliwer and Dresselhaus, 2010), is expressed in the generative cell and can be used as a germ cell marker (Figure 3D). Eight to thirteen percent of the mab1 (RNAi) mutant pollen of heterozygous plants (see Supplemental Table 2 online) lacked ACD, generating in most cases (4%) two vegetative-like nuclei of identical size containing large nucleoli (Figures 3E and 3F). The level of DNA condensation is similar in both nuclei (Figure 3G) and different from that of germ cells (Figure 3C). Germ cell cytoplasm is not generated around these nuclei, indicated by the lack of the marker protein YFP-α-tubulin (Figure 3H). Less frequently (0.5%), we observed pollen containing two vegetative-like nuclei separated from each other (Figure 3I) or even rarely (0.3%) pollen with one large nucleus containing two nucleoli (Figure 3J). The presence of two nucleoli indicates that DNA replication and mitosis was completed, as maize contains only one nucleolus organizer region on chromosome 6 (Givens and Phillips, 1976). We assume that mitotic spindles were too short and lacking the asymmetry required for separating the daughter chromosomes to a large vegetative and a small generative cell, respectively (Twell, 2011). The second most abundant mutant phenotype observed in mab1 (RNAi) mutant pollen was a defect in cytokinesis (4.6%). Although ellipsoid germ-like cells seemed to form, cellularization is not completed and these cells never migrated to the opposite pole (Figure 3K). These phenotypes have never been observed in wild-type pollen (see Supplemental Table 2 online). Compared with ∼12% wild-type pollen, a large portion of mab1 (RNAi) pollen was collapsed and degenerated during the progression of anther development in some plants (50 to 90%; Figure 3L) indicating that the majority of mab1 (RNAi) mutant pollen degenerate during further development, which may also affect the development of wild-type pollen in heterozygous anthers.
Figure 3.
The Male Germline of Maize Is Arrested in mab1 (RNAi) Mutants after the First Mitotic Division.
(A) A male germ cell containing a small and condensed nucleus has differentiated after asymmetric division at the bicellular stage in wild-type pollen. Note that its nucleus (arrow) is much smaller than the vegetative nucleus (arrowhead).
(B) Immediately after formation, the germ cell migrates toward the pole opposite to the vegetative nucleus in wild-type pollen.
(C) DAPI staining of wild-type pollen displays highly condensed chromatin of the germ cell.
(D) The cytoplasm of wild-type germ cells is labeled by the male germline marker YFP-α-tubulin.
(E) and (F) Two examples of the most frequent mab1 (RNAi) mutant phenotypes each showing two attached nuclei of comparable size of vegetative nuclei but lacking cellularization.
(G) DAPI staining of mutant pollen shows a similar chromatin condensation level of both nuclei.
(H) The germ cell marker YFP-α-tubulin is not expressed in the cytoplasm surrounding the mutant vegetative-like nuclei.
(I) Another mutant group showed two separated nuclei of identical size lacking cellularization and marker gene expression.
(J) In rare cases, mutant pollen showed one large nucleus containing two nucleoli.
(K) The second most abundant mutant phenotype showed a cellularization defect. Note the ellipsoid germ cell–like cell (yellow arrow).
(L) Mutant pollen grains collapsed and degenerated frequently.
Asterisks mark the germination pore, arrowheads point toward vegetative (-like) nuclei, and arrows point toward nuclei of germ cells. See Supplemental Table 2 online for statistics. Bars = 20 µm.
MAB1 Is Expressed during Male and Female Meiosis and Meiosis-to-Mitosis Transition and Is Strongly Upregulated in Zygotes
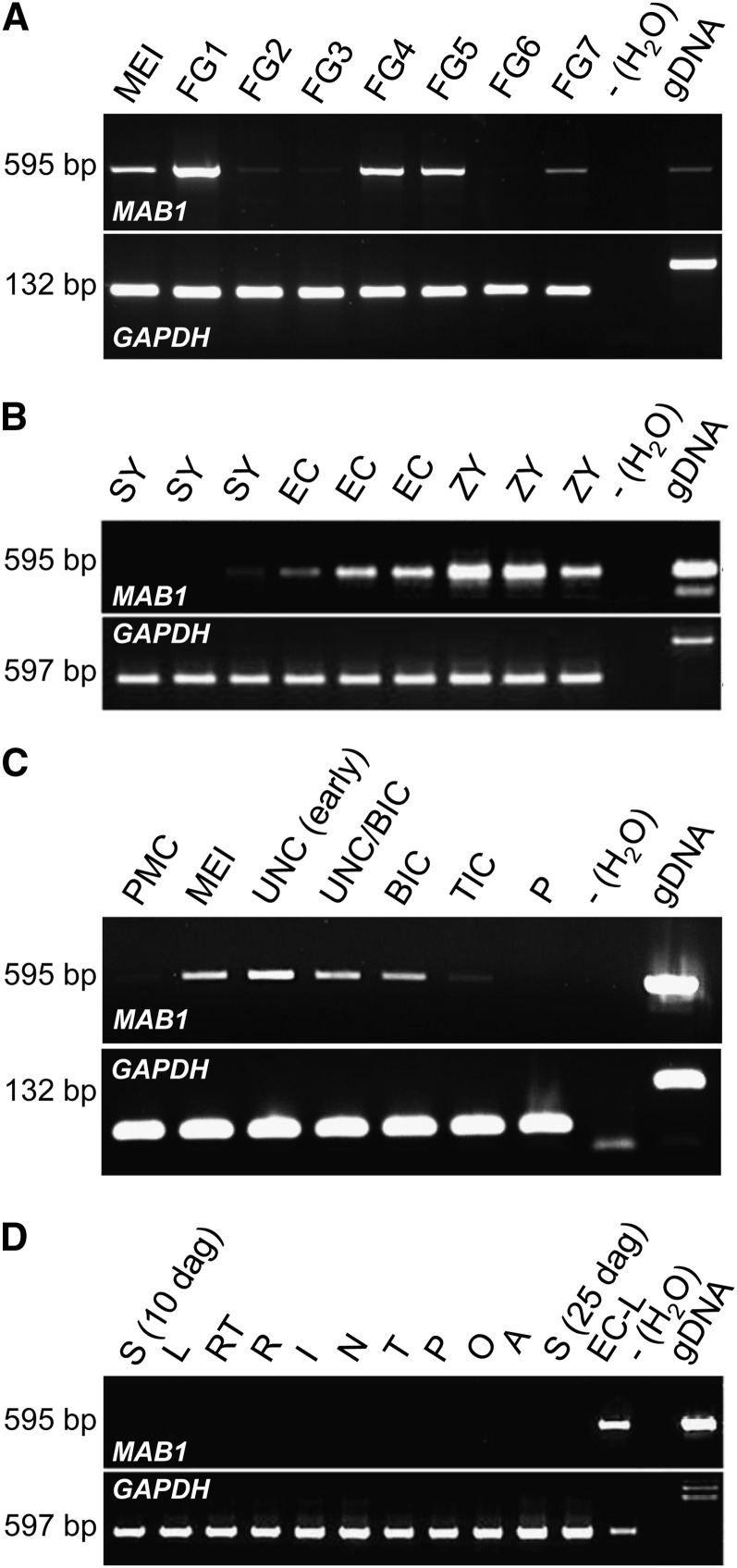
Expression of MAB1 was analyzed by quantitative RT-PCR and found to coincide with the observed phenotypes. DNase treatment was performed with all samples, as MAB1 represents an intronless gene. As shown in Figure 4A, transcripts were first detected in ovules containing female meiocytes undergoing meiosis, and the strongest expression was observed in ovules containing spores (female gametophyte stage FG1; stages after Evans and Grossniklaus, 2009). After mitosis I, gene activity is strongly downregulated and significantly upregulated again during the last free mitotic division (mitosis III) at stage FG4 and during the cellularization that takes place at stage FG5. During maturation of the female gametophyte, MAB1 is first downregulated at FG6 and transcripts appear again in mature gametophytes at stage FG7. Single-cell RT-PCR of dissected egg apparatus cells was performed to study MAB1 expression before and after fertilization. Synergid cells show weak or lack of expression levels in gels. More sensitive DNA gel blot analysis confirmed signals in all synergid cells analyzed (data not shown). Strong expression was detected in the egg cell, and gene expression was even upregulated in zygotes (Figure 4B). Notably, the expression pattern of MAB1 during male germline development is similar to the corresponding stages of female development (Figure 4C): Transcripts were first detected during meiosis, highest transcript levels are detected in microspores, and gene expression is downregulated after PMI. MAB1 is not expressed during vegetative development (Figure 4D).
Figure 4.
Expression of MAB1 Is Restricted to Male and Female Germlines and Is Strongest after Meiosis and in the Zygote of Maize.
RT-PCR after 38 PCR cycles using equal amounts of template mRNA. Intron-flanking primers for GAPDH served as a control, as MAB1 lacks introns.
(A) Expression during female gametophyte development from meiosis (MEI) to mature female gametophytes (stage FG7). Stages after Evans and Grossniklaus (2009). gDNA, genomic DNA.
(B) Single-cell RT-PCR of manually dissected cells of the mature female gametophyte (stage FG7), including synergids (SY), egg cells (EC), and zygotes (ZY).
(C) Expression during male gametophyte development from pollen mother cell (PMC), meiosis (MEI), unicellular (UNC), bicellular (BIC), and tricellular (TIC) toward mature pollen (P).
(D) Tissue expression analysis of MAB1, including seedlings (S) 10 and 25 d after germination, leaves (L), root tips (RT), roots without tips (R), internodes (I), nodes (N), mature tassels (T), mature pollen (P), mature ovaries (O), and mature anthers (A). An egg cell library (EC-L; Dresselhaus et al., 1994) served as a positive control. RT-PCRs with water served as negative, and a genomic PCR (genomic DNA) as positive controls in each experiment.
MAB1 Activity Is Required for Chromosome Separation during Meiosis
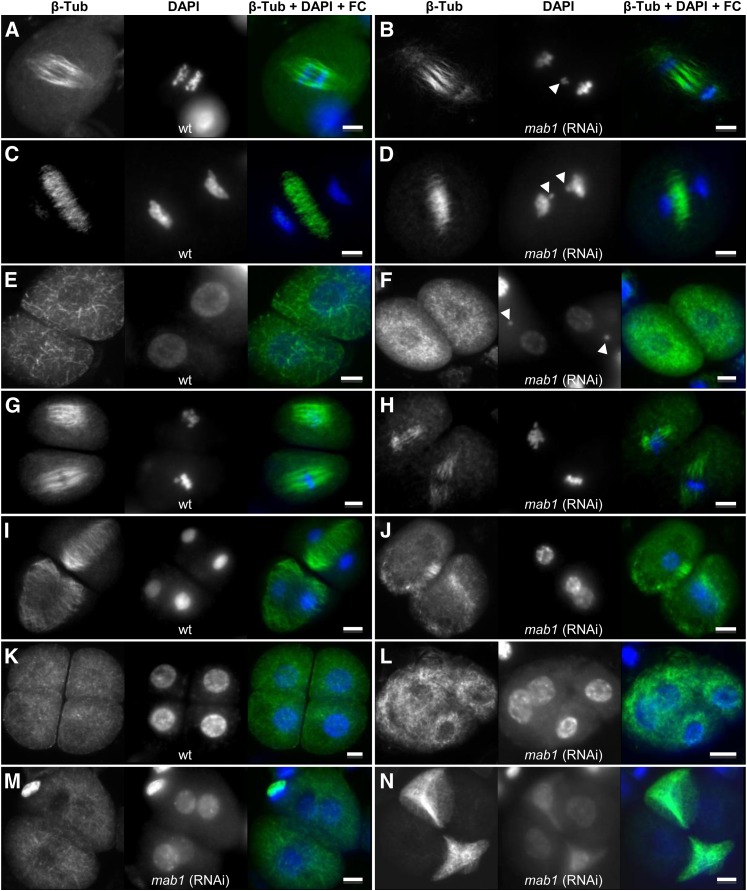
The expression pattern described above indicates that MAB1 activity might also be required during meiosis. Due to technical difficulties associated with studies on female meiosis, we compared microsporogenesis in dissected wild-type and mab1 (RNAi) anthers. Chromosomes and DNA were stained by DAPI and MTs using an anti-β-Tub-Cy3 antibody. Abnormalities were rarely found during meiosis in wild-type microsporocytes (3%; n = 65) but increased 11-fold to 45.5% (n = 104) in mutant microsporocytes (Table 2). As shown in Figures 5A to 5D, some chromosomes were not properly pulled toward spindle poles and were lost during the anaphase–telophase transition at meiosis I in ∼17.4% of analyzed microsporocytes of mab1 (RNAi) plants (compared with 1.5% in wild-type cells). In dyads, lost chromosomes are visible as micronuclei (Figure 5F). During meiosis II, the spindle apparatus was short in mutant dyads (Figure 5H) and ∼35% shorter than in wild-type dyads (∼40 μm in the wild type and 25 μm in mab1 [RNAi); Figure 5G). Moreover, the meiosis II spindles in RNAi dyads were often positioned with an angle of around 40° between them, whereas the two spindles in wild-type dyads are always positioned in parallel to each other. As a consequence, the resulting tetrads are neither identical nor nicely separated as in wild-type anthers (Figure 5K) but instead consist each of two pairs of microspores containing nuclei not properly separated from each other (Figures 5J, 5L, and 5M). Some meiosis-defective mab1 (RNAi) tetrads underwent apoptosis as indicated by weak DAPI staining, showing DNA degradation and the complete dissolution of MTs (Figure 5N). Whereas 28.1% of the analyzed microspores of mab1 (RNAi) showed these meiosis II abnormalities, only 1.5% of the wild-type microspores had such defects (Table 2).
Table 2. Immunohistological analysis of male meiosis in MAB1-RNAi lines.
| Meiosis I |
Meiosis II |
∑ Meiosis | ||||
| Genotype | na | Normal | Abnormalb | Normal | Abnormalc | defects |
| RNAi (line 143) | 44 (100%) | 18 (40.9%) | 8 (18.2%) | 5 (11.4%) | 13 (29.5%) | 47.7% |
| RNAi (line 1149) | 60 (100%) | 20 (33.3%) | 10 (16.7%) | 14 (23.3%) | 16 (26.7%) | 43.4% |
| Wild-type (A188) | 65 (100%) | 34 (52.3%) | 1 (1.5%) | 29 (44.7%) | 1 (1.5%) | 3% |
DAPI-stained microsporocytes with fluorescently labeled microtubules were counted from three wild-type and three RNAi plants.
Total No. of microspores counted.
Abnormalities at meiosis I refer to chromosome segregation defects and occurance of micronuclei after cytokinesis.
Abnormalities at meiosis II refer to shorter and misoriented spindles leading to abnormal tetrads or in the case of wt only to abnormal tetrads.
Figure 5.
In Contrast with the Wild Type, Maize mab1 (RNAi) Mutant Lines Show MT Organization Defects during Male Meiosis I and II.
MTs were stained using an anti-β-Tub-Cy3 antibody, DNA was counterstained by DAPI, and both images were merged using false color (FC) as indicated at the top of image rows. Arrowheads point toward unsegregated chromosomes. Left image row, the wild type (wt); right image row, mutant lines. Bars = 10 µm.
(A) and (B) Anaphase I of meiosis I.
(C) and (D) Telophase I of meiosis I.
(E) and (F) Dyads.
(G) and (H) Metaphase II of meiosis II.
(I) and (J) Telophase II of meiosis II.
(K) to (N) Tetrads.
MAB1 Shows a Cell Cycle–Dependent Nucleoplasm/Cytoplasm Localization Pattern
To investigate the subcellular localization of MAB1 during male and female germline development, we generated seven independent transgenic maize lines expressing a MAB1-EGFP (for enhanced green fluorescent protein) fusion protein under the control of the endogenous MAB1 promoter. Surprisingly, we have not been able to detect EGFP signals in any of these seven lines during male and female germline development or zygotic division (see Supplemental Figures 3A to 3G online), although the EGFP fusion protein is generated from the identical construct in transient transformation assays using maize BMS suspension cells (see Supplemental Figures 3H and 3I online). Our findings indicate that the MAB1-EGFP is expressed only at low levels and/or is highly unstable or that it is only transiently present at very precise developmental stages that we were not able to catch in our microscopy studies. Importantly, we could also show that the MAB1 promoter was functional, as MAB1-EGFP transcripts could be detected in ovules and anthers of transgenic maize plants (see Supplemental Figures 3J and 3K online).
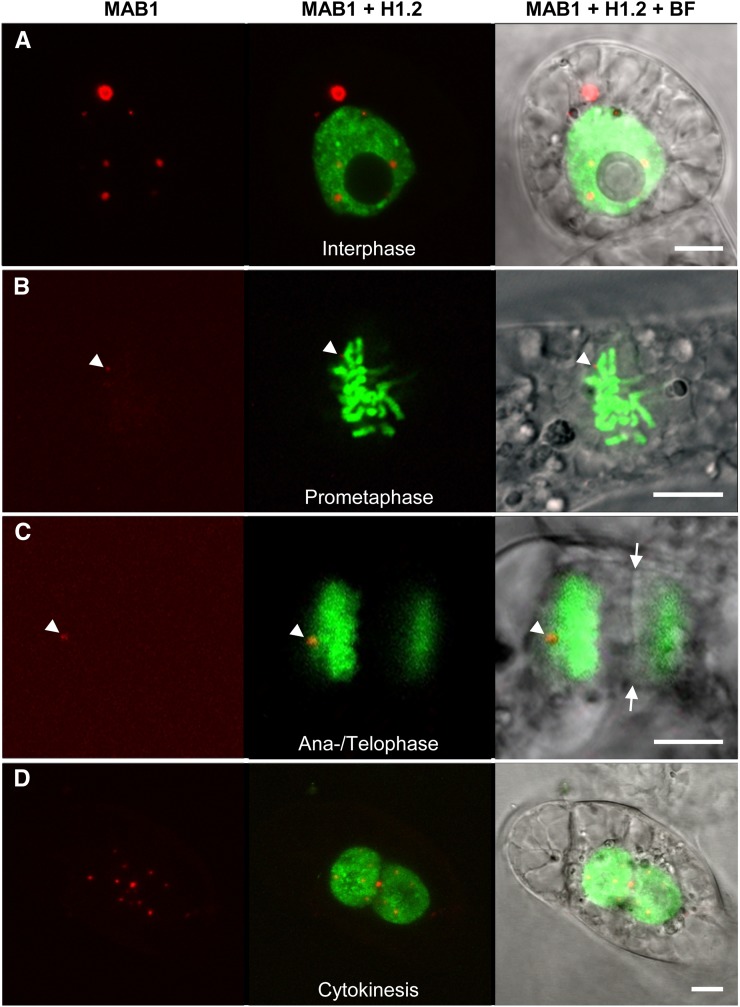
The finding that we could also express MAB1 in transiently transformed tobacco BY-2 cells indicated that it might be possible to learn more about its subcellular localization during the mitotic cell cycle using this heterologous system. We therefore first cotransformed monomeric red fluorescent protein (mRFP)-MAB1 and the histone marker EGFP-H1.2 from Arabidopsis (Launholt et al., 2006). As shown in Figure 6, mRFP-MAB1 is distributed during interphase in the nucleoplasm and cytoplasm in a speckled pattern. In the cytoplasm, MAB1 speckles occasionally form larger aggregates. During mitosis, the fusion protein almost completely disappears and is visible only at the spindle pole region during the anaphase–telophase transition (Figure 6C). mRFP-MAB1 signals reform into speckle-like structures during cytokinesis and are first present around and inside newly formed daughter nuclei after cellularization was completed (Figure 6D).
Figure 6.
Localization of MAB1 and Histone H1.2 during the Cell Cycle in Tobacco BY-2 Cells.
MAB1 was N-terminally fused to mRFP, and histone H1.2 of Arabidopsis was fused C-terminally to EGFP. The left image of each panel shows mRFP-MAB1 alone, a merged image of both fusion proteins is shown in the middle, and the right image is the corresponding merged bright-field (BF) and fluorescence image to visualize cell structures. Interphase (A), prometaphase (B), late anaphase/telophase (C), and cytokinesis (D). Note that the nuclear envelopes of both daughter cells have been formed already in (D). Arrowheads show mRFP-MAB1 in (B) and (C), and arrows indicate the phragmoplast in (C). Bars = 10 µm.
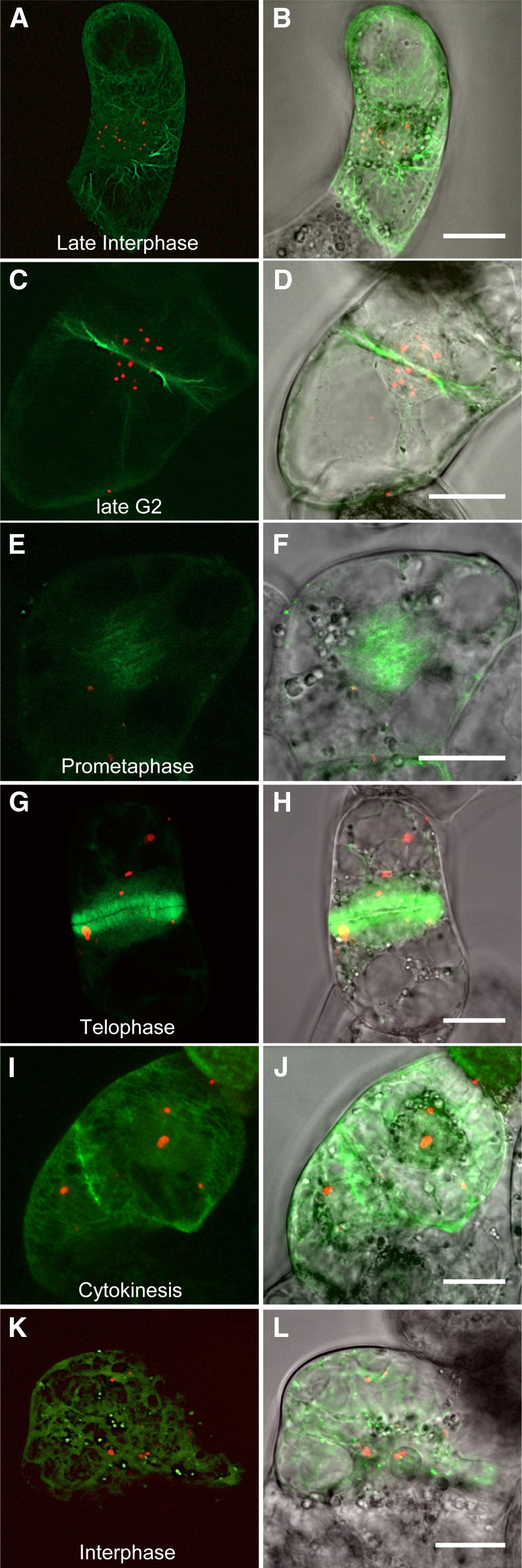
The observed localization pattern and phenotype observed during meiosis II (Figure 5) indicated that MAB1 might be associated with MTs or MT nucleation sites during spindle apparatus formation and function. To study this possibility, we cotransformed mRFP-MAB1 and the MT marker 2xEGFP-MBD (microtubule binding domain of microtubule-associated protein MAP4 from mouse) in BY-2 cells. At late interphase and especially during late G2 mRFP-MAB1, signals are visible mainly in the nucleoplasm and disappear during prometaphase (Figures 7A to 7F). Association with the PPB could not be found. At late telophase (Figures 7G and 7H), MAB1 reappears in the form of speckles of various sizes without any obvious specific subcellular localization. To study its interaction with MT nucleation sites, a maize γ-tubulin gene was cloned and fused with EGFP (EGFP-TubG1: Z. mays γ-tubulin 1). Constructs were cotransformed in BY-2 cells to investigate the presence of mRFP-MAB1 at the minus ends of MTs. As shown in Figures 7K and 7L, colocalization could not be detected. In summary, these observations show that MAB1 does not directly interact with the spindle apparatus but might be involved in regulating the stability of the spindle apparatus regulator(s).
Figure 7.
Colocalization Studies of MAB1 and MTs during the Cell Cycle.
Transient transformation of tobacco BY-2 cells with mRFP-MAB1 and 2xEGFP-MBD both N-terminally fused to EGFP to visualize MT strands ([A] to [J]). EGFP-TubG1 was cotransformed with mRFP-MAB1 to show the minus ends of MTs ([K] and [L]). (A), (C), (E), (G), (I), and (K) are merged fluorescence images, and (B), (D), (F), (H), (J), and (L) are fluorescence images merged with bright-field images. Interphase ([A] and [B]), late G2 showing PPB ([C] and [D]), prometaphase ([E] and [F]), telophase ([G] and [H]), shortly after cytokinesis ([I] and [J]). Bars = 20 µm.
MAB1 Interacts with Itself, with CUL3a, and with a Candidate Substrate Protein p60 of Katanin
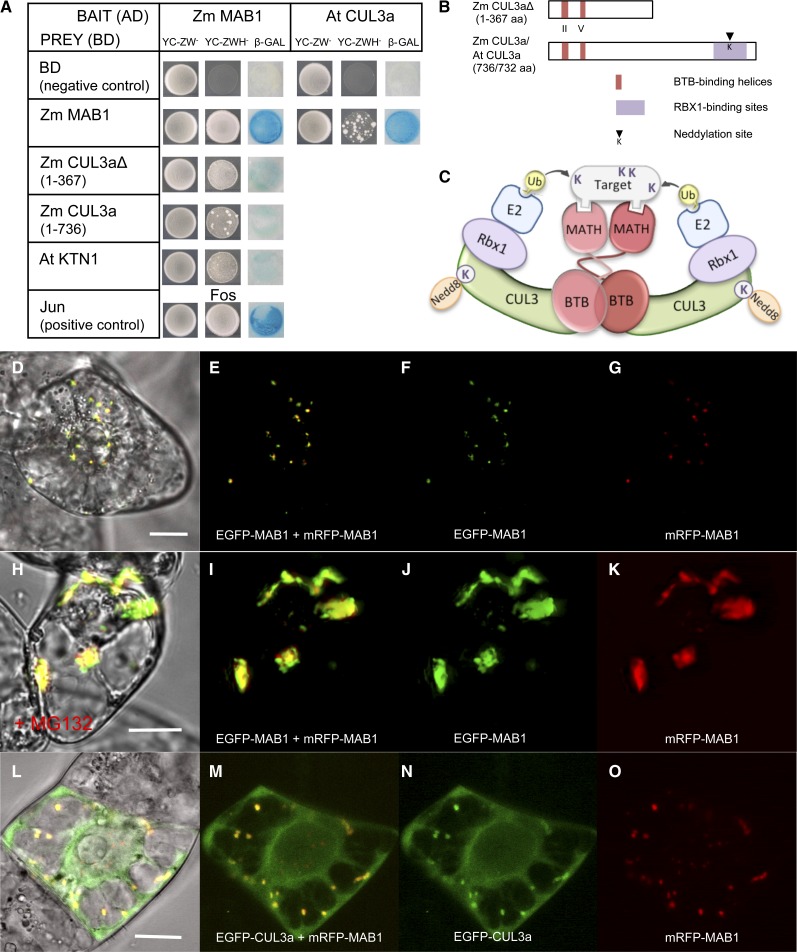
The observation that MAB1 forms aggregates in plant suspension cells together with reports about other MATH-BTB proteins as substrate-specific adaptors in CUL3 E3 ligase complexes prompted us to test the self-interaction of MAB1 and its interaction with CUL3 proteins. Yeast two-hybrid experiments (Figure 8A) showed that MAB1 is capable of interacting with itself and additionally is capable of interacting with Arabidopsis CUL3a (Dieterle et al., 2005). To study whether MAB1 is also able to interact with maize CUL3 proteins, we analyzed the maize genome and identified three CUL3 genes that we named Zm CUL3a-c (see Supplemental Figure 4 online). A C-terminal truncated version of CUL3a containing the predicted BTB binding helices at the remaining N terminus (Figure 8B) as well as the whole protein showed interaction with MAB1 (Figure 8A), indicating that the N terminus of CUL3a is required for binding. Based on these findings as well as on structural investigations with human SPOP (Zhuang et al., 2009), we suggest that MAB1 takes center stage in a dimeric E3 ligase interaction model that we present in Figure 8C: MAB1 dimerizes via its BTB domain that simultaneously interacts with CUL3a-c. CUL3 proteins of maize contain, as do other CUL3 proteins, a neddylation site for posttranslational modification by the Nedd8/Rub1 protein complex (see Supplemental Figure 4 online), and they may also bind Rbx1, which itself interacts with an E2 ubiquitin carrier protein. The MATH domains of dimeric MAB1 may coordinately interact with either one larger substrate (Figure 8C) or two smaller substrates (data not shown) and mediate their ubiquitinylation.
Figure 8.
MAB1 Interaction Studies Indicate That It Is an Unstable Protein and Functions as a Dimeric Substrate-Specific Adaptor of a CUL3-Based E3 Ligase in the Proteasomal Degradation Pathway.
(A) Yeast two-hybrid interaction studies show that MAB1 is able to form homodimers in vivo and to directly interact with truncated and full-length maize CUL3a as well as CUL3a from Arabidopsis. MAB1 is additionally able to interact with At KTN1 (Arabidopsis p60 subunit of katanin). Specificity of the bait constructs (MAB1-AD and At Cul3a-AD) was confirmed by cotransformation with empty prey vector (negative control). Jun and Fos interaction served as a positive control.
(B) Diagram of truncated and full-length maize CUL3a proteins used in yeast two-hybrid analysis. The regions predicted for interactions within the E3 complex are indicated.
(C) Schematic model of the CUL3-MATH-BTB-E3 ligase complex based on our data and the three-dimensional structure of human Hs SPOP and SCFSkp2 E3 complexes (Zheng et al., 2002; Zhuang et al., 2009).
(D) to (G) MAB1 forms homodimers in both the cytoplasm and nucleus of BY-2 cells.
(H) to (K) Proteasome inhibitor MG132 treatment leads to accumulation of MAB1 in large aggregates mainly in the cytoplasm of BY-2 cells.
(L) to (O) CUL3a and MAB1 colocalize in speckles in the cytoplasm but not in the nucleus of BY-2 cells. Bars = 20 μm.
To confirm these findings and to study the subcellular localization of these interactions in plant cells, mRFP-MAB1/EGFP-MAB1 and mRFP-MAB1/EGFP-CUL3a were cotransformed in tobacco BY-2 cells. During interphase, fusion proteins were found in small speckles both in the cytoplasm and nucleus. While both fusion proteins colocalize in the majority of speckles, some speckles predominately contain either the mRFP or EGFP version of MAB1 (Figures 8D to 8G; see Supplemental Figures 5A to 5H online). Application of the proteasome inhibitor MG132 led to the accumulation of both fusion proteins in large aggregates, predominately in the cytoplasm (Figures 8H to 8K; see Supplemental Figures 5I to 5P online), confirming the hypothesis that MAB1 is an unstable protein that is itself subjected to ubiquitin-mediated degradation. Colocalization studies with MAB1 and CUL3a showed that both proteins colocalize in the cytoplasm but not in the nucleoplasm (Figures 8L to 8O). As shown in Supplemental Figures 5Q and 5R online, EGFP-CUL3a signals are evenly distributed in the cytoplasm and occur in cytoplasmic speckles only in the presence of MAB1. This finding is surprising as CUL3a contains a putative nuclear localization site at its C terminus (see Supplemental Figure 4 online). In summary, these observations indicate that (1) MAB1 and CUL3a interact in the cytoplasm, (2) that MAB1 recruits CUL3a or vice versa in higher order cytoplasmic complexes, (3) that CUL3a either prevents MAB1 from nuclear localization or stabilizes the complex in the cytoplasm, and (4) that it may possess a CUL3a-independent function in the nucleus. Notably, MAB1 without CUL3a was enriched in nuclear speckles in late interphase and G2 as well as after cytokinesis (Figures 6D and 7A to 7D; see Supplemental Figures 5S and 5T online), supporting the hypothesis that it may be less stable in the cytoplasm. Interestingly human SPOP also recruits CUL3 into larger aggregates, but in the nucleus (Kwon et al., 2006).
Finally, we studied whether MAB1 is capable of interacting with the MT-severing protein katanin. In Arabidopsis, the p60 subunit At KTN1 of katanin has been reported (Burk et al., 2001; Stoppin-Mellet et al., 2002) and was used for yeast two-hybrid studies. As shown in Figure 8A, MAB1 is able to weakly interact with KTN1 in vivo.
DISCUSSION
The MATH-BTB protein family is common to both animals and plants. However, it is a phenomenon that Arabidopsis and human genomes encode only a few members (six and two, respectively) of the MATH-BTB protein family that has largely expanded more than 10-fold in other organisms with at least 31 genes in maize (this report), 68 genes in rice, and 46 genes in C. elegans (Stogios et al., 2005; Gingerich et al., 2007). All 31 maize genes are annotated as protein-coding genes in searched databases, but information about gene expression pattern is available for only 19 genes. According to the rice database, all 68 genes are expressed, indicating that active proteins are generated. All six Arabidopsis MATH-BTB proteins group with six maize and four rice proteins in a conserved core clade, suggesting that these proteins might possess similar functions. The expanded group lacks Arabidopsis proteins and contains at least 64 members in rice and 25 members in maize. Gingerich et al. (2007) showed that members of the expanded group display a significantly faster evolution of the substrate recognition MATH domain. They concluded that their substrates might have changed rapidly themselves, such as genes involved in reproduction, speciation, or defense. We reported here a MATH-BTB protein from the expanded clade (Zm MAB1) that plays an important role in both meiotic and mitotic divisions during plant reproduction, which have not been previously described for any plant MATH-BTB protein or CUL3-based E3 ligase.
One mechanism ensuring faithful cell division is selective degradation of key cell cycle regulators. Indeed, CUL-based E3 ligases have emerged as crucial regulators of the cell cycle in mammalian cells, among which CUL3-based E3 ligases are required during mitosis. Different BTB domain proteins serve as substrate-specific adaptors in CUL3-based E3 ligase complexes, increasing the number and variety of potential substrates. In human, it has been shown that KLHL/CUL3 E3 ligases, which contain BTB-Kelch substrate-specific adaptors that lack a MATH domain, are needed for chromosome segregation and mitotic progression via targeting the chromosome passenger protein kinase AURORA B for degradation (Sumara et al., 2007). In C. elegans, the female germline-specific MATH-BTB protein MEL-26 is required for the formation of the first mitotic spindle in the early embryo by downregulating the MT-severing protein MEI-1 (p60 subunit of katanin) activity at the meiosis-to-mitosis transition (Clark-Maguire and Mains, 1994). Failure to eliminate MEI-1 during mitosis results in short, disoriented mitotic spindles (Clark-Maguire and Mains, 1994) similar to spindles found in our mab1 (RNAi) meiosis mutants. Spindle length occurring during meiosis in maize significantly extends the length of mitotic spindles (Franklin and Cande, 1999), thus requiring enzymatic activities that will reduce their length during the meiosis-to-mitosis transition. Moreover, the first mitotic spindle after meiosis is asymmetric in plants, generating daughter nuclei and cells of different identity (Twell, 2011). MEL-26 accomplishes the downregulation of MEI-1 by acting as a substrate-specific adaptor that recruits MEI-1 to a CUL3-based E3 ubiquitin ligase, resulting in MEI-1 ubiquitination and subsequent degradation (Furukawa et al., 2003; Pintard et al., 2003; Xu et al., 2003). We found that the p60 subunit of katanin from Arabidopsis (At KTN1) is able to interact with MAB1, further suggesting that MAB1 possesses similar targets and may function in a similar manner to animal MEL-26. Loss of KTN1 activity has recently been shown to result in cell division plane defects, loss of growth heterogeneity, and response of mechanical forces (Uyttewaal et al., 2012). Similarly, cul3a cul3b double homozygous mutants in Arabidopsis showed enlarged and abnormal embryonic cells as well as abnormal cell divisions, in particular hypophyseal cell plate deviations. However, increased levels of the katanin-like protein were not detected (Thomann et al., 2005). Thus, katanin-like protein levels might also be regulated independently of CUL3 E3 ligases. In C. elegans, MEL-26 also interacts in a CUL3-independent mechanism with the actin binding protein POD-1 (Luke-Glaser et al., 2005) and a CUL3-dependent mechanism with the katanin p60 subunit-related protein FIGL-1 (Luke-Glaser et al., 2007). Like KTN1, both MEI-1 and FIGL-1 belong to a family of AAA-ATPases that are degraded at different developmental stages. The CUL3/MEL-26 complex degrades mitotic FIGL-1 during the mitosis-to-meiosis transition and meiosis. By contrast, meiotic MEI-1 is degraded at the meiosis-to-mitosis transition. FIGL-1 is stable inside nuclei and is thus protected from cytoplasmic MEL-26 during early embryo development and able to contribute to further mitotic divisions (Luke-Glaser et al., 2007; Johnson et al., 2009). The observation that MAB1 and CUL3a do not interact in the nucleus of tobacco BY-2 cells further suggests that unknown nuclear substrates may be degraded in the cytoplasm and are protected in the nucleus or that MAB1 may posses a CUL3a-independent function in the nucleus. By database searching, we identified a large number of similar, but uncharacterized AAA-ATPases in maize that cannot be clearly predicted as either MEI-1 or FIGL-1 orthologs. A systematic search will now be necessary to identify CUL3-dependent and -independent substrates of MAB1, such as AAA-ATPases and other proteins.
In C. elegans, it was further reported that MEL-26 is induced during meiosis and that the protein is present only at low levels until the completion of meiosis, after which protein levels increase substantially (Johnson et al., 2009). It was shown that the level of MEL-26 itself is kept low during meiosis by the action of a CUL2-containing E3 ubiquitin ligase until it is required for the postmeiotic degradation of MEI-1. Whether MAB1 stability is as tightly regulated at the protein level remains to be shown. However, the observations that MAB1-EGFP fluorescence cannot be detected during male and female meiosis and gametogenesis in maize and strongly accumulates in MG132-treated cells indicate that MAB1 is an unstable protein that is strongly regulated at the protein level itself and thus probably required only in minor amounts at short and defined stages during meiosis and gametogenesis. RNAi approaches in maize usually do not show mutant phenotype numbers above 40% in a heterozygous situation (Dresselhaus et al., 2005; Srilunchang et al., 2010; Krohn et al., 2012), further supporting the hypothesis that the gene is strongly regulated and RNAi transcript amounts are sufficient to abolish gene activity.
Little is known about the substrates of other MATH-BTB domain proteins. Human SPOP, for example, serves as a CUL3-ubiquitin ligase adaptor for targeting several proteins. One of its targets, the multifunctional protein Daxx, plays a major role in regulating apoptosis induction (Kwon et al., 2006). DNA degradation and cell death were also found in mab1 (RNAi) male tetrads after meiosis. Additionally, many female gametophytes were collapsed, suggesting that MAB1 may also be required for cell viability. However, because of the presence of a MATH domain, MATH-BTB proteins might be generally involved in apoptosis (Aravind et al., 1999) and the observed cell death phenotype might be rather unspecific. Until now only two classes of MATH-BTB interacting proteins have been identified as candidate substrates in Arabidopsis: (1) the class I homeobox-leucine zipper transcription factor ATHB6, a negative regulator of ABA responses; and (2) members of the ERF/AP2 transcription factor family, which regulate the ethylene response (Weber and Hellmann, 2009; Lechner et al., 2011). Depending on our greenhouse conditions (heat and low air humidity), we also observed a strong environmental effect on the occurrence of aborted pollen in mab1 (RNAi) plants compared with wild-type plants that may hint at a role for MAB1 activity during increased stress sensitivity. Downregulation of Arabidopsis MATH-BTB genes also resulted in male sterility (Lechner et al., 2011), indicating that the corresponding proteins are involved in a variety of biological processes, including stress responses and development.
It would thus be interesting to find out how Arabidopsis or human cells manage to regulate a large variety of biological processes with such a small number of MATH-BTB/CUL3 substrate adaptors and whether the six Arabidopsis proteins are also involved in the fundamental process of regulating spindle length during reproduction. A possible key to this enigma is the number of splicing variants or additional BTB domain proteins containing domains other than MATH that might have been recruited for the same functions described above. Hs SPOP (for Speckle-type POZ protein), for example, generates 16 functional splicing variants likely resulting in proteins with different activities and targets. The expression pattern of Arabidopsis MATH-BTB/CUL3 E3 ligase components is less specific than in maize. Both At CUL3a and At CUL3b as well as MATH-BTB (At BPM1-6) genes are expressed in all parts of the plant (Weber et al., 2005; Weber and Hellmann, 2009). However, interaction studies in yeast two-hybrid assays have shown that Arabidopsis CUL3a and CUL3b can both interact with BPM1 and BPM3 but not with BPM5 and BPM6 (Weber et al., 2005). Additionally, MATH-BTB proteins may form heterodimers to increase complexity and, thus, specificity to target a larger number of substrates during various biological processes.
MAB1 deficiency disturbs ACD, which is of indisputable importance for proper male germline development and embryogenesis. Even though ACD is not as obvious during female gametogenesis, the polarity of the female gametophyte is established along its micropylar-chalazal axis and likely is already determined after meiosis during the first mitotic division of the functional megaspore (Webb and Gunning, 1990; Bajon et al., 1999). This polarity is clearly reflected by the successive establishment of two different gametic cells (egg and central cell), which are enclosed by highly specialized accessory cells at both poles (Huang and Russell, 1992; Pagnussat et al., 2009; Krohn et al., 2012). ACD and cell fate specification are closely related: First, the establishment of an internal cell asymmetry is required; and second, the positioning of the division plane and the spindle apparatus with respect to the axis is defined by the polarity of the cell. Accordingly, cytokinesis results in the asymmetric distribution of cellular components (Bisgrove et al., 2003) and presumably cell fate determinants. In mammalian cells, it has been shown that asymmetric mitosis is achieved by asymmetric labeling of peripheral centrosomal proteins for degradation when centrioles separate and migrate to opposite poles (Fuentealba et al., 2008). Moreover, in cases where the site of cytokinesis is not predetermined by a PPB, the cell division plane will bisect the spindle axis (Bisgrove et al., 2003). A PPB marks the position of the first mitotic division of the zygote (Webb and Gunning, 1991). However, a PPB is absent in meiocytes and gametophytic cells (reviewed in Otegui and Staehelin, 2000), indicating that the division site is specified by unknown signals or polarity determinants in the cytoplasm. MAB1 is a candidate regulator of correct PPB-independent spindle positioning by regulating katanin activity during the meiosis-to-mitosis transition or the katanin-mediated displacement of spindles in meiosis II to generate longer spindle assemblies.
To conclude, we discovered a fundamental role of a maize MATH-BTB protein of the expanded clade of plant MATH-BTBs that lacks Arabidopsis proteins. The observed phenotypes and interaction partners suggest a comparable role of MAB1 to MEL-26 of C. elegans. Due to the early arrest of mab1 (RNAi) mutants in both germlines, we could not determine the function of MAB1 during the zygotic division. However, the observation that MTs are shorter and disorientated in mel-26 mutant zygotes (Pintard et al., 2003; Xu et al., 2003) suggests that mab1 (RNAi) mutants in maize may also affect the future ACD plane of the zygote. The third interactor of MEL-26, POD-1, supports this notion as it is not a substrate of MEL-26, but rather binds to its MATH domain to support MEL-26 localization to the cell cortex and to promote cytokinesis in early embryos in a CUL3-independent manner (Luke-Glaser et al., 2005). Zygote- and/or egg cell–specific promoters could now be used to elucidate the role of MAB1 during ACD in the maize zygote. In summary, our results provide a functional link between the cell division machinery and the molecular mechanism that regulates ACD by ubiquitination during germline and probably early embryo development of plants.
METHODS
Plant Material and Isolation of Cells from Maize Gametophytes
Maize (Zea mays) plants were grown under standard greenhouse conditions at 26°C with 16 h of supplementary light during the day period and a relative air humidity of 40 to 60%. Single embryo sac cells of maize were isolated according to Kranz et al. (1991). Maize zygotes were isolated according to Cordts et al. (2001). Male meiocytes and immature and mature pollen grains were manually collected from the three larger anthers of each spikelet. Briefly, fresh anthers were cut transversely with a razor blade under a binocular microscope. Male germ cells were released by immersing open anthers in a droplet of 0.3 M mannitol solution or extracted from anthers by gentle pushing using curved tip tweezers. Cells were either transferred with a pipette onto a microscope slide for subsequent phenotypic analysis or into an Eppendorf tube for immunolabeling. Microscopy of living pollen in mannitol solution was done within 1 h of isolation. Tobacco (Nicotiana tabacum) BY-2 suspension cells were cultivated in liquid MS (Murashige and Skoog, 1962) medium containing 4.4 g/L of MS salts (Duchefa), 30 g/L Suc, 100 mg/L myo-inositol, 1 mg/L thiamin, and 255 mg/L KH2PO4, pH 5.0, supplemented with 0.2 mg/L 2,4-D, and kept in the dark at 26°C with shaking at 60 to 70 rpm.
Transient and Stable Transformation
Tobacco BY-2 cells were transiently transformed by particle bombardment using plasmids encoding EGFP and/or mRFP1 fusion proteins under the control of the MAB1 and constitutively active 35S promoters (a detailed description of constructs is given in Supplemental Methods 1 online). A fresh tobacco BY-2 culture was initiated and grown for 3 d. One to two hours before transformation, cells were collected by filtering through a 50-µm nylon mesh and spread as a thin layer onto 35-mm Petri dishes with solid MS medium (Kirihara, 1994). For preparation of plasmid-coated gold particles, 5 µg of plasmid solution (1 µg/µL) was added to 50-μL aliquots of a 60-mg/mL gold suspension (0.4 to 1.2 µm; Bio-Rad) and prepared as described previously (Dresselhaus et al., 2006). Cells were bombarded with 7-μL aliquots of plasmid-coated gold particles using the PDS1000/He particle delivery system (Bio-Rad), 1100-p.s.i. rupture discs, a partial vacuum of 28-inch Hg, and a 6-cm target distance. After transformation, plates were incubated for 6 h to overnight in the dark at 26°C. At least 4 h before microscopy observation, cells were rinsed from the plates into 35-mm Petri dishes using liquid MS medium and placed in the dark with shaking (110 rpm, 26°C). To study the effect of the proteasome inhibitor MG132, BY-2 cells were treated with 50 µM MG132 12 h after bombardment. Following the addition of MG132 stock solution (10 mM in DMSO), cells were further cultured at 24°C in a dark chamber with shaking at 80 rpm until microscopy observations. Control cell samples were treated with the equivalent concentration of DMSO. For microscopy, 100 μL of suspension culture was transferred onto cover slips fixed to metal slides provided with an opening diameter of 20 mm in the center.
For stable transformation of maize, immature hybrid embryos (cross of inbred lines A188 and H99) were isolated under sterile conditions, 11 to 13 d after pollination. Culture, osmotic pretreatment, and particle bombardment of explants were performed according to Brettschneider et al. (1997), with the exception that 0.6% phytagel (Sigma-Aldrich), which was used instead of agarose. For cotransformation with the selection marker phosphinothricin acetyltransferase (PAT) (Becker et al., 1994), a plasmid mixture of 3 µg Zm MAB1-RNAi or 3 µg PZmMAB1:ZmMAB1-EGFP and 2 µg P35S:PAT (for generation of constructs, see Supplemental Methods 1 online) was precipitated onto gold particles (1 µm; Bio-Rad) as described (Becker et al., 1994). Subsequent culture and selection of PAT-resistant maize explants were performed as described (Brettschneider et al., 1997). One week after transfer to soil, plantlets were sprayed two times within a week with an aqueous solution of 250 mg/L BASTA in 0.1% Tween 20.
Collection and Preparation of Maize Ovules and Anthers for Microscopy
Defined stages of immature and mature maize embryo sacs were identified according to the ear silk length, which was used as a morphological feature with respect to the corresponding stage of female gametophyte development (according to Huang and Sheridan, 1994). At a silk length of 0 to 0.5 mm, we found ovules containing either megaspore mother cells at meiosis or four megaspores. The functional megaspore during the first mitotic division (female gametophyte stage 1 [FG1]) corresponded to a silk length in the range of 0.5 to 1 mm. Two-nucleate (FG2) and four-nucleate (FG4) embryo sacs were found in ovules with a silk length of 1 to 2.5 mm. At later stages (silk length ranging from 2.5 to 7 mm), most ovules contained eight-nucleate or mature embryo sacs. For phenotypic analysis of maize embryo sacs, immature and mature cobs were harvested and treated with a fixing/clearing method followed by Kasten’s fluorescent periodic acid-Schiff staining (Srilunchang et al., 2010). Samples were mounted in methyl salicylate on glass slides under a cover slip and analyzed by confocal laser scanning microscopy.
Anther length is a reliable parameter for developmental stages of the male gametophyte (Chang and Neuffer, 1994), and its development can be followed from primordium formation within developing spikelets at a tassel inflorescence to pollen shed. However, anther length strongly depends on the genotype and plant cultivation place. In our study, inbred line A188 and A188xH99-derived transgenic lines showed similar anther lengths and corresponding developmental stages. From each tassel, at least 10 spikelet samples were collected and measured. The three biggest anthers from each spikelet were measured and used to determine stages of developing male gametophytes. The observed developmental stages were grouped into seven classes: (1) pollen mother cells before meiosis with an anther size of 0.5 to 1.5 mm, (2) microsporocytes at meiosis with an anther size of 2 to 2.6 mm, (3) early uninucleated microspores with a corresponding anther size of 2.5 to 3.2 mm, (4) late uninucleated microspores until early binucleate pollen (PMI) with an anther size of 3 to 4 mm, (5) late bicellular stage with an anther size of 3.8 to 4.6 mm, (6) early tricellular stage with an anther size of around 5 mm, and (7) mature pollen with an anther size of around 5 to 6 mm. To ensure that developing spores and pollen were collected at the correct stages, anthers were fixed in 3:1 ethanol:acetic acid at room temperature for 2 to 24 h, squashed, and stained with 10 µg/mL DAPI.
Immunofluorescence of Spindles during Male Meiosis
Immunofluorescence using male meiocytes was performed after modifying a previously described protocol of Singh et al. (2011). Freshly isolated anthers of the appropriate stages were fixed for 1 h in fixative solution (4% [v/v) paraformaldehyde in PHEMS buffer [60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, and 0.32 M sorbitol, pH 7.2]) and washed several times in PHEMS buffer. Meiocytes were isolated from anthers in a drop of PHEMS buffer and collected in Eppendorf tubes. Embedding of meiocytes in polyacrylamide was performed as described by Bass et al. (1997). All other steps were conducted according to Singh et al. (2011) with the only difference of the enzyme digestion step. The callosic cell wall of meiocytes was digested in freshly made 1% β-glucanase (Sigma-Aldrich) supplemented with proteinase inhibitor cocktail cOmplete, Mini, EDTA-free (Roche) in PHEMS buffer at pH 5.3 for 1 h at 37°C. Samples were labeled with a 1:150 dilution of a monoclonal anti-β-Tubulin-Cy3 Clone TUB 2.1 antibody (Sigma-Aldrich) during overnight incubation at 4°C and double stained afterwards for DNA with 10 µg/mL DAPI for 30 min at 4°C.
Microscopy
Images were either taken with the LSM 510 META confocal laser scanning microscope (Zeiss) using the Zeiss LSM image browser software version 3.5.0.359 or an Axiovert 200M fluorescence microscope using the AxioVision software 4.5 (Zeiss). Phenotypic analysis of microspores and pollen was done using bright field with differential interference contrast microscopy. Fluorescence of periodic acid-Schiff-stained nuclei of maize embryo sacs was detected with 488-nm excitation and a long-pass 505 filter. Excitation wavelengths and emission filters for other fluorophores were: 488 nm and band-pass 505 to 530 nm for EGFP, 514 nm and long-pass 530 for YFP, 543 nm and band-pass 560 to 615 for mRFP, and 543 nm and long-pass 560 for Cy3. DAPI was excited with the 365-nm line of a HBO mercury arc lamp. Images were recorded using AxioCam cameras (MrC; Zeiss) and processed with ImageJ (http://rsbweb.nih.gov/ij) as maximum-intensity projections of selected optical sections.
Quantitative RT-PCR
All primer sequences are provided in Supplemental Table 3 online. To study the expression of Zm MAB1 by quantitative RT-PCR, total RNA was extracted from different maize tissues and organs using TRIzol reagent (Invitrogen) or TriFast reagent (PEQLab), according to the manufacturers’ recommendations. The quality of RNA preparations was analyzed by denaturing agarose gel electrophoresis. Prior to first-strand cDNA synthesis, RNA samples were quantified and treated with 1 unit of RNase-free DNase I (MBI Fermentas) according to the manufacturer’s instructions. mRNA was isolated from whole manually isolated single egg cells, synergids, or zygotes using the Dynabeads mRNA DIRECT Micro kit (Dynal) and a magnetic particle transfer device (PickPen; BioNobile) as previously described (Sprunck et al., 2005). First-strand cDNA synthesis was performed using 0.5 µg oligo(dT)18 primer and 1 µg of DNase-treated total RNA or isolated poly(A)* mRNA and RevertAid H Minus M-MuLV reverse transcriptase (MBI Fermentas) following the manufacturer’s protocol with the addition of 40 units of RNaseOUT (RNaseOut recombinant ribonuclease inhibitor; Invitrogen). After the RT reaction, the quality and quantity of the individual cDNA populations was tested by control PCRs using specific primer pairs for GAPDH and gene-specific primer pairs for MAB1. See Supplemental Table 2 online for primer sequences. Thirty-eight PCR cycles were performed, ensuring that PCR reactions were in the linear amplification phase and not saturated. Gels were subjected to DNA gel blot hybridization (see below).
Quantitative RT-PCR reactions were performed using a Mastercycler ep realplex 2 system (Eppendorf) in 20-μL reaction volumes using KAPA SYBR Fast Universal qPCR MasterMix (Peqlab) and 100 nM final primers concentration. The following primer pairs were used: Zm GAP-F2 and Zm GAP-R2 for the maize housekeeping gene GAPDH and qMAB1-F and qMAB1-R for MAB1. PCR efficiency for each primer pair was estimated from a 10-fold dilution series of MAB1-containing plasmid DNA. Nontemplate controls and positive controls (genomic DNA) were additionally assayed by quantitative PCR for each gene. Each biological replicate was analyzed in technical triplicates. Statistical processing of raw quantitative RT-PCR data was performed as described in the manual for the GeNorm software (http://medgen.ugent.be/genorm/) by the ΔCt (cycle threshold) method to calculate relative MAB1 expression in MAB1-RNAi lines.
DNA Gel Blot Hybridization
Extraction of genomic DNA from maize leaves was performed according to Pallotta et al. (2000). Ten micrograms of genomic DNA from each mab1 (RNAi) plant was digested overnight with NotI and BspTI, while genomic DNA of PZmMAB1:ZmMAB1-EGFP plants was digested with SfiI. Restricted genomic DNA was separated in 0.8% agarose gels. Genomic DNA or PCR amplified DNA was transferred onto Hybond NX nylon membranes (Amersham) by capillary transfer (20× SSC) and fixed to membranes with 120 mJ using the Stratalinker 1800UV cross-linker (Stratagene) or transferred by alkaline capillary transfer (0.4 M NaOH) without fixation. Further procedure was performed according to Dresselhaus et al. (1999).
Yeast Two-Hybrid Protein Interaction Studies
A lexA-based yeast two-hybrid system (Hybrid Hunter; Invitrogen) containing pYESTrp as prey and pHybLex/Zeo as bait vectors was used together with the yeast reporter strain L40 [MATα his3∆200 trp1-901 leu2-3112 ade2 LYS2::(4lexAop-HIS3) URA3::(8lexAop-lacZ) GAL4], according to the manufacturer’s recommendations. A description of the constructs used for yeast two-hybrid analysis is given in Supplemental Methods 1 online. Successful cloning of open reading frames containing fragments fused with the B42 activation or LexA DNA binding domains was verified by sequencing the fusion products with primers pYESTrp-F and pYESTrp-R, or pHybLex/Zeo-F and pHybLex/Zeo-R, provided with the Hybrid Hunter Kit. L40 yeast cells were grown and cotransformed with bait and prey constructs using a standard lithium acetate technique described in the manufacturer’s protocol. To assay for His prototrophy, individual colonies of transformants were diluted 1:500 in autoclaved distilled water. Twenty-five microliters of each diluted yeast transformant was spotted on selective YC-ZWH− medium lacking Trp and His. To confirm the interactions, a β-galactosidase activity test was conducted: Another 25 μL of the diluted transformants was spotted in a grid pattern onto YC-ZW− plates. All plates were incubated at 30°C for 2 to 4 d until colonies appeared. In a β-gal filter assay, a strong interaction yielded detectable blue color in <30 min. Longer incubations of up to 5 h were required for weaker interactions. The controls pHybLex/Zeo-Fos and pYESTrp-Jun included in the Hybrid Hunter kit were used as a positive control for bait and prey plasmids, respectively. To test for nonspecific activation of the bait plasmids, all transcripts were cloned into pHybLex/Zeo and cotransformed together with pYESTrp without insertion into yeast reporter strain L40.
Database Query, Alignments, and Phylogenetic Analysis
Querying the databases of ESTs and nonredundant nucleotides of the National Center for Biotechnology Information’s GenBank and Phytozome v5.0 using BLASTp assembled a data set of CUL3 and MATH-BTB sequences of maize. In these searches, amino acid sequences of Zm MAB1 and At Cul3a proteins were employed as queries. Similar searches were performed in a database of the completely sequenced genome of maize B73 (www.maizesequence.org). The sequences obtained from all databases were aligned with MUSCLE v3.8.31 (Edgar, 2004), and only nonredundant full-length sequences were taken into further consideration. A total of three maize CUL3 and 31 MATH-BTB amino acid sequences were identified. Sequences were aligned and compared with known CUL3 and MATH-BTB proteins from rice (Gingerich et al., 2007), Arabidopsis thaliana (Dieterle et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005; Thomann et al., 2005; Weber et al., 2005), Caenorhabditis elegans (Pintard et al., 2003; Xu et al., 2003), and human (Zhuang et al., 2009). The resulting alignments were subsequently manually optimized in SeaView v4 (Gouy et al., 2010) and displayed using Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The phylogenic tree of MATH-BTB sequences was generated in SeaView by the maximum likelihood method with the program PhyML 3.0 (Guindon et al., 2010) implemented in SeaView using the GTR model and four categories of rate substitution. Branch lengths and model parameters were optimized, and the tree topology was obtained using both strategies of nearest-neighbor interchange and subtree pruning and regrafting. The approximate likelihood test was computed to perform a Shimodaira-Hasegawa–like statistic to support every bifurcation.
Accession Numbers
MATH-BTB accession numbers are listed in Supplemental Table 1 online. The Zm MAB1 genomic sequence can be found in GenBank under accession number EU344973 and Zm TubG1 under GenBank accession number NM_001111998. Accession numbers of CUL proteins are as follows: Zm CUL3a (GRMZM2G380184), Zm CUL3b (GRMZM2G126253), Zm CUL3c (GRMZM2G027750), Os CUL3a (LOC_Os02g51180), Os CUL3b (LOC_Os04g55030), Os CUL3c (LOC_Os08g07400), At CUL3a (AT1G26830), At CUL3b (AT1G69670), Ce CUL3 (NP_503151), Hs CUL3 (NP_003581), and Sc CUL3 (AAQ91375). Sequence data from Arabidopsis histone H1.2 gene At2g30620 can be found at GenBank under NM_128614.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of Selected Plant and Animal MATH-BTB Proteins.
Supplemental Figure 2. Relative Expression Levels of MAB1 in Indicated Reproductive Tissues of Five mab1 (RNAi) Plants Derived from Two Independent RNAi Lines (no.143 and no.1149).
Supplemental Figure 3. In Vivo Instability of MAB1 Protein in Transgenic Maize.
Supplemental Figure 4. The Conservation among Three Identified Maize CUL3 Proteins and Their Orthologs in Different Organisms.
Supplemental Figure 5. Additional Examples Showing MAB1 Homodimerization, Stability, and Subcellular Localization of Its Interaction with CUL3a in Tobacco BY-2 Cells.
Supplemental Table 1. Designated Gene Names and Identifiers for MATH-BTB Genes Used in the Phylogenetic Analysis and Protein Alignment.
Supplemental Table 2. Phenotypic Classification of Male Gametophyte Defects of Heterozygous mab1 (RNAi) Silencing Lines.
Supplemental Table 3. Primers Used for cDNA Synthesis, RT-PCR, and Cloning.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Figure 1.
Acknowledgments
We thank Günther Peissig for plant care, Klaus Grasser for providing the EGFP-H1.2 construct, and Dave Jackson for the YFP-α-tubulin marker line. We acknowledge financial support by a postgraduate scholarship to N.G.K. (Coodenação de Aperfeiçoamento de Pessoal de Nível Superior–Brasília/Brazil), the Alexander von Humboldt-Foundation for a postdoctoral fellowship to D.L.-L. (IV-KRO/1113833STP), and an institute partnership to D.L.-L., S.S., and T.D. as well as the Croatian Ministry of Science, Education, and Sport for a grant to D.L.-L. (Project 119-1191196-1225).
AUTHOR CONTRIBUTIONS
T.D. and S.S. designed the research experiments. M.J., K.S., and N.G.K. performed research and together with T.D., S.S., and D.L.-L. analyzed the data. T.D. and M.J. wrote the article.
Glossary
- ACD
asymmetric cell division
- MT
microtubule
- MTOC
microtubular organizing center
- PPB
preprophase band
- RNAi
RNA interference
- BY-2
Bright Yellow-2
- PMI
pollen mitosis I
- DAPI
4′,6-diamidino-2-phenylindole
- EGFP
enhanced green fluorescent protein
- YFP
yellow fluorescent protein
- mRFP
monomeric red fluorescent protein
- MS
Murashige and Skoog
References
- Ambrose J.C., Cyr R. (2008). Mitotic spindle organization by the preprophase band. Mol. Plant 1: 950–960 [DOI] [PubMed] [Google Scholar]
- Aravind L., Dixit V.M., Koonin E.V. (1999). The domains of death: Evolution of the apoptosis machinery. Trends Biochem. Sci. 24: 47–53 [DOI] [PubMed] [Google Scholar]
- Bajon C., Horlow C., Motamayor J.C., Sauvanet A., Robert D. (1999). Megasporogenesis in Arabidopsis thaliana L.: An ultrastructural study. Sex. Plant Reprod. 12: 99–109 [Google Scholar]
- Baskin T., Cande W.Z. (1990). The structure and function of the mitotic spindle in flowering plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 277–315 [Google Scholar]
- Bass H.W., Marshall W.F., Sedat J.W., Agard D.A., Cande W.Z. (1997). Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D., Brettschneider R., Lörz H. (1994). Fertile transgenic wheat from microprojectile bombardment of scutellar tissue. Plant J. 5: 299–307 [DOI] [PubMed] [Google Scholar]
- Bisgrove S.R., Henderson D.C., Kropf D.L. (2003). Asymmetric division in fucoid zygotes is positioned by telophase nuclei. Plant Cell 15: 854–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Kurz T. (2006). Degrade to create: Developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development 133: 773–784 [DOI] [PubMed] [Google Scholar]
- Brettschneider R., Becker D., Lörz H. (1997). Efficient transformation of scutellar tissue of immature maize embryos. Theor. Appl. Genet. 94: 737–748 [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., Ye Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Canaday J., Stoppin-Mellet V., Mutterer J., Lambert A.M., Schmit A.C. (2000). Higher plant cells: Gamma-tubulin and microtubule nucleation in the absence of centrosomes. Microsc. Res. Tech. 49: 487–495 [DOI] [PubMed] [Google Scholar]
- Chang M.T., Neuffer M.G. (1994). Chromosomal behavior during microsporogenesis. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 460–475.
- Christensen C.A., King E.J., Jordan J.R., Drews G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10: 49–64 [Google Scholar]
- Clark-Maguire S., Mains P.E. (1994). mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S., Bantin J., Wittich P.E., Kranz E., Lörz H., Dresselhaus T. (2001). ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 25: 103–114 [DOI] [PubMed] [Google Scholar]
- Dieterle M., Thomann A., Renou J.P., Parmentier Y., Cognat V., Lemonnier G., Müller R., Shen W.H., Kretsch T., Genschik P. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41: 386–399 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Amien S., Márton M.L., Strecke A., Brettschneider R., Cordts S. (2005). TRANSPARENT LEAF AREA1 encodes a secreted proteolipid required for anther maturation, morphogenesis, and differentiation during leaf development in maize. Plant Cell 17: 730–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T., Cordts S., Lörz H. (1999). A transcript encoding translation initiation factor eIF-5A is stored in unfertilized egg cells of maize. Plant Mol. Biol. 39: 1063–1071 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Lörz H., Kranz E. (1994). Representative cDNA libraries from few plant cells. Plant J. 5: 605–610 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Srilunchang K.O., Leljak-Levanic D., Schreiber D.N., Garg P. (2006). The fertilization-induced DNA replication factor MCM6 of maize shuttles between cytoplasm and nucleus, and is essential for plant growth and development. Plant Physiol. 140: 512–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D.W. (2007). Straighten up and fly right: Microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr. Opin. Cell Biol. 20: 107–116 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Shaw S.L. (2006). Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Evans M.M.S., Grossniklaus U. (2009). The maize megagametophyte. In Handbook of Maize: Its Biology. J.L. Bennetzen and S.C. Hake, eds. (New York: Springer), pp. 79–104.
- Fabritius A.S., Ellefson M.L., McNally F.J. (2011). Nuclear and spindle positioning during oocyte meiosis. Curr. Opin. Cell Biol. 23: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L.G., Shen Y.P., Feng S.H., Bostick M., Callis J., Hellmann H., Deng X.W. (2005). Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A.E., Cande W.Z. (1999). Nuclear organization and chromosome segregation. Plant Cell 11: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba L.C., Eivers E., Geissert D., Taelman V., De Robertis E.M. (2008). Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc. Natl. Acad. Sci. USA 105: 7732–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., He Y.J., Borchers C., Xiong Y. (2003). Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Geyer R., Wee S., Anderson S., Yates J., Wolf D.A. (2003). BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12: 783–790 [DOI] [PubMed] [Google Scholar]
- Gingerich D.J., Gagne J.M., Salter D.W., Hellmann H., Estelle M., Ma L.G., Vierstra R.D. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Gingerich D.J., Hanada K., Shiu S.H., Vierstra R.D. (2007). Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19: 2329–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens J.F., Phillips R.L. (1976). The nucleolus organizer region of maize (Zea mays L.). Chromosoma 57: 103–117 [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Huang B.-Q., Russell S.D. (1992). Female germ unit: Organization, isolation, and function. Int. Rev. Cytol. 140: 233–292 [Google Scholar]
- Huang B.Q., Sheridan W.F. (1994). Female gametophyte development in maize - microtubular organization and embryo sac polarity. Plant Cell 6: 845–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Lu C., Raharjo E., McNally K., McNally F.J., Mains P.E. (2009). Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis. Dev. Biol. 330: 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirihara J.A. (1994). Selection of stable transformants from Black Mexican Sweet maize suspension cultures. In The Maize Handbook, M. Freeling and V. Walbot, eds. (New York: Springer-Verlag), pp. 690–694.
- Kliwer I., Dresselhaus T. (2010). Establishment of the male germline and sperm cell movement during pollen germination and tube growth in maize. Plant Signal. Behav. 5: 885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz E., Bautor J., Lörz H. (1991). In vitro fertilization of single, isolated gametes of maize mediated by electrofusion. Sex. Plant Reprod. 4: 12–16 [Google Scholar]
- Krohn N.G., Lausser A., Juranić M., Dresselhaus T. (2012). Egg cell signaling by the secreted peptide ZmEAL1 controls antipodal cell fate. Dev. Cell 23: 219–225 [DOI] [PubMed] [Google Scholar]
- Kwon J.E., La M., Oh K.H., Oh Y.M., Kim G.R., Seol J.H., Baek S.H., Chiba T., Tanaka K., Bang O.S., Joe C.O., Chung C.H. (2006). BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 281: 12664–12672 [DOI] [PubMed] [Google Scholar]
- Launholt D., Merkle T., Houben A., Schulz A., Grasser K.D. (2006). Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 18: 2904–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E., Leonhardt N., Eisler H., Parmentier Y., Alioua M., Jacquet H., Leung J., Genschik P. (2011). MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 21: 1116–1128 [DOI] [PubMed] [Google Scholar]
- Li R., Gundersen G.G. (2008). Beyond polymer polarity: How the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9: 860–873 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2004). Microtubules and the shape of plants to come. Nat. Rev. Mol. Cell Biol. 5: 13–22 [DOI] [PubMed] [Google Scholar]
- Lu C., Mains P.E. (2007). The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev. Biol. 302: 438–447 [DOI] [PubMed] [Google Scholar]
- Lüders J., Stearns T. (2007). Microtubule-organizing centres: A re-evaluation. Nat. Rev. Mol. Cell Biol. 8: 161–167 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Lu C.G., Mains P.E., Peter M. (2005). The BTB protein MEL-26 promotes cytokinesis in C. elegans by a CUL-3-independent mechanism. Curr. Biol. 15: 1605–1615 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Tyers M., Peter M. (2007). The AAA-ATPase FIGL-1 controls mitotic progression, and its levels are regulated by the CUL-3(MEL-26) E3 ligase in the C. elegans germ line. J. Cell Sci. 120: 3179–3187 [DOI] [PubMed] [Google Scholar]
- McCarthy E.K., Goldstein B. (2006). Asymmetric spindle positioning. Curr. Opin. Cell Biol. 18: 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy Campbell E.K., Werts A.D., Goldstein B. (2009). A cell cycle timer for asymmetric spindle positioning. PLoS Biol. 7: e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Otegui M., Staehelin L.A. (2000). Cytokinesis in flowering plants: More than one way to divide a cell. Curr. Opin. Plant Biol. 3: 493–502 [DOI] [PubMed] [Google Scholar]
- Paciorek T., Bergmann D.C. (2010). The secret to life is being different: Asymmetric divisions in plant development. Curr. Opin. Plant Biol. 13: 661–669 [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Alandete-Saez M., Bowman J.L., Sundaresan V. (2009). Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J.H., Willems A., Johnson J.L.F., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., Peter M. (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316 [DOI] [PubMed] [Google Scholar]
- Ranganath R.M. (2005). Asymmetric cell divisions in flowering plants - One mother, “two-many” daughters. Plant Biol (Stuttg) 7: 425–448 [DOI] [PubMed] [Google Scholar]
- Sawin K.E., Tran P.T. (2006). Cytoplasmic microtubule organization in fission yeast. Yeast 23: 1001–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B., Benfey P.N. (1999). Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 505–537 [DOI] [PubMed] [Google Scholar]
- Singh M., Goel S., Meeley R.B., Dantec C., Parrinello H., Michaud C., Leblanc O., Grimanelli D. (2011). Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N.A., Singh S.P., Wang M.B., Stoutjesdijk P.A., Green A.G., Waterhouse P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320 [DOI] [PubMed] [Google Scholar]
- Sprunck S., Baumann U., Edwards K., Langridge P., Dresselhaus T. (2005). The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J. 41: 660–672 [DOI] [PubMed] [Google Scholar]
- Sprunck S., Gross-Hardt R. (2011). Nuclear behavior, cell polarity, and cell specification in the female gametophyte. Sex. Plant Reprod. 24: 123–136 [DOI] [PubMed] [Google Scholar]
- Srilunchang K.O., Krohn N.G., Dresselhaus T. (2010). Di-SUMO-like DSUL is required for nuclei positioning, cell specification and viability during female gametophyte maturation in maize. Development 137: 333–345 [DOI] [PubMed] [Google Scholar]
- Stogios P.J., Downs G.S., Jauhal J.J.S., Nandra S.K., Privé G.G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V., Gaillard J., Vantard M. (2002). Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem. J. 365: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Maerki S., Peter M. (2008). E3 ubiquitin ligases and mitosis: Embracing the complexity. Trends Cell Biol. 18: 84–94 [DOI] [PubMed] [Google Scholar]
- Sumara I., Quadroni M., Frei C., Olma M.H., Sumara G., Ricci R., Peter M. (2007). A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev. Cell 12: 887–900 [DOI] [PubMed] [Google Scholar]
- Thomann A., Brukhin V., Dieterle M., Gheyeselinck J., Vantard M., Grossniklaus U., Genschik P. (2005). Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 43: 437–448 [DOI] [PubMed] [Google Scholar]
- Twell D. (2011). Male gametogenesis and germline specification in flowering plants. Sex. Plant Reprod. 24: 149–160 [DOI] [PubMed] [Google Scholar]
- Ueda M., Zhang Z., Laux T. (2011). Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 20: 264–270 [DOI] [PubMed] [Google Scholar]
- Uyttewaal M., Burian A., Alim K., Landrein B., Borowska-Wykręt D., Dedieu A., Peaucelle A., Ludynia M., Traas J., Boudaoud A., Kwiatkowska D., Hamant O. (2012). Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Webb M.C., Gunning B.E.S. (1990). Embryo sac development in Arabidopsis thaliana. I. Megasporogensis, including the microtubular cytoskeleton. Sex. Plant Reprod. 3: 244–256 [Google Scholar]
- Webb M.C., Gunning B.E.S. (1991). The microtubular cytoskeleton during development of zygote, proembryo and free-nuclear endosperm in Arabidopsis thaliana (L.) Heynh. Planta 184: 187–195 [DOI] [PubMed] [Google Scholar]
- Weber H., Bernhardt A., Dieterle M., Hano P., Mutlu A., Estelle M., Genschik P., Hellmann H. (2005). Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Hellmann H. (2009). Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 276: 6624–6635 [DOI] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T.H., Vidal M., Elledge S.J., Harper J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321 [DOI] [PubMed] [Google Scholar]
- Yang W.C., Shi D.Q., Chen Y.H. (2010). Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 61: 89–108 [DOI] [PubMed] [Google Scholar]
- Zhang H., Dawe R.K. (2011). Mechanisms of plant spindle formation. Chromosome Res. 19: 335–344 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Laux T. (2011). The asymmetric division of the Arabidopsis zygote: From cell polarity to an embryo axis. Sex. Plant Reprod. 24: 161–169 [DOI] [PubMed] [Google Scholar]
- Zheng N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zhuang M., et al. (2009). Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]