Photosystem I (PSI) is one of the largest and most complex macromolecular assemblies known in nature. However, the molecular mechanisms of the assembly process of PSI remain largely unknown. This study reports that PSBP-DOMAIN PROTEIN1, a nuclear-encoded chloroplast protein found in the thylakoid lumen, plays an important role in the assembly of the PSI reaction center heterodimer.
Abstract
To gain insights into the molecular details of photosystem I (PSI) biogenesis, we characterized the PsbP-domain protein1 (ppd1) mutant of Arabidopsis thaliana that specifically lacks PSI activity. Deletion of PPD1 results in an inability of the mutant to grow photoautotrophically and a specific loss of the stable PSI complex. Unaltered transcription and translation of plastid-encoded PSI genes indicate that PPD1 acts at the posttranslational level. In vivo protein labeling experiments reveal that the rate of synthesis of PSI reaction center proteins PsaA/B in ppd1 is comparable to that of wild-type plants, whereas the rate of turnover of PsaA/B proteins is higher in ppd1 than in wild-type plants. With increasing leaf age, PPD1 content decreases considerably, while PSI content remains constant. PPD1 is a nuclear-encoded thylakoid lumenal protein and is associated with PSI but is not an integral subunit of PSI. Biochemical and molecular analyses reveal that PPD1 interacts directly and specifically with PsaB and PsaA. Yeast two-hybrid experiments show that PPD1 interacts with some lumenal loops of PsaB and PsaA. Our results suggest that PPD1 is a PSI assembly factor that assists the proper folding and integration of PsaB and PsaA into the thylakoid membrane.
INTRODUCTION
Photosystem I (PSI) is a highly conserved multisubunit pigment-protein complex embedded in the thylakoid membrane of cyanobacteria and photosynthetic eukaryotes. It represents one of the largest and most complex macromolecular assemblies known in nature (Amunts and Nelson, 2009). It catalyzes the final step of photosynthetic electron transport: the oxidation of plastocyanin in the thylakoid lumen and the reduction of ferredoxin in the chloroplast stroma. In higher plants, PSI consists of at least 15 core subunits (PsaA to PsaL and PsaN to PsaP), four different light-harvesting antenna proteins (Lhca1-4), and a large number of cofactors. Among 15 core subunits, the subunits PsaA, PsaB, PsaC, PsaI, and PsaJ are chloroplast encoded, whereas the remaining subunits PsaD-H, PsaK-L, and PsaN-P are nuclear encoded. The two key subunits PsaA and PsaB form a heterodimer that binds a majority of the chlorophyll molecules, the primary electron donor P700 (a chlorophyll a dimer), the primary acceptors A0 (a chlorophyll a), the secondary acceptor A1 (a phylloquinone), and the [4Fe-4S] cluster FX. The terminal acceptors, FA and FB, both [4Fe-4S] clusters, are bound to the PsaC subunit. The crystal structure of PSI from pea (Pisum sativum) at 3.3-Å resolution reveals the presence of 173 chlorophyll molecules, 15 carotenoid molecules, three iron-sulfur clusters, and two phylloquinones (Amunts et al., 2007, 2010). The biogenesis of the PSI complex depends on the coordinated expression of both nuclear and chloroplast genes and the targeting of subunits and cofactors to proper location within the chloroplast. Therefore, the biogenesis of the PSI complex requires a spatially and temporally coordinated assembly process with a highly ordered sequence to ensure the proper incorporation of subunits and the proper association of the various cofactors (Wollman et al., 1999; Hippler et al., 2002; Schöttler et al., 2011).
To understand the mechanisms involved in PSI assembly, a set of protein factors involved in PSI assembly has been identified in cyanobacteria, algae, and higher plants either by screening of photosynthesis-deficient mutants, by reverse genetics, or by a biochemical approach (reviewed in Schöttler et al., 2011). PSI assembly factors can be divided into two functional classes: (1) chaperones that function as a scaffold for the assembly of the protein complex and/or mediate the establishment of protein–protein interactions within the complex, and (2) proteins that are involved in synthesis of cofactors and/or transfer into the nascent complex (reviewed in Schöttler et al., 2011). Among PSI assembly chaperones, HYPOTHETICAL CHLOROPLAST READING FRAME NUMBER3 (Ycf3) and Ycf4, encoded by chloroplast genes, have been demonstrated to play an important role in the assembly of the PSI complex in the alga Chlamydomonas reinhardtii (Boudreau et al., 1997). Ycf3 appears to act as a chaperone that interacts directly and specially with at least PsaA and PsaD during assembly of the PSI complex (Naver et al., 2001). Ycf4 plays an essential role either in the subsequent assembly of the PSI intermediate complex PsaA to PsaF or during formation of the stromal ridge in C. reinhardtii (Ozawa et al., 2009; Schöttler et al., 2011). However, it has been reported recently that, different from in C. reinhardtii, Ycf4 is not essential for photosynthesis in tobacco (Nicotiana tabacum) but is still involved in PSI assembly (Krech et al., 2012). PALE YELLOW GREEN7 (Pyg7) is indispensable for PSI accumulation at posttranslational level in Arabidopsis thaliana (Stöckel et al., 2006). YCF3-INTERACTING PROTEIN1 (Y3IP1), a nucleus-encoded thylakoid protein, cooperates with Ycf3 in the assembly of the stable PSI complex in higher plants (Albus et al., 2010).
In addition, several PSI assembly factors or auxiliary proteins that are involved in PSI cofactor biogenesis and attachment have been identified. Phylloquinone is the only cofactor specific to PSI and not found in other photosynthetic complexes. In Arabidopsis, disruption of phylloquinone synthesis results in drastic reduction or abolishment of the PSI complex (Gross et al., 2006; Lohmann et al., 2006; Kim et al., 2008). However, in C. reinhardtii, loss of phylloquinone has no effect on the accumulation of PSI subunits but leads to a decrease in the plastoquinone pool size and the synthesis of photosystem II (PSII) subunits (Lefebvre-Legendre et al., 2007). Besides phylloquinone synthesis, impairment in the biogenesis of [4Fe-4S] clusters or in the transfer of [4Fe-4S] clusters into targeted proteins may lead to defective PSI accumulation. RUBREDOXIN A is essential for insertion of FX into the PsaA/PsaB heterodimer and for the formation of the stromal ridge in the cyanobacterium Synechocystis sp PCC 6803 (Shen et al., 2002). HIGH CHLOROPHYLL FLUORESCENCE 101 is involved in assembly of [4Fe-4S] cluster–containing complexes in Arabidopsis by transiently binding [4Fe-4S] clusters and functioning as a scaffold for [4Fe-4S] cluster insertion (Lezhneva et al., 2004; Stöckel and Oelmüller, 2004; Schwenkert et al., 2010). In addition, ACCUMULATION OF PHOTOSYSTEM ONE1 was originally identified to affect [4Fe-4S] cluster–containing proteins but is suggested recently to affect chloroplast gene expression by promoting the splicing of chloroplast group II introns (Amann et al., 2004; Watkins et al., 2011).
Although a set of PSI assembly factors has been identified and a model of PSI biogenesis has also been proposed recently (reviewed in Schöttler et al., 2011), the mechanism for PSI assembly is still not well understood compared with the detailed knowledge of the biogenesis of PSII (Rokka et al., 2005; Mulo et al., 2008). Identification of novel factors for PSI biogenesis will provide a deeper understanding of the molecular mechanism for PSI biogenesis. Because PSI acts a light-driven plastocyanin-ferredoxin oxidoreductase by mediating electron transfer from reduced plastocyanin in the lumenal side of thylakoid membrane to oxidized ferredoxin in the stromal side of the chloroplast, PSI assembly factors should exist not only in the stroma side of chloroplasts but also in the lumenal side of thylakoid membrane to ensure proper functioning of PSI. Until now, however, no PSI assembly factors located in the lumenal side of thylakoid membrane have been identified (reviewed in Schöttler et al., 2011).
Here, we report on identification and characterization of a novel nuclear-encoded PSI assembly factor PSBP-DOMAIN PROTEIN1 (PPD1), a thylakoid lumenal protein, which is essential for accumulation of the PSI complex in Arabidopsis. Our results reveal that PPD1 represents a novel factor for PSI assembly that assists the proper folding and integration of PsaB and PsaA into the thylakoid membrane by interacting directly and specifically with some lumenal loops of PsaB and PsaA during the assembly of the PSI complex.
RESULTS
PSI Activity Is Abolished in ppd1
The PsbP is a well-characterized PSII subunit that is required for optimal oxygen-evolving activity in Arabidopsis (Ifuku et al., 2010). Plants contain a family of PsbP-related proteins termed PsbP-like (PPL) and PPD proteins. In Arabidopsis, there are two PPL genes and seven PPD genes (Ifuku et al., 2010). It has been reported that the PPL1 protein is involved in PSII repair process under high light conditions and that the PPL2 protein contributes to NDH complex accumulation and function (Ishihara et al., 2007), suggesting that PsbP-related proteins may be involved in other photosynthetic processes. Recently, it has been suggested that the PPD5 protein is involved in strigolactone biosynthesis (Roose et al., 2011). However, the functions of the remaining six PPD proteins are largely unknown. Thus, we initiated a project to systematically characterize the functions of the other six PPD proteins. Here, the function of the PPD1 protein is characterized. The T-DNA insertion line of At4g15510/PPD1 (ppd1-1) obtained from the Arabidopsis Biological Source Center (ABRC) was identified (see Supplemental Figure 1 online). When grown in soil, homozygous ppd1-1 seedlings were lethal. Cultivation on Suc-containing medium rescued mutant seedlings. However, they were still pale yellowish and growth-retarded compared with wild-type plants and were not able to flower (see Supplemental Figure 1A online).
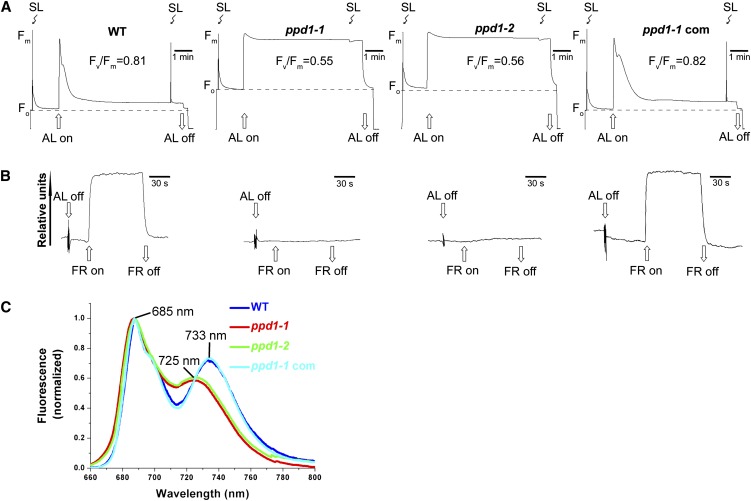
To gain insight into the primary target of PPD1 mutation, noninvasive fluorometric analyses were conducted (Figure 1A). The ratio of variable fluorescence to maximal fluorescence (Fv/Fm), which is an indicator of the maximum efficiency of PSII photochemistry, was reduced in ppd1-1 (0.55 ± 0.05 versus the wild type 0.81 ± 0.01). Photochemical quenching (qP), nonphotochemical quenching (NPQ), and the actual PSII efficiency (ΦPSII) were 0.89 ± 0.01, 0.14 ± 0.01, and 0.70 ± 0.01, respectively, in wild-type plants. However, qP, NPQ, and ΦPSII could not be measured in ppd1-1, which is a behavior typical for mutants blocked in photosynthetic electron transport downstream of PSII (Meurer et al., 1996).
Figure 1.
Spectroscopic Characterization of the Wild Type, the Mutant (ppd1-1 and ppd1-2), and the Complemented Mutant.
(A) Chlorophyll a fluorescence induction analyses. AL, actinic light; SL, saturating light; WT, the wild type.
(B) Redox kinetics of P700 at 820 nm induced by far-red light (FR; 720 nm). Absorbance of P700 at 820 nm is a measurement of the P700 redox state. FR, far-red light.
(C) 77K fluorescence emission spectra of thylakoid membranes after excitation at 436 nm. The fluorescence emission signals were normalized to the PSII emission maximum at 685 nm.
To further investigate whether and how PSI is affected in ppd1-1, the light-induced redox kinetics of PSI were examined. The mutant did not show any detectable absorbance changes of P700 at 820 nm (Figure 1B), indicating that PSI was not functional in ppd1-1. In addition, 77K fluorescence emission spectra were also characterized in ppd1-1 (Figure 1C). The maximum fluorescence emission band peaking at 733 nm in the wild type, characteristic for a functional PSI, was shifted to 725 nm in ppd1-1. As no PSI core complex was detectable but Lhca1-4 proteins were still detectable in ppd1-1 (Figure 2), this blue shift in ppd1-1 could be attributable to the free, uncoupled light-harvesting complex I (LHCI) proteins. Similar observations have been shown in several PSI-deficient Arabidopsis mutants (Stöckel and Oelmüller, 2004; Stöckel et al., 2006; Albus et al., 2010).
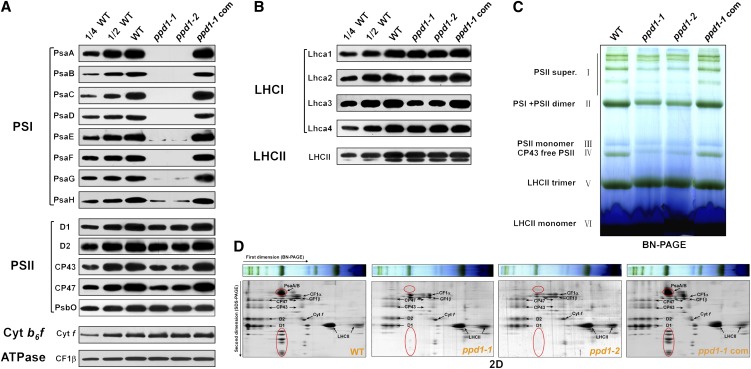
Figure 2.
Analyses of Thylakoid Membrane Proteins from the Wild Type, the Mutant (ppd1-1 and ppd1-2), and the Complemented Mutant.
(A) Immunoblot analysis of thylakoid proteins loaded on the basis of equal total leaf proteins (15 μg). Designations of thylakoid membrane protein complexes and their diagnostic components are labeled at left. Three independent biological replicates were performed and a representative one is shown. WT, the wild type.
(B) Immunoblot analysis of PSI and PSII antenna proteins. Equivalent total leaf proteins (15 μg) were loaded.
(C) BN-PAGE analysis of chlorophyll-protein complexes. Equal thylakoid membrane (10 μg chlorophyll) from the leaves of the wild type, mutants (ppd1-1 and ppd1-2), and the complemented mutant were solubilized by treatment with 1% (w/v) DM and separated by BN-PAGE. The assignments of macromolecular protein complexes of thylakoid membranes indicated at left were identified according to Peng et al. (2006). PSII super, PSII supercomplexes.
(D) 2D BN/SDS-PAGE fractionation of thylakoid membrane protein complexes. After separation in the first dimension in a nondenaturing gel, the protein lanes were subjected to a denaturing 2D gel (2D BN/SDS-PAGE) followed by Coomassie blue staining. The identity of relevant proteins is indicated by arrows. The major PSI proteins, PsaA/B, as well as minor proteins, which appeared only in the wild type, are circled in red.
To verify the observed phenotype in the ppd1-1 mutant is specific for the knockout of PPD1, a second line with a T-DNA insertion in PPD1, ppd1-2, was obtained from the Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat). The ppd1-2 line behaved like the ppd1-1 line with respect to growth and spectroscopic characterization (Figure 1; see Supplemental Figure 1 online). To confirm that the PPD1 gene disruption was responsible for the mutant phenotype, complementation for the ppd1-1 line was performed with At4g15510 genomic DNA. The complemented plant showed normal wild-type behavior (Figure 1). Thus, it can be concluded that disruption of the PPD1 gene is responsible for the observed ppd1 phenotypes.
Because of the nonfunctional PSI, photosynthetic electron transport is blocked; thus PSII becomes the major target of excessive light. To test this possibility, the wild type and the mutant were grown under either higher or lower light conditions and then the changes in Fv/Fm and the amount of subunits of PSII were investigated. Our data show that under higher light conditions, there was a greater decrease in Fv/Fm and the amount of subunits of PSII in the mutant, but no significant changes were observed in Fv/Fm and the amount of subunits of PSII in wild-type plants (see Supplemental Figure 2 online), suggesting that the decrease in Fv/Fm in the mutant may be caused by photoinhibition. Taken together, these data suggest that electron transport through PSI in ppd1 was primarily affected.
ppd1 Mutants Are Specifically Impaired in Accumulation of the PSI Complex
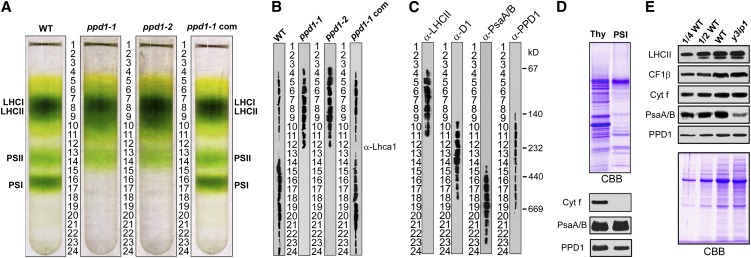
We then investigated the impaired PSI function in ppd1 at the molecular level. Thus, thylakoid proteins were analyzed immunobiochemically using a set of antibodies against diagnostic subunits of four multiprotein complexes in the thylakoid membranes (i.e., PSII, Cyt b6f complex, PSI, and ATP synthase) (Figures 2A and 2B). None of the PSI subunits PsaA, PsaB, PasC, PsaD, and PsaF were detectable, and only traces, if any, of PSI subunits PsaE, PsaG, and PsaH could be detected in ppd1. This was independent of the light intensity in which the seedlings were grown (see Supplemental Figure 2B online). However, considerable PSI antenna proteins were still present in ppd1. Lhca1 and Lhca4 were slightly reduced in ppd1 compared with those in the wild type. Lhca2 and Lhca3 in the mutant decreased to ∼50 and 25% of those in the wild type, respectively. The PSII subunits PsbO and light-harvesting complex II (LHCII) were largely unchanged in the mutant compared with the wild type, whereas there was a considerable decrease in the amount of other PSII subunits (D1, D2, CP43, and CP47). The diagnostic subunits of Cyt b6f complex (Cyt f) and ATP synthase (CF1β) in the mutant accumulated to similar levels compared with the wild type. To investigate the possible changes in structure of thylakoid proteins, chlorophyll-protein complexes were solubilized from thylakoid membranes using dodecyl-β-d-maltopyranoside (DM) and separated by blue native PAGE (BN-PAGE). Figure 2C shows that after the first-dimensional separation in the presence of Coomassie blue dye, six major bands were resolved, representing PSII supercomplexes (band I), monomeric PSI and dimeric PSII (band II), monomeric PSII (band III), CP43 minus PSII (band IV), trimeric LHCII (band V), and monomeric LHCII (band VI) according to our previous study (Peng et al., 2006). The BN-PAGE analysis demonstrates that the amount of PSII and PSI per unit of chlorophyll was significantly lower in thylakoid membranes isolated from ppd1 than that from the wild type. Analyses of the two-dimensional (2D) SDS-urea-PAGE gels after Coomassie blue staining confirms that PSI core proteins were not detectable in ppd1, and the relative amounts of the PSII core subunits CP47, CP43, D1, and D2 were considerably reduced in ppd1, while no significant changes were observed in the amounts of Cyt b6f and ATP synthase (Figure 2D).
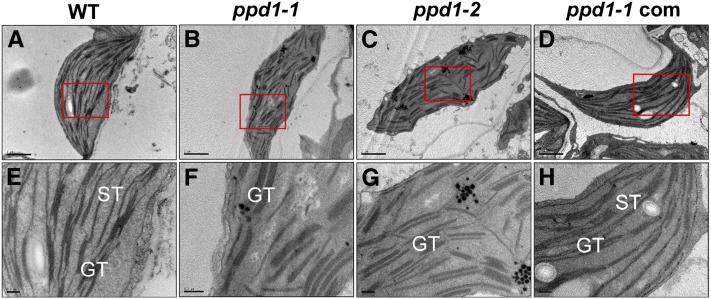
Lack of PSI in ppd1 is accompanied by alterations in chloroplast ultrastructure (Figure 3). Only few stroma lamellae could be observed in ppd1. Moreover, the grana stacks, located in parallel bundles in the wild-type, appeared to be less organized in ppd1 probably due to the lack of connecting stroma lamellae.
Figure 3.
Electron Micrographs of Chloroplasts from the Leaves of the Wild Type, the Mutant (ppd1-1 and ppd1-2), and the Complemented Mutant.
The chloroplast from the wild type (A), ppd1-1 (B), ppd1-2 (C), and the ppd1-1 complemented mutant (D). The close-up view obtained from the wild type (E), ppd1-1 (F), ppd1-2 (G), and the ppd1-1 complemented mutant (H). GT, grana thylakoids; ST, stroma thylakoids; WT, the wild type. Bars = 1 μm in (A) to (D) and 0.2 μm in (E) to (H).
PPD1 Regulates PSI Accumulation at the Posttranslational Level
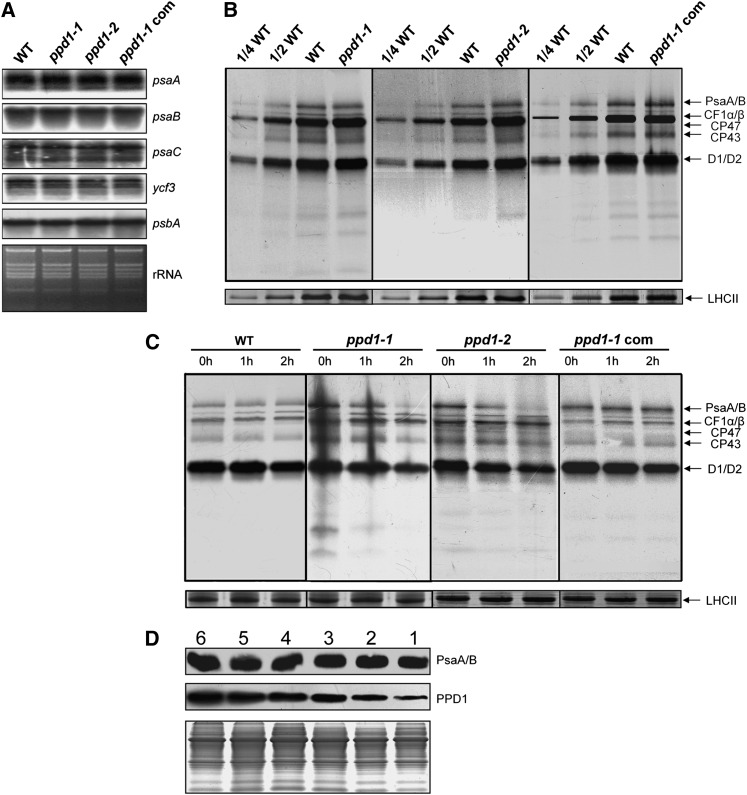
There are alternative possibilities for the lack of PSI accumulation in ppd1. For example, PPD1 may be involved in the transcription of PSI genes, accumulation of stable PSI mRNAs, and/or translation of PSI subunits. Since PPD1 is localized to plastids (Peltier et al., 2002), only the PSI genes in the plastid genome are relevant to any possible function of PPD1 prior to PSI assembly (i.e., gene expression). The plastid genome of higher plants encodes three essential subunits of PSI: PsaA and PsaB (the two reaction center proteins of PSI encoded by the psaA/B operon and translated from a polycistronic mRNA) and PsaC (an essential iron-sulfur protein) (Meng et al., 1988; Takahashi et al., 1991). As plastid-encoded assembly factor Ycf3 is essential for PSI accumulation (Ruf et al., 1997), we also included the ycf3 gene in the analysis of the expression of plastid PSI genes in the ppd1 mutant. We first examined PSI transcript patterns and accumulation in the wild type and ppd1 (Figure 4A). The levels of transcripts of psaA/B, psaC, ycf3, and psbA (encoding the D1 subunit of PSII) were unchanged in ppd1, suggesting that PPD1 acts neither at the transcriptional level nor at the level of mRNA maturation or stability.
Figure 4.
PPD1 Acts on PSI Accumulation at the Posttranslational Level.
(A) RNA gel blot analysis for representative plastid genes of the wild type (WT), ppd1-1, ppd1-2, and the complemented plant. Shown below is the ethidium bromide–stained rRNA prior to blotting to confirm integrity of RNA samples.
(B) In vivo pulse labeling analysis of thylakoid membrane proteins of the wild-type, ppd1-1, ppd1-2, and the complemented mutant. After a 20-min pulse labeling of Arabidopsis young seedlings in the presence of cycloheximide for 30 min to inhibit synthesis of nuclear-encoded proteins, thylakoid membrane proteins were isolated and fractionated by SDS-PAGE and visualized autoradiographically. Nonlabeled LHCII visualized by staining with CBB served as loading control. Four independent pulse-labeling experiments were performed and the same results were obtained each, and a representative one is shown.
(C) Pulse-chase analyses of thylakoid membrane proteins of the wild type, ppd1-1, ppd1-2, and the complemented mutant. A 20-min pulse labeling in Arabidopsis young seedlings in the presence of cycloheximide was followed by a chase of unlabeled Met for 1 or 2 h, respectively. As loading control, nonlabeled LHCII was visualized by staining with CBB. Signal detection was performed as in (B). Three independent experiments were performed, similar results were obtained, and a representative autoradiogram is shown.
(D) Changes in the amounts of PsaA/B and PPD1 proteins during leaf development. Thirty-day-old wild-type plants just prior to flowering were used in this study (see Supplemental Figure 4 online). Shown is a representative developmental series of leaves numbered from old to young (from 1 to 6). Equal loading is shown by Coomassie blue staining of total leaf proteins at the bottom half of the panel.
To investigate the possible involvement of PPD1 in the translation of PSI mRNAs, the association of psaA/B, psaC, ycf3, and psbA with ribosome in the wild type and ppd1 was compared by polysome-loading experiments (see Supplemental Figure 3 online). Total leaf lysates were fractionated, and RNA purified from the gradient fractions was analyzed with probes specific for psaA/B psaC, ycf3, and psbA transcripts. No significant differences in the association with polysomes between the wild type and ppd1 were observed for the psaA/B, psaC, ycf3, and psbA genes, suggesting that PSI mRNA translation proceeds at comparable efficiency and that PPD1 is not a translation factor for plastid genome-encoded PSI mRNAs.
To further investigate whether the lack of PSI accumulation in ppd1 is associated with either impaired protein synthesis or accelerated degradation of PSI component proteins, the synthesis of thylakoid membrane proteins was examined by in vivo pulse labeling experiments. To this end, the leaf proteins were pulse labeled with [35S]Met in the presence of cycloheximide, which inhibits the synthesis of the nuclear-encoded proteins (Figure 4B). Our results show that the rate of synthesis of PSI reaction center PsaA/B proteins remained largely unchanged in ppd1. In addition, the rates of synthesis of the PSII subunits CP43, the D1 and D2 proteins, and the α- and β-subunits of the chloroplast ATP synthase (CF1α/β) remained almost unchanged in ppd1. This indicates that accelerated degradation rather than a block in protein synthesis is responsible for the lack of PSI accumulation in ppd.
The impairment in stability and/or assembly of PSI complex may result in the accelerated degradation of PSI complex, which leads to the absence of PSI reaction center polypeptides in ppd1. To investigate this possibility, pulse labeling for 20 min was followed by a chase with unlabeled cold Met in the presence of cycloheximide to monitor the turnover rates of plastid-encoded proteins (Figure 4C). Our results show that the turnover rate of PsaA/B proteins was higher in ppd1 than in the wild type. Also, the turnover rates of PSII subunits CP43, D1, and D2 were higher in ppd1 than in the wild type. These results suggest that the absence of PSI reaction center polypeptides in ppd1 is due to the impairment in stability and/or assembly of PSI complex.
Differential Accumulation of PSI and PPD1 during Leaf Development
Recently, the relationship between PSI assembly factor Ycf4 and PSI accumulation during tobacco leaf ontogenesis was investigated, and it was found that the amount of Ycf4 decreases strongly while PSI contents remain constant with increasing leaf age (Krech et al., 2012). It is thus suggested that Ycf4 is not required for PSI stability but acts an assembly factor for PSI (Krech et al., 2012). Therefore, we also investigated the relationship between PPD1 and PSI accumulation during leaf ontogenesis to examine whether PPD1 is involved in stability or assembly of PSI. We compared the accumulation of PPD1 with that of the PSI reaction center subunits PsaA/B in a series of leaves harvested from 30-d-old wild-type plants. In these plants, leaf number 1 represents the oldest leaves and leaf number 6 represents the youngest leaves (see Supplemental Figure 4A online). There were no significant differences in chlorophyll contents between these young and old leaves (see Supplemental Figure 4B online), indicating that leaf chlorophyll did not begin to degrade and that senescence did not start yet in these leaves. Figure 4D shows that the accumulation of PsaA/B remained largely unchanged with increasing leaf age. However, the PPD1 protein exhibited an age-dependent decrease in protein accumulation. The accumulation of PPD1 was highest in very young leaves and continuously decreased with leaf age. Since PSI levels remain unchanged with leaf age, this in turn suggests a high stability of the PSI complex. Thus, our results suggest that PPD1 is not required for PSI stability and that PPD1 may act as an assembly factor for PSI.
PPD1 Is a Thylakoid Lumenal Protein Associated Primarily with Stroma-Exposed Thylakoids
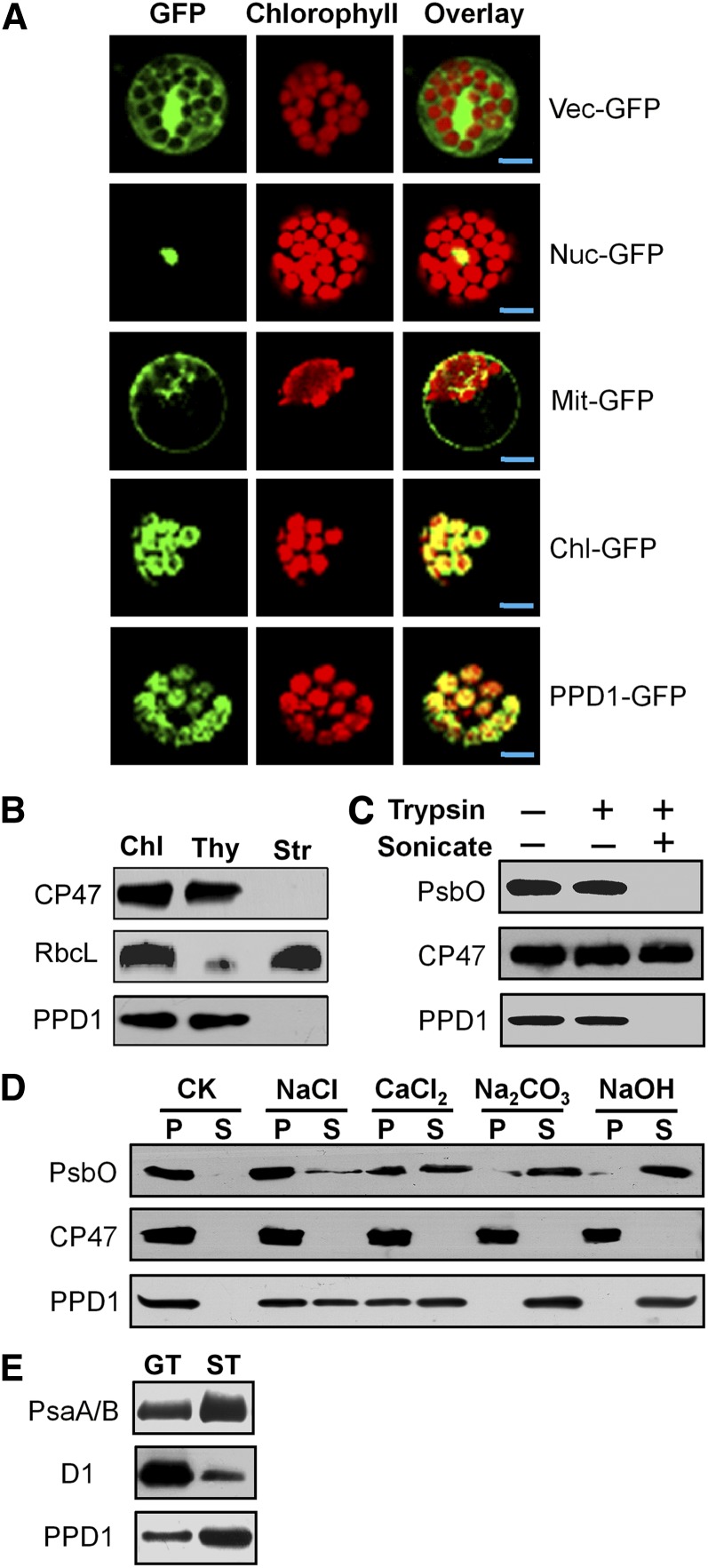
To determine the subcellular localization of PPD1, PPD1–green fluorescent protein (GFP) fusion was introduced into Arabidopsis protoplasts by transient transformation. The analyses of the subcellular localization of the PPD1-GFP fusion proteins by confocal laser scanning microscopy reveal that PPD1 is localized to the chloroplast (Figure 5A). To further determine the localization of PPD1 in the chloroplast, polyclonal antiserum was raised against PPD1 protein. A 20-kD protein was detected in total leaf proteins in wild-type plants, but no signal was detected in total leaf proteins prepared from the mutant (see Supplemental Figure 1F online). The immunoblot analyses of soluble and membrane fractions from Percoll-purified chloroplasts demonstrate that PPD1 is associated with the thylakoid membranes of isolated chloroplasts (Figure 5B). Protease digestion assays of intact thylakoid membranes show that PPD1 was protected from trypsin treatment, as is the oxygen-evolving lumenal PsbO protein of PSII. However, after sonication of the thylakoid membranes, PPD1 and PsbO both were sensitive to trypsin digestion (Figure 5C). These results indicate that PPD1 is a thylakoid lumenal protein, which is in agreement with the results of previous proteomic studies (Peltier et al., 2002). To assess whether PPD1 is a peripheral or intrinsic thylakoid membrane protein, sonicated thylakoid membranes were incubated with chaotropic salts or alkaline known to release the peripheral proteins from thylakoid membranes. A considerable amount of PPD1 was released from thylakoid membranes by treatment with 1 M NaCl or 1 M CaCl2 and effectively removed by 0.2 M Na2CO3 or 0.1 M NaOH (Figure 5D), indicating that PPD1 is peripherally associated with the thylakoid membrane. To further investigate whether PPD1 is localized in grana and/or lamellae thylakoid membrane, the fractions of grana membranes and stroma lamellae were isolated from thylakoid membranes by differential ultracentrifugation. Our results show that PPD1 was detected mainly in the stroma lamellae–enriched fraction (Figure 5E). Taken together, our results reveal that PPD1 is a thylakoid lumenal protein but peripherally and mainly associated with stroma-exposed thylakoid membranes.
Figure 5.
Subcellular Localization and Bindings of PPD1 Protein to the Lumenal Thylakoid Membranes.
(A) Localization of PPD1 protein within the chloroplast by GFP assay. The fluorescence of PPD1-GFP specifically matched with that of Chl-GFP, confirming chloroplast targeting of PPD1 exclusively. Chl-GFP, chloroplast control; Mit-GFP, mitochondrial control; Nuc-GFP, nuclear control; PPD1-GFP, PPD1-GFP fusion; Vec-GFP, control with empty vector. Bars = 5 μm.
(B) PPD1 localizes to the thylakoid membrane fractions. Intact chloroplasts (Chl) were isolated from wild-type leaves and then separated into thylakoid (Thy) membrane and stroma (Str) fractions. Polyclonal antisera were used against the integral membrane protein CP47, the abundant stroma protein ribulose biphosphate carboxylase large subunit (RbcL), and PPD1.
(C) Protease protection assays reveal that PPD1 is a lumenal protein. Thylakoids were subjected to trypsin digestion; when necessary, brief sonication was performed to expose their lumenal parts. PsbO and CP47 served as controls for the thylakoid lumenal protein and the intrinsic thylakoid membrane protein, respectively.
(D) Chaotropic agent washing assays further show that PPD1 is associated with thylakoid membrane. After brief sonication, thylakoids were extracted with various chaotropic agents as indicated, and thylakoid-associated proteins in pellets (P) and proteins released in supernatants (S) were immunodetected. PsbO and CP47 were used as controls for extrinsic membrane protein and integral membrane protein, respectively.
(E) PPD1 resides predominantly in stroma-exposed thylakoids. Thylakoids were solubilized with digitonin and subfractionated by differential ultracentrifugation into grana thylakoids (GT) and stroma lamellae thylakoids (ST) and subjected to immunoblot analysis. The lanes on each gel were loaded on an equal chlorophyll basis.
PPD1 Is Partially Associated with PSI Complexes but Is Not an Integral Part
To test whether PPD1 is part of a multiprotein complex or an integral subunit of PSI, isolated thylakoid membranes were solubilized by DM and separated by Suc gradient sedimentation. Fractions of the gradient were subjected to immunoblot analysis. We observed that the PSI complex was completely missing in ppd1, but PSI antenna proteins were detected as monomers at the top of the Suc gradient (Figures 6A and 6B), indicating that PPD1 has little effect on the expression of the outer antenna but specifically fails to assemble mature PSI core complexes. Probing of gradient fractions with anti-PPD1 antiserum reveals the presence of two distinct protein complexes. While the bulk of the PPD1 protein was present in a complex peaking at ∼200 kD, a considerable fraction was associated with much larger complexes, peaking at ∼500 kD (Figure 6C). Detection of PPD1 in gradient fractions corresponding to much higher masses than free PPD1 indicate that PPD1 may not act alone but seems to be part of a larger assembly complex for PSI. Moreover, these larger complexes were found in fractions that contained the PSI reaction center (represented by PsaA/B) (Figure 6C). In addition, PPD1 was also found in the PSI preparations (Figure 6D). These results demonstrate that PPD1 is associated with PSI.
Figure 6.
Association of the PPD1 Protein with the PSI Complex.
(A) Suc density gradient sedimentation of thylakoid membranes isolated from the leaves of the wild type (WT), ppd1-1, ppd1-2, and the complemented mutant. Thylakoids (1 mg chlorophyll/mL) were solubilized with 1% (w/v) DM and separated by centrifugation in a linear 0.1 to 1 M Suc gradient. After ultracentrifugation, 24 fractions of equal volume were collected (numbered from the top to the bottom). The identities of various chlorophyll-protein complexes were identified by probing with appropriate antibodies. Four independent sedimentations were done with similar results, and the result of a representative experiment is shown.
(B) Distributions of PSI antenna proteins within the gradient fractions. Fractions of the gradient from the wild type, ppd1-1, ppd1-2, and the complemented mutant were immunoblotted with antibody directed against PSI antenna protein Lhca1.
(C) Detection of PPD1-containing complexes after Suc gradient centrifugation of wild-type thylakoids. The fractionated fractions were immunodetected with anti-LHCII, anti-D1, anti-PsaA/B, or anti-PPD1 antisera.
(D) The PSI complex contains PPD1 protein with substoichiometric amounts relative to PSI subunit. PSI complex was isolated and fractionated on 10 to 17% gradient SDS-PAGE and visualized by Coomassie blue (CBB) staining or followed by immunoblotting with anti-Cyt f, anti-PsaA/B, or anti-PPD1 antisera.
(E) Stable accumulation of PPD1 in the PSI-deficient mutant y3ip1. Equivalent total leaf protein samples were loaded prior to immunoblotting with anti-PPD1. In the bottom half of the panel, a Coomassie blue–stained gel image of the same samples is shown.
We further explored whether PPD1 is a subunit of PSI. To this purpose, the content of PPD1 was compared between the wild type and PSI deficiency mutant y3ip1 (Albus et al., 2010). Our results show that the content of the PPD1 protein in y3ip1 was comparable to that in the wild type (Figure 6E), suggesting that PPD1 is independent of the accumulation of PSI complexes and not an integral part of PSI complexes. Taken together, our results indicate that PPD1 is partially associated with PSI complexes but not a subunit of the PSI complex.
PPD1 Interacts with PSI Reaction Center PsaA and PsaB Proteins
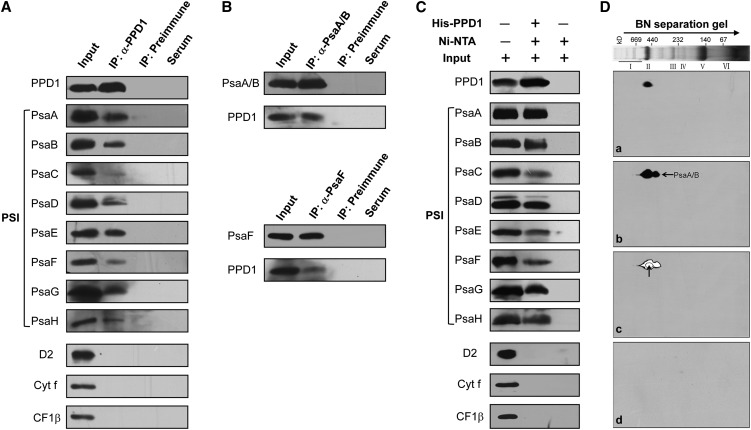
The above results show that PPD1 is required for the accumulation of PSI and is associated with the PSI complex. It is thus expected that it may interact with some subunits of the PSI complex. To test this possibility, coimmunoprecipitation assays were performed on DM-solubilized thylakoid membranes. Figures 7A and 7B show that the PSI complex could be precipitated by anti-PPD1 antiserum. By contrast, the proteins D2, Cyt f, and CF1β were not precipitated under such conditions. In addition, pull-down assays provided further support for such interactions between PPD1 and the PSI complex (Figure 7C). Additional evidence came from the results obtained by immunoblotting with anti-PPD1 and anti-PsaA/B antisera of 2D BN/SDS-PAGE gels, showing colocalization of PPD1 with the PSI core complex both in the wild type and y3ip1 mutant (see Supplemental Figure 5 online).
Figure 7.
Assays of the Interaction between PPD1 Protein and the PSI Complex.
(A) Coimmunoprecipitation assays showing the interactions between PPD1 and the PSI complex. Solubilized thylakoid membranes were incubated with preimmune serum and an excess of anti-PPD1 antiserum. The immunoprecipitates were probed with specific antibodies, as indicated to the left. A sample of thylakoid membranes equivalent to 2 µg of chlorophyll was loaded simultaneously (input).
(B) PPD1 can be reciprocally coimmunoprecipitated with anti-PsaA/B or anti-PsaF antisera. Input, loading of thylakoid membranes equivalent to 2 µg chlorophyll.
(C) Pull-down assays. His-PPD1 recombinant proteins coupled to nickel-nitrilotriacetic acid (Ni-NTA) resin were incubated with solubilized thylakoid membranes, and the bound proteins eluted were analyzed immunobiochemically employing the antibodies as in (A). Input denotes loading of thylakoid membranes equivalent to 2 µg of chlorophyll.
(D) PPD1 interacts directly with PsaA/B proteins revealed by overlay assays. Thylakoid membrane proteins were resolved by 2D BN/SDS-PAGE and transferred to nitrocellulose membranes and immunodetected. (a) The nitrocellulose membrane was incubated with His-PPD1 fusion proteins and subsequently immunodetected with anti-His tag. (b) After blotting, the membrane was subjected to immunodetection with anti-PsaA/B as indicated by the arrow. (c) Superposition of (a) and (b) as indicated by the arrow. (d) As a negative control, after blotting, the membrane without incubation with His-PPD1 fusion protein was subjected to immunodetection with anti-His tag antibody. Positions of molecular mass markers are indicated at the top of the BN-PAGE image.
Overlay assays were performed further to explore which subunits of PSI complexes interact with PPD1. The chlorophyll-protein complexes were separated by BN-PAGE followed by 2D fractionations and then resolved proteins were blotted onto nitrocellulose membranes. The membranes were first incubated with purified His-PPD1 fusion protein and the possible proteins binding to PPD1 were subsequently detected by immunoblotting with the His tag–specific antibody. Figure 7D shows that one spot was detected to comigrate with the distinct signal of the PsaA/B proteins in the PSI complex as identified by probe of a duplicate membrane with anti-PsaA/B antibodies. As a negative control, probing the nitrocellulose membrane without addition of His-PPD1 with anti-His tag antibody did not yield any signal. This suggests that PPD1 interacts with only PsaA/B proteins of PSI complexes.
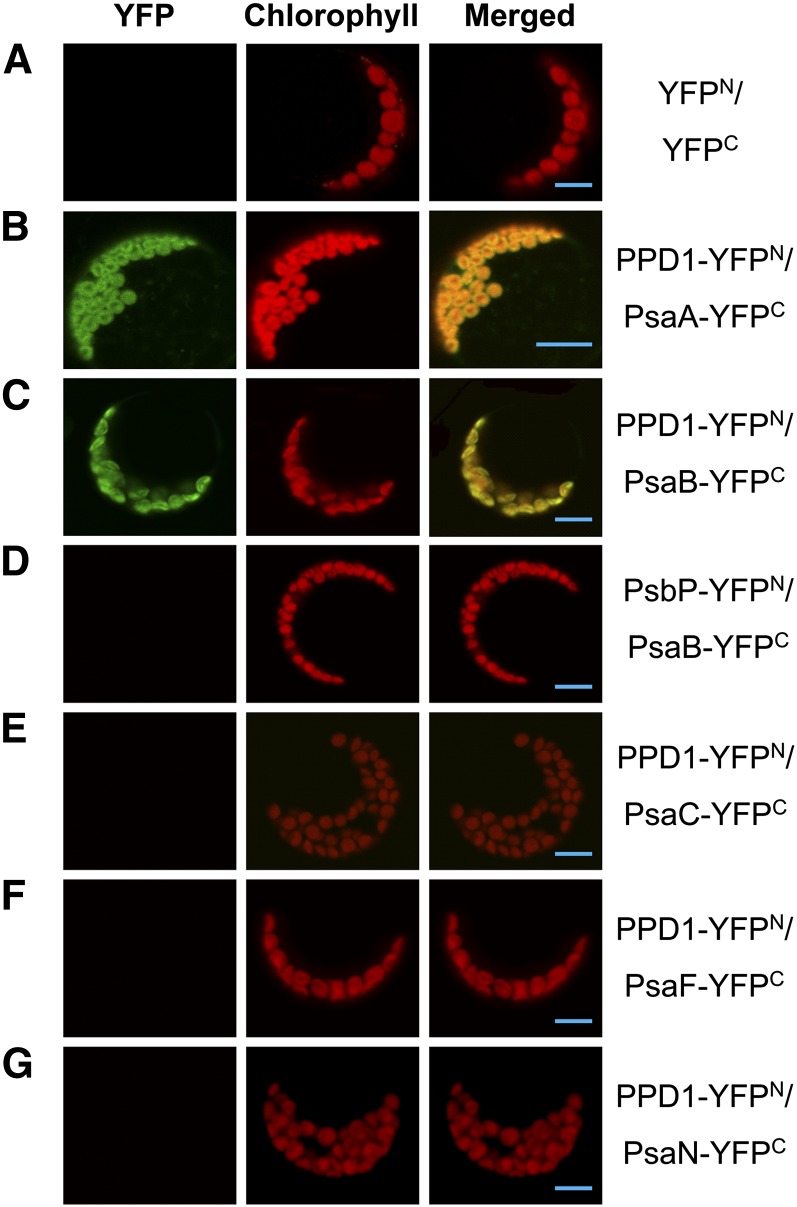
To determine whether PPD1 interacts with PsaA or PsaB alone or both, bimolecular fluorescence complementation (BiFC) analyses were performed. Our results reveal that PPD1 interacts directly with both PsaA and PsaB, but not with PsaC, PsaF, or PsaN (the only subunit located entirely in the thylakoid lumen), and that PsbP does not interact with PsaB (Figure 8). Far protein gel blot assays also demonstrate that PPD1 interacts with both PsaA and PsaB (see Supplemental Figure 6 online). Thus, our results clearly show that PPD1 interacts directly and specifically with both PsaA and PsaB.
Figure 8.
In Vivo Interactions of PPD1 with PsaA and PsaB Examined by BiFC.
(A) YFPN and YFPC transiently coexpressed in protoplasts as a negative control, showing no signal.
(B) Cotransformation of PPD1-YFPN and PsaA-YFPC shows reconstitution of YFP as an indicator of interaction.
(C) Coexpression of PPD1-YFPN and PsaB-YFPC shows YFP, indicative of interaction.
(D) PsbP-YFPN and PsaB-YFPC coexpression showing no signal.
(E) PPD1-YFPN and PsaC-YFPC cotransformation showing no signal.
(F) PPD1-YFPN and PsaF-YFPC cotransformation showing no signal.
(G) PPD1-YFPN and PsaN-YFPC cotransformation showing no signal.
Bars = 5 μm
Localization of PsaA- and PsaB-Interacting Domains with PPD1
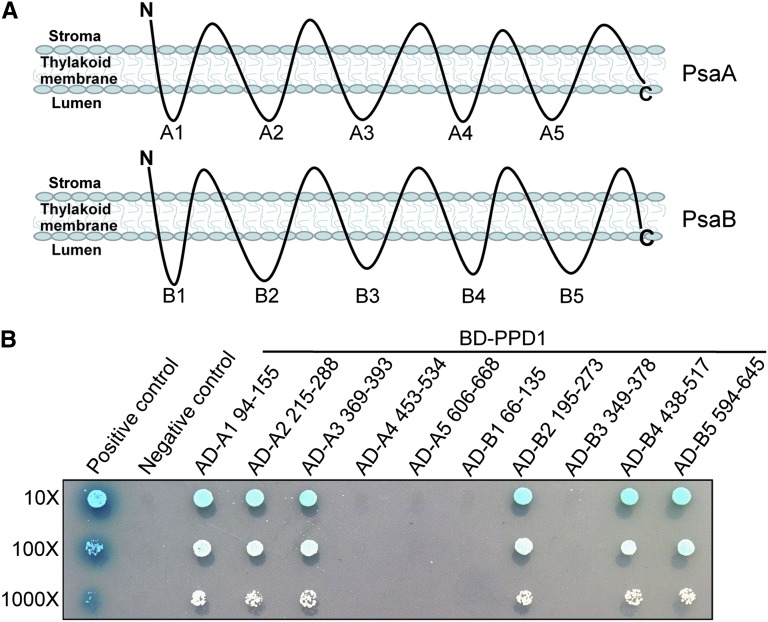
Since our results show that PPD1 is a thylakoid lumenal protein (Figure 5), we further examined which lumenal domains of PsaA and PsaB are responsible for the direct interactions with PPD1 by yeast two-hybrid assay. The lumenal loops of PsaA were designated as A1 to A5, respectively. Similarly, the lumenal loops of PsaB were designated as B1 to B5, respectively (Figure 9A). Our results show that PPD1 interacted with three loops of PsaA corresponding to A1 (amino acids 94 to 155), A2 (amino acids 215 to 288), and A3 (amino acids 369 to 393), but not with A4 (amino acids 453 to 534) and A5 (amino acids 606 to 668). PPD1 also interacted with three loops of PsaB corresponding to loop B2 (amino acids 195 to 273), B4 (amino acids 438 to 517), and B5 (amino acids 594 to 645), but not with B1 (amino acids 66 to 135) and B3 (amino acids 349 to 378) (Figure 9B). These data demonstrate that PPD1 interacts specifically with distinct lumenal loops of PsaA and PsaB.
Figure 9.
Mapping of PsaA and PsaB-Interacting Lumenal Domains with PPD1 Probed by Yeast Two-Hybrid Experiments.
(A) Schematic illustration of the thylakoid topology of PsaA or PsaB employed to detect candidate interaction lumenal loops. PsaA and PsaB each harbors five lumenal domains denoted as A1 to A5 and B1 to B5, respectively. Note that the diagram was not drawn in scale.
(B) Interaction mapping to distinct lumenal domains of PsaA and PsaB. PPD1 was fused to bait construct, PsaA and PsaB lumenal side fragments were fused to prey constructs. AD, the Gal4 DNA activation domain; BD, the Gal4 DNA binding domain. Numbers indicate amino acid positions.
Since plant PsaF is a transmembrane protein with a large N-terminal lumenal domain, containing an additional 18–amino acid residue extension but not found in cyanobacteria and because this domain is close to the lumenal loops of PsaB (Amunts et al., 2010), we further investigated whether there is an interaction between PPD1 and the lumenal domain of PsaF by yeast two-hybrid assay. Our results show that there was no interaction between PPD1 and the lumenal domain of PsaF (see Supplemental Figure 7 online).
DISCUSSION
PSI represents one of the largest and most complex macromolecular assemblies in nature. The biogenesis of PSI requires the coordinated assembly of nuclear-encoded and chloroplast-encoded protein subunits as well as the insertion of nearly 200 cofactors (Amunts et al., 2010). Although a set of factors involved in PSI assembly, cofactor biogenesis, and cofactor attachment has been identified, we are still far from understanding the molecular mechanisms underlying the assembly of PSI (reviewed in Schöttler et al., 2011). Identification of new assembly factors will obviously provide new insights into the mechanism of PSI biogenesis. Moreover, all factors involved in PSI assembly, cofactor synthesis, and cofactor attachments reported until now are either potential intrinsic thylakoid membrane proteins or are localized in the stromal side of thylakoid membranes. The electron transport of PSI is involved not only in the stroma side of thylakoid membranes but also in the lumenal side of thylakoid membranes. Understandably, some factors involved in the assembly of PSI should be localized in the lumenal side of thylakoid membrane in order to ensure efficient functions of PSI. Here, we report a novel nuclear-encoded protein, PPD1, a thylakoid lumenal protein, which is essential for PSI assembly in higher plants. The functions of PPD1 during the assembly of PSI are discussed as follows.
Chlorophyll fluorescence measurements show that Fv/Fm was reduced in ppd1 (Figure 1A). The decrease in Fv/Fm is due to secondary effects resulting from lesions within PSI (see Supplemental Figure 2 online). In addition, we found that no qP and NPQ were detected in ppd1, indicating that the electron flow downstream of PSII is inhibited. Determination of absorbance changes of P700 at 820 nm demonstrates that PSI in ppd1 is not functional (Figure 1B). Moreover, immunoblot analysis confirmed that essential subunits of PSI complex are either absent or severely reduced in ppd1 (Figure 2). Furthermore, the blue shift in the 77K fluorescence emission spectrum of the mutant (Figure 1C) indicates that the 77K fluorescence emission signals are attributable to the free, uncoupled LHCI proteins as no PSI core complex was detectable (Figure 2). This blue shift was also observed in other PSI-deficient mutants (Haldrup et al., 2000; Stöckel and Oelmüller, 2004; Stöckel et al., 2006; Albus et al., 2010). Finally, loss of PSI in the mutant had strong effects on the organization of the thylakoid membrane. Stacking of grana thylakoids is comparable to the wild type, whereas stroma thylakoids are barely detectable (Figure 3). Taken together, our data on spectroscopy measurements, biochemical analyses, and chloroplast ultrastructure observation clearly reveal a specific impairment in PSI accumulation in the ppd1 mutant, thus providing a straightforward explanation for its pigment-deficient and growth-retarded phenotypes (see Supplemental Figure 1A online).
Inactivation of PPD1 leads to a loss in PSI accumulation and an inability of the mutant to grow photoautotrophically (see Supplemental Figure 1A online). Our analyses of transcript patterns, RNA accumulation levels, and translation of plastid PSI genes exclude the possibility that PPD1 represents a specific transcription or translation factor for plastid-encoded PSI genes (Figure 4A; see Supplemental Figure 3 online). Furthermore, in vivo labeling experiments of thylakoid proteins revealed that major subunits of PSI reaction center are synthesized in ppd1 (Figure 4B), indicating that PPD1 is unlikely to be involved in the synthesis of PSI subunits. Therefore, our results indicate that PPD1 acts at the posttranslational level. The pulse-chase analyses confirmed that PPD1 appears to be required for the stability and/or assembly of the PSI complex (Figure 4C).
To investigate whether PPD1 is involved in the stability and/or assembly of the PSI complex, we examined the relationship between the expression of PPD1 and PSI accumulation during leaf development. We observed that PSI was still present and maintained in high amounts in mature and old leaves, whereas the level of PPD1 declined continuously with leaf aging (Figure 4D; see Supplemental Figure 4 online). A similar result has been reported recently that with increasing leaf age in tobacco, the levels of Ycf4 and Y3IP1, two PSI assembly factors, decrease significantly, while PSI contents remain unchanged (Krech et al., 2012). The high requirement for PSI synthesis in young developing leaves is expected to coincide with a high demand for the PSI assembly factor, thus explaining the high abundance of PPD1 in very young leaves (Figure 4D). Accumulation of a high amount of PSI in old leaves accompanied by a significant decrease of PPD1 suggests that the function of PPD1 is involved in PSI assembly rather than in PSI stability. In addition, it was interesting to observe that accumulation of the PPD1 protein was not reduced in the PSI-deficient y3ip1 mutant (Figure 6E). This demonstrates that PPD1 accumulated in the thylakoid membrane independently of PSI. This finding again suggests that PPD1 plays an important role as an assembly factor in PSI biogenesis. Moreover, we observed that in contrast with PSI core subunits, there were substantial amounts of PSI antenna proteins in the ppd1 mutant (Figures 2A and 2B), suggesting that PSI antenna proteins accumulate independently of the PSI reaction center (Stöckel and Oelmüller, 2004; Stöckel et al., 2006). Thus, our results indicate that the function of PPD1 in PSI biogenesis is involved in the assembly of the core complex.
If PPD1 is required for PSI assembly, it is likely that it interacts with some of the PSI subunits. Suc gradient sedimentation and 2D BN/SDS-PAGE coupled with immunoblots show that a subfraction of PPD1 correlated in migration with PSI, suggesting that PPD1 is associated with the PSI complex (Figure 6C; see Supplemental Figure 5 online). The presence of PPD1 in isolated PSI complexes further confirmed that PPD1 is associated with PSI complex (Figure 6D). The pull-down and coimmunoprecipitation assays provide further evidence for an interaction of PPD1 with PSI complex (Figures 7A to 7C). No interaction between PPD1 and the thylakoid proteins D2 of PSII, Cyt f of Cyt b6f, and CF1β of ATPase was observed, suggesting that the observed interactions between PPD1 and the PSI complex was specific (Figures 7A and 7C). Protein overlay and BiFC experiments show that PPD1 interacts directly and specifically with PsaA and PsaB in PSI complexes (Figures 7D and 8). Yeast two-hybrid analyses further reveal that PPD1 interacts specifically with distinct lumenal loops of PsaA and PsaB (Figure 9).
A series of studies have shown that PSI in cyanobacteria, algae, and plants can be assembled and remains functional in vivo without each of these subunits, PsaE, PsaF, PsaH, PsaI, PsJ, PsaK, PsaL, and PsaN (Schöttler et al., 2011). PsaB and PsaA are strongly coupled and PsaB could act as an anchor protein during PSI assembly (Wollman et al., 1999). The formation of the heterodimeric core PsaA/PsaB initiates the assembly of PSI and provides the scaffold for other subunits of PSI (Rochaix, 1997; Redding et al., 1999). In this regard, it is conceivable that the stable formation of the heterodimer PsaA/PsaB regulates the cooperated stable assembly of the entire PSI. In this study, our results show that inactivation of PPD1 resulted in the complete loss of PsaA and PsaB, thus leading to loss of PSI accumulation and PSI function (Figures 1 to 3). Our results also show that PPD1 specifically interacts with PsaA and PsaB (Figures 7 to 9; see Supplemental Figure 6 online). Moreover, we observed that the turnover rates of PsaA/B proteins were accelerated in ppd1 relative to the wild type (Figure 4C). These results suggest that PPD1 plays an essential role in the formation of the PSI reaction center heterodimer PsaA/PsaB. Based on the results in this study, we tentatively propose that PPD1 assists the proper folding and integration of PsaB into the thylakoid membrane and that it may also participate in the subsequent integration of PsaA and perhaps of some of the other subunits associated with PSI core complex.
PSI catalyzes the last step of the light-driven photosynthesis electron transport, the oxidation of the soluble electron carrier plastocyanin located in the lumenal side of thylakoid membrane and the reduction of ferredoxin located in the stromal side. The oxidized P700 chlorophyll a dimer is reduced by plastocyanin, which binds to PSI via interactions with PsaB and the lumenal domain of PsaF. It has been shown that PsaF is required for efficient plastocyanin oxidation (Haldrup et al., 2000). Moreover, the proper orientation of the plastocyanin binding PsaF region is stabilized by the lumenal loop of PsaB (Sommer et al., 2002). Considering that PPD1 is a thylakoid lumenal protein (Figure 5) and interacts with the specific lumenal loops of PsaB but not with the lumenal domain of PsaF (Figures 8 and 9; see Supplemental Figure 7 online), we propose that PPD1 may also play an important role in assisting the proper folding of PsaB to ensure proper interactions with plastocyanin and PsaF, thus maintaining the oxidation of plastocyanin in the thylakoid lumen.
PPD1 belongs to the PsbP protein family. This family consists of 11 members, two authentic PsbP proteins, and nine PsbP homologs. PsbP homologs are classified into two PPL and seven PPD proteins (Ifuku, et al., 2010). It seems that PPL proteins have functions that are different from those of PsbP. PPL1 functions in the efficient repair of PSII after photodamage, whereas PPL2 is a novel thylakoid lumenal factor required for the assembly of the NAD(P)H dehydrogenase complex (Ishihara et al., 2007). Although it has been reported recently that PPD5 may have an analogous function with respect to the strigolactone biosynthesis pathway (Roose et al., 2011), the molecular functions of other PPD proteins in higher plants remain unknown. In this study, our results revealed the function of PPD1 that plays an essential role in PSI assembly in Arabidopsis. The PPD1 protein is conserved in green algae and higher plants, and a putative homolog has not been identified in cyanobacteria (Ifuku, et al., 2010), suggesting that PPD1 is newly evolved in photosynthetic eukaryotes. Thus, our results suggest that PPD1 is essential for PSI biogenesis in the thylakoid membrane in higher plants and probably not required for PSI assembly in photosynthetic prokaryotes.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana (ecotype Columbia) T-DNA insertion line ppd1-1 (SALK_143493C) and ppd1-2 (GABI_853A11) were obtained from the ABRC and the GABI-KAT collection, respectively. The precise T-DNA insertion site was confirmed and sequenced by PCR (see Supplemental Figure 1 online; for primers used, see Supplemental Table 1 online). The homozygous mutants were maintained through heterozygous plants. The wild type and mutants (ppd1-1, ppd1-2, and y3ip1) were grown on Murashige and Skoog (MS)–supplemented medium containing 3% Suc and 0.8% agar in a growth chamber under short-day conditions (12-h-light/12-h-dark cycle) with a photon flux density of 50 µmol m−2 s−1 at 22°C. To ensure synchronized germination, the seeds were incubated in darkness for 2 d at 4°C prior to sowing. For growth on agar plates, seeds were surface sterilized and sown on MS medium.
To test the effects of light intensity on the stability of thylakoid membrane proteins, the wild type and ppd1 were grown in parallel under low-light (10 µmol m−2 s−1), moderate-light (50 µmol m−2 s−1), and high-light (120 µmol m−2 s1) conditions before protein extraction.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence was measured using a PAM-2000 portable chlorophyll fluorometer (Heinz Walz). After a dark adaptation for 30 min, minimum fluorescence (Fo) was determined by weak red light. Maximum fluorescence of dark-adapted state (Fm) was measured during a subsequent saturating pulse of white light (8000 μmol m−2 s−1 for 0.8 s). The leaf was then continuously illuminated with actinic light at an intensity of 50 μmol m−2 s−1 (which is equivalent of growth light intensity) for ∼5 min. The steady state fluorescence (Fs) was thereafter recorded, and a second saturating pulse of white light (8000 μmol m−2 s−1 for 0.8 s) was imposed to determine the maximum fluorescence level in the light-adapted state (Fm′). The actinic light was removed, and the minimal fluorescence level in the light-adapted state (Fo′) was determined by illuminating the leaf with a 3-s pulse of far-red light. The maximal efficiency of PSII photochemistry in the dark-adapted state and the actual PSII efficiency were calculated as Fv/Fm = (Fm − Fo)/Fm and ΦPSII = (Fm′ − Fs)/Fm′, respectively. The qP coefficient and NPQ were calculated as qP = (Fm′ − Fs)/(Fm′ − Fo′) and NPQ = (Fm/Fm′) − 1, respectively.
P700 Absorbance Changes
The light-induced in vivo absorbance changes of P700 at 820 nm were measured using the PAM chlorophyll fluorometer equipped with an ED 800T emitter-detector unit (Walz), and the measurements were performed according to Meurer et al. (1996). Absorbance changes induced by saturating far-red light (720 nm) represent the relative amounts of photooxidizable P700.
77K Fluorescence Emission Spectra
Fluorescence emission spectra of thylakoid membranes (10 μg chlorophyll mL−1) at 77K were recorded from 600 to 800 nm after excitation at 436 nm using the spectrofluorometer (Hitachi F-4500). The spectra distributions of the emission monochromator and detector (photomultiplier) were corrected by the program designed for the model F-4500 fluorescence spectrophotometer by Hitachi Company.
Transmission Electron Microscopy
Transmission electron microscopy was performed basically according to our previous study (Peng et al., 2006). Leaves from 3-week-old plants maintained on an MS-supplemented plate were collected. Micrographs were taken using a transmission electron microscope (JEM-1230; JEOL).
Isolation of Chloroplasts, Total Thylakoid Membranes, and Soluble Proteins
Chloroplasts and total thylakoid membranes and soluble proteins were isolated from 3-week-old plants according to Stöckel and Oelmüller (2004).
Preparations of Intact Thylakoid Membranes, Grana-Enriched Thylakoid Membranes, Stroma Lamellae–Enriched Thylakoid Membranes, and PSI Complexes
Intact thylakoid membranes were prepared according to our previous study (Peng et al., 2006). Subfractionation of grana-enriched and stroma lamellae–enriched thylakoid was performed as described by Lu et al. (2011). The isolation of PSI complexes was performed according to Jensen et al. (2000), and then isolated PSI complexes were fractioned by 10 to 17% gradient SDS-PAGE according to Naver et al. (2001).
Immunoblot, SDS-PAGE, and BN-PAGE Analyses
For immunoblot analysis, total protein samples of seedling leaves were prepared as previously described (Martínez-García et al., 1999). Immunoblotting was performed according to standard techniques by probing with specific antibodies after electroblotting SDS-PAGE or Tricine-SDS-PAGE–separated proteins (Schägger, 2006) onto nitrocellulose membranes (GE Healthcare), and signals from secondary conjugated antibodies were visualized by the enhanced chemiluminescence method. BN-PAGE was performed according to our previous study (Peng et al., 2006). The PsaA, PsaB, PsaC, PsaD, PsaE, PsaF, PsaG, PsaH, and Lhca1-4 antibodies were purchased from Agrisera. All other antibodies were produced in our laboratory (Peng et al., 2006).
In Vivo Pulse-Chase Labeling of Chloroplast Proteins
In vivo protein labeling was performed essentially as previously described by Meurer et al. (1998). For pulse labeling, primary leaves from 12-d-old young seedlings were preincubated in 20 µg mL−1 cycloheximide for 30 min prior to radiolabeling with 1µ Ci µ−1 [35S]Met (specific activity >1000 Ci mmol; Amersham Pharmacia Biotech) in the presence of cycloheximide for 20 min at 22°C. Pulse labeling of the leaves was followed by a chase in the presence of 1 mM cold unlabeled Met. After labeling, the thylakoid membranes were isolated and the proteins were subjected to SDS-PAGE before autoradiography.
RNA Gel Blot and Polysome Association Analyses
Total RNA was extracted from fresh leaf tissues using Trizol reagent and separated on a 1.0% (w/v) agarose-formaldehyde gel, blotted to positively charged nylon membrane (Amersham Pharmacia Biotech), and hybridized with 32P-labeled cDNA probes prepared according to our previous study (Peng et al., 2006) (for primers used, see Supplemental Table 1 online). Polysomes were isolated from leaf tissues according to Barkan (1988). RNA in each fraction was isolated, separated, and transferred onto nylon membranes, which were probed following high-stringency hybridization and washing; the blots were exposed to x-ray film for 1 to 3 d.
Antiserum Production
For the production of polyclonal antibodies against PPD1, the nucleotide sequences encoding the soluble part of PPD1 (amino acids 101 to 287 corresponding to nucleotide positions 301 to 861 of the PPD1 gene) were amplified from cDNA (for primers used for the His-tag construct, see Supplemental Table 1 online). The resulting DNA fragment was cleaved with EcoRI and HindIII and fused in frame with the N-terminal His affinity tag of pET28a (Novagen), and the resulting plasmids were transformed into Escherichia coli strain BL21 (DE3). The fusion protein was purified on a nickel-nitrilotriacetic acid agarose resin matrix and raised in rabbit with purified antigen.
Subcellular Localization of GFP Proteins, BiFC, and Immunolocalization Assays
For subcellular localization of GFP protein, full-length PPD1 was subcloned into the pBI121-35s-sGFP vector with the GFP at C terminus. The constructs for nuclear, chloroplast, and mitochondria localization were constructed according to our previous study (Cai et al., 2009). The resulting fusion constructs and the empty vector were transformed into the protoplasts of Arabidopsis. BiFC assays were performed according to Walter et al. (2004). Full-length cDNAs were cloned into pUC-SPYNE and pUC-SPYCE, and plasmids were cotransformed into protoplasts (for primers used for fusion constructs, see Supplemental Table 1 online). Yellow fluorescent protein (YFP) was imaged using a confocal laser scanning microscope (LSM 510 Meta; Zeiss). The intracellular localization of PPD1 was determined essentially according to Lennartz et al. (2001). The protease protection assays were performed essentially as previously reported by Meurer et al. (1998).
Suc Gradient Fractionation of Thylakoid Membranes
Intact thylakoid membranes (1 mg chlorophyll mL−1) were washed twice with 5 mM EDTA, pH 7.8, centrifuged (5 min, 10,000g, 4°C), resuspended in double-distilled sterile water, and then solubilized with 1% DM for 10 min on ice and centrifuged (5 min, 20,000g, 4°C). Aliquots of the supernatant were loaded onto a linear 0.1 to 1 M Suc gradient in 5 mM MgCl2, 10 mM NaCl, 0.06% (w/v) DM, and 25 mM MES-NaOH, pH 5.7. The gradient was centrifuged for 22 h at 160,000g at 4°C on an SW40 Ti rotor (Beckman). After centrifugation, 24 fractions were collected from the top to the bottom of the gradient. For further analysis, the proteins in each fraction were separated by SDS-PAGE and characterized by immunoblot analysis.
Pull-Down, Coimmunoprecipitation, and Protein Overlay Assays
Coimmunoprecipitation, pull-down, and protein overlay assays were performed basically according to our previous studies (Sun et al., 2007; Ouyang et al., 2011).
Yeast Two-Hybrid Assay
Yeast two-hybrid analyses were performed using the GAL4 two-hybrid system according to the manufacturer’s instructions (TaKaRa). PPD1 coding region was cloned in frame downstream of the DNA binding domain in the bait vector pGBKT7, and the coding fragments of the candidate interaction proteins were fused in frame to prey vector pGADT7 containing DNA activation domain. The primers used for yeast two-hybrid assays are listed in Supplemental Table 1 online. Direct interaction of two proteins was investigated by cotransformation of the respective plasmid constructs into yeast reporter strain Y187. The positive clones were arrayed with serial dilutions on SD/-Leu/-Trp/-His selection medium containing 40 μg mL−1 X-α-Gal for testing the LacZ gene activity.
Complementation of the ppd1 Mutant
To complement ppd1-1, the genomic DNA was subcloned into the pCAMBIA1301 vector (Cambia) under the control of cauliflower mosaic virus 35S promoter. The resultant construct was transformed into Agrobacterium tumefaciens GV3101 strain and introduced into ppd1 heterozygous plants by the method of Zhang et al. (2006). Individual transgenic plants were selected on the basis of resistance to 50 mg L−1 hygromycin in half-strength MS medium and 0.8% agar. The resistant ones were transferred to soil and grown in the growth chamber to produce seeds. The success of complementation was confirmed by immunoblot analyses, measurements of chlorophyll fluorescence, P700 absorbance changes, chloroplast ultrastructure, immunoblot and BN-PAGE analyses, RNA gel blot analysis, in vivo pulse labeling, and polysome loading experiments.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: PPD1 (AT4G15510), ycf3 (ATCG00360), y3ip1 (AT5G44650), PsaA (ATCG00350), PsaB (ATCG00340), PsaC (ATCG01060), PsaD (AT4G02770), PsaE (AT2G20260), PsaF (AT1G31330), PsaG (AT1G55670), PsaH (AT1G52230), PsaN (AT5G64040), Lhca1 (AT3G54890), Lhca2 (AT3G61470), Lhca3 (AT1G61520), Lhca4 (AT3G47470), LHCII (AT1G29920), PsbP (AT1G06680), PsbA/D1 (ATCG00020), D2 (ATCG00270), CP43 (ATCG00280), CP47 (ATCG00680), PsbO (AT5G66570), Cyt f (ATCG00540), CF1α (ATCG00120),CF1β (ATCG00480), and RbcL (ATCG00490); PPD1 T-DNA mutants, SALK_143493C (ppd1-1) and GABI_853A11 (ppd1-2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotype, Identification and Complementation of the ppd1 Mutant.
Supplemental Figure 2. Stability of Thylakoid Membrane Proteins in Relation to Light Intensity in the Wild Type, the ppd1-1 and ppd1-2 Mutants, and the Complemented Plant.
Supplemental Figure 3. Association of psaA/B, psaC, ycf3, and psbA mRNAs with Polysomes.
Supplemental Figure 4. Phenotype and Measurements of Chlorophyll Contents of Young and Old Leaves in the Wild Type.
Supplemental Figure 5. Diagnostic Immunoblot Analyses of 2D BN/SDS-PAGE Gels in the Wild Type, ppd1-1, ppd1-2, and y3ip1 Mutants, and the Complemented Plant.
Supplemental Figure 6. Far Protein Gel Blot Assays of the Interactions of PPD1 Protein with PsaA and PsaB Proteins.
Supplemental Figure 7. Yeast Two-Hybrid Assay of the Interaction between PPD1 and the Lumenal Domain of PsaF.
Supplemental Table 1. A List of Primers Used in This Study.
Acknowledgments
We thank the ABRC and GABI-KAT for the seed stocks. This work was supported by the State Key Basic Research and Development Plan of China (2009CB118503), the Solar Energy Initiative of the Chinese Academy of Sciences, and the National Natural Science Foundation of China (30725024).
AUTHOR CONTRIBUTIONS
J.L., H.Y., and C.L. designed the study. J.L. and H.Y. performed the research. Q.L., X.W., F.C., L.P., L.Z., and C.L. analyzed the data. J.L., H.Y., and C.L. wrote the article.
Glossary
- PSI
photosystem I
- PSII
photosystem II
- Fv/Fm
ratio of variable fluorescence to maximal fluorescence
- qP
photochemical quenching
- NPQ
nonphotochemical quenching
- ΦPSII
PSII efficiency
- DM
dodecyl-β-d-maltopyranoside
- BN
blue native
- 2D
two-dimensional
- GFP
green fluorescent protein
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- Fo
minimum fluorescence
- Fs
steady state fluorescence
- YFP
yellow fluorescent protein
References
- Albus C.A., Ruf S., Schöttler M.A., Lein W., Kehr J., Bock R. (2010). Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22: 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann K., Lezhneva L., Wanner G., Herrmann R.G., Meurer J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A., Nelson N. (2009). Plant photosystem I design in the light of evolution. Structure 17: 637–650 [DOI] [PubMed] [Google Scholar]
- Amunts A., Drory O., Nelson N. (2007). The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447: 58–63 [DOI] [PubMed] [Google Scholar]
- Amunts A., Toporik H., Borovikova A., Nelson N. (2010). Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.-D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Ji D., Peng L., Guo J., Ma J., Zou M., Lu C., Zhang L. (2009). LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J., Cho W.K., Lezhneva L., Falk J., Krupinska K., Shinozaki K., Seki M., Herrmann R.G., Meurer J. (2006). A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J. Biol. Chem. 281: 17189–17196 [DOI] [PubMed] [Google Scholar]
- Haldrup A., Simpson D.J., Scheller H.V. (2000). Down-regulation of the PSI-F subunit of photosystem I in Arabidopsis thaliana. The PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J. Biol. Chem. 275: 31211–31218 [DOI] [PubMed] [Google Scholar]
- Hippler M., Rimbault B., Takahashi Y. (2002). Photosynthetic complex assembly in Chlamydomonas reinhardtii. Protist 153: 197–220 [DOI] [PubMed] [Google Scholar]
- Ifuku K., Ishihara S., Sato F. (2010). Molecular functions of oxygen-evolving complex family proteins in photosynthetic electron flow. J. Integr. Plant Biol. 52: 723–734 [DOI] [PubMed] [Google Scholar]
- Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Sato F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.E., Gilpin M., Knoetzel J., Scheller H.V. (2000). The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J. Biol. Chem. 275: 24701–24708 [DOI] [PubMed] [Google Scholar]
- Kim H.U., van Oostende C., Basset G.J., Browse J. (2008). The AAE14 gene encodes the Arabidopsis o-succinylbenzoyl-CoA ligase that is essential for phylloquinone synthesis and photosystem-I function. Plant J. 54: 272–283 [DOI] [PubMed] [Google Scholar]
- Krech K., Ruf S., Masduki F.F., Thiele W., Bednarczyk D., Albus C.A., Tiller N., Hasse C., Schöttler M.A., Bock R. (2012). The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre-Legendre L., Rappaport F., Finazzi G., Ceol M., Grivet C., Hopfgartner G., Rochaix J.D. (2007). Loss of phylloquinone in Chlamydomonas affects plastoquinone pool size and photosystem II synthesis. J. Biol. Chem. 282: 13250–13263 [DOI] [PubMed] [Google Scholar]
- Lennartz K., Plücken H., Seidler A., Westhoff P., Bechtold N., Meierhoff K. (2001). HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L., Amann K., Meurer J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Lohmann A., Schöttler M.A., Bréhélin C., Kessler F., Bock R., Cahoon E.B., Dörmann P. (2006). Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 281: 40461–40472 [DOI] [PubMed] [Google Scholar]
- Lu Y., Hall D.A., Last R.L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Monte E., Quail P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Meng B.Y., Tanaka M., Wakasugi T., Ohme M., Shinozaki K., Sugiura M. (1988). Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: Determination of the transcriptional initiation site by in vitro capping. Curr. Genet. 14: 395–400 [DOI] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. (1996). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J., Plücken H., Kowallik K.V., Westhoff P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.-M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Naver H., Boudreau E., Rochaix J.-D. (2001). Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13: 2731–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M., Li X., Ma J., Chi W., Xiao J., Zou M., Chen F., Lu C., Zhang L. (2011). LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat. Commun. 2: 227. [DOI] [PubMed] [Google Scholar]
- Ozawa S.-I., Nield J., Terao A., Stauber E.J., Hippler M., Koike H., Rochaix J.-D., Takahashi Y. (2009). Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21: 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.B., Emanuelsson O., Kalume D.E., Ytterberg J., Friso G., Rudella A., Liberles D.A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K.J. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Cournac L., Vassiliev I.R., Golbeck J.H., Peltier G., Rochaix J.-D. (1999). Photosystem I is indispensable for photoautotrophic growth, CO2 fixation, and H2 photoproduction in Chlamydomonas reinhardtii. J. Biol. Chem. 274: 10466–10473 [DOI] [PubMed] [Google Scholar]
- Rochaix J.-D. (1997). Chloroplast reverse genetics: New insights into the function of plastid genes. Trends Plant Sci. 2: 419–425 [Google Scholar]
- Rokka A., Suorsa M., Saleem A., Battchikova N., Aro E.-M. (2005). Synthesis and assembly of thylakoid protein complexes: Multiple assembly steps of photosystem II. Biochem. J. 388: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J.L., Frankel L.K., Bricker T.M. (2011). Developmental defects in mutants of the PsbP domain protein 5 in Arabidopsis thaliana. PLoS ONE 6: e28624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Kössel H., Bock R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. (2006). Tricine-SDS-PAGE. Nat. Protoc. 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Albus C.A., Bock R. (2011). Photosystem I: Its biogenesis and function in higher plants. J. Plant Physiol. 168: 1452–1461 [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Netz D.J., Frazzon J., Pierik A.J., Bill E., Gross J., Lill R., Meurer J. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Zhao J., Reimer S.K., Antonkine M.L., Cai Q., Weiland S.M., Golbeck J.H., Bryant D.A. (2002). Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277: 20343–20354 [DOI] [PubMed] [Google Scholar]
- Sommer F., Drepper F., Hippler M. (2002). The luminal helix l of PsaB is essential for recognition of plastocyanin or cytochrome c6 and fast electron transfer to photosystem I in Chlamydomonas reinhardtii. J. Biol. Chem. 277: 6573–6581 [DOI] [PubMed] [Google Scholar]
- Stöckel J., Bennewitz S., Hein P., Oelmüller R. (2006). The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141: 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J., Oelmüller R. (2004). A novel protein for photosystem I biogenesis. J. Biol. Chem. 279: 10243–10251 [DOI] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. (2007). Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Goldschmidt-Clermont M., Soen S.-Y., Franzen L.G., Rochaix J.-D. (1991). Direct chloroplast transformation in Chlamydomonas reinhardtii: Insertional inactivation of the psaC gene encoding the iron-sulfur protein destabilizes photosystem I. EMBO J. 10: 2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Watkins K.P., Rojas M., Friso G., van Wijk K.J., Meurer J., Barkan A. (2011). APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.-A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim. Biophys. Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]