GALS1, GALS2, and GALS3 are members of glycosyltransferase family GT92 in Arabidopsis thaliana. Loss-of-function mutants in the three corresponding genes are deficient in pectic β-1,4-galactan. GALS1 is shown to function as a β-1,4-galactan synthase in vitro, and GALS1 overexpressors have a 50% increased content of β-1,4-galactan in the cell walls.
Abstract
β-1,4-Galactans are abundant polysaccharides in plant cell walls, which are generally found as side chains of rhamnogalacturonan I. Rhamnogalacturonan I is a major component of pectin with a backbone of alternating rhamnose and galacturonic acid residues and side chains that include α-1,5-arabinans, β-1,4-galactans, and arabinogalactans. Many enzymes are required to synthesize pectin, but few have been identified. Pectin is most abundant in primary walls of expanding cells, but β-1,4-galactan is relatively abundant in secondary walls, especially in tension wood that forms in response to mechanical stress. We investigated enzymes in glycosyltransferase family GT92, which has three members in Arabidopsis thaliana, which we designated GALACTAN SYNTHASE1, (GALS1), GALS2 and GALS3. Loss-of-function mutants in the corresponding genes had a decreased β-1,4-galactan content, and overexpression of GALS1 resulted in plants with 50% higher β-1,4-galactan content. The plants did not have an obvious growth phenotype. Heterologously expressed and affinity-purified GALS1 could transfer Gal residues from UDP-Gal onto β-1,4-galactopentaose. GALS1 specifically formed β-1,4-galactosyl linkages and could add successive β-1,4-galactosyl residues to the acceptor. These observations confirm the identity of the GT92 enzyme as β-1,4-galactan synthase. The identification of this enzyme could provide an important tool for engineering plants with improved bioenergy properties.
INTRODUCTION
Plant cell walls are predominantly composed of different polysaccharides, which can be classified into cellulose, hemicelluloses, and pectin. Pectin consist of two major types of polysaccharides: homogalacturonan, which is entirely composed of α-1,4-linked d-galacturonosyl residues, and rhamnogalacturonan I (RGI), which has a backbone of alternating α-1,2-linked l-rhamnose and α-1,4-linked galacturonic acid residues with side chains of arabinan and β-1,4-d-galactan attached to the rhamnosyl residues (Mohnen, 2008; Harholt et al., 2010). Commonly, the β-1,4-d-galactan side chains are substituted with Ara and branched arabinans in structures known as Type I arabinogalactans. Other domains of pectin include rhamnogalacturonan II (RGII), a complex structure with a homogalacturonan backbone and four side chains with numerous different and rare sugars, and xylogalacturonan, which is a type of homogalacturonan substituted with xylosyl residues. The galacturonic acid residues are methyl esterified to various degrees in homogalacturonan and can be esterified with acetate at O-2 or O-3 in both homogalacturonan and RGI (Mohnen, 2008). Ferulate esterification of galactans and arabinans occurs in certain species. It has been estimated that as many as 67 different transferases are required for the biosynthesis of pectin (Mohnen, 2008; Harholt et al., 2010), and so far only one has been unambiguously identified, namely, the homogalacturonan galacturonosyltransferase GAUT1 from Arabidopsis thaliana (Sterling et al., 2006). A xylogalacturonan xylosyltransferase designated XYLOGALACTURONAN DEFICIENT1 (XGD1) has also been identified in Arabidopsis, but activity of the isolated protein in vitro has yet to be demonstrated (Jensen et al., 2008). Three Arabidopsis xylosyltransferases, RGII XYLOSYLTRANSFERASE1 (RGXT1), RGXT2, and RGXT3, transfer Xyl onto Fuc in vitro and are involved in RGII biosynthesis (Egelund et al., 2006, 2008). However, the acceptor substrate in vivo has not yet been clearly determined. Finally, two putative arabinosyltransferases in Arabidopsis named ARABINAN DEFICIENT1 (ARAD1) and ARAD2 are known to be involved in the biosynthesis of arabinan side chains of RGI (Harholt et al., 2006, 2012). However, this notion has not been substantiated with in vitro activity data. β-1,4-Galactan constitutes a large part of pectin and of the total cell wall (e.g., ∼30 to 40% of potato [Solanum tuberosum] pectin) (Jarvis et al., 1981; Oxenboll Sørensen et al., 2000). In general, most if not all of the β-1,4-linked Gal in cell walls is bound to an RGI backbone. Part of the galactans is tightly bound to the wall, and after extraction, some may not be bound to an acidic backbone. However, it is not clear if such neutral galactans are generated during the extraction or are present in the wall, where they most likely would originate from enzymatic action on RGI. The specific function of β-1,4-galactan in primary walls is poorly understood. β-1,4-Galactan has been implicated in cell elongation (McCartney et al., 2003) and has been demonstrated to be a water retaining viscoelastic component with a likely role in modulating the mechanical properties of cell walls (Tang et al., 1999; McCartney et al., 2000; Ha et al., 2005). However, potato tubers with reduced galactan content did not show any growth or developmental phenotype (Oxenboll Sørensen et al., 2000; Martín et al., 2005; Ulvskov et al., 2005), although the galactan deficient potato tubers were found to be slightly more brittle, indicating that galactan may play a role in transmitting stresses to cellulose microfibrils (Ulvskov et al., 2005). In Arabidopsis, reduced galactan content did not lead to any strong growth phenotype, but the stems were thinner than in the wild type (Obro et al., 2009). Other studies have shown that β-1,4-galactan is not uniformly distributed in the primary wall and for example is absent from pit fields in Solanum lycopersicum (tomato; Orfila and Knox, 2000). Secondary walls generally have a low content of pectin, but β-1,4-galactan is a major wall component in gelatinous fibers, which are abundant in secondary walls in reaction wood (tension wood and compression wood) and in certain plants such as Linum usitatissimum (flax, Andersson-Gunnerås et al., 2006; Gorshkova and Morvan, 2006; Arend, 2008; Mellerowicz and Gorshkova, 2012). Turnover of β-1,4-galactan in flax during development is essential for the mechanical properties of the fibers (Roach et al., 2011).
β-1,4-Galactan synthase activity in plant extracts was demonstrated more than 40 years ago (McNab et al., 1968), and several subsequent studies have characterized the activity but not led to the identification of the enzyme (see recent reviews for a discussion of earlier studies of β-1,4-galactan synthesis) (Mohnen, 2008; Harholt et al., 2010). In this article, we report the identification of β-1,4-galactan synthase, which we designate GALS1. The enzyme belongs to glycosyltransferase family GT92, which has three members in Arabidopsis. Loss-of-function mutants in all three genes are galactan deficient, and the isolated GALS1 protein catalyzes β-1,4-galactan synthesis in vitro.
RESULTS
Glycosyltransferase Family GT92 Contains β-1,4-Galactosyltransferases
To identify candidate enzymes for β-1,4-galactan synthase, we explored the CAZy (Carbohydrate Active enZYmes) database (www.cazy.org) (Cantarel et al., 2009). Glycosyltransferases in CAZy are divided into 91 families, 42 of which have angiosperm members. The β-1,4-galactan synthases were expected to be found in a family of enzymes that invert the configuration of the anomeric carbon during catalysis, which limits the number of candidate families. Many glycosyltransferases from various families have already been investigated by analyzing loss-of-function mutants; however, in most cases, no clear indication of the role of the specific glycosyltransferases could be shown, and previous studies have not led to the identification of candidates for a β-1,4-galactan synthase. Recently, a new family of glycosyltransferases, GT92, was added to CAZy with the identification of novel β-1,4-galactosyltransferases in animals. The GT92 proteins are reported from pigeon (Columba livia), where they catalyze the transfer of β-1,4-linked Gal onto β-1,4-linked Gal in N-glycan structures (Suzuki and Yamamoto, 2010). However, genes encoding GT92 proteins are not found in all birds or in mammals. A GT92 protein in Caenorhabditis elegans has been shown to be a β-1,4-galactosyltransferase that adds Gal onto a core Fuc in N-linked glycans (Titz et al., 2009). All plants that have had their genomes sequenced have members of GT92, but these are likely to have a different role than in animals, since β-1,4-Gal is not known from plant N-glycans. Furthermore, increased expression of GT92 genes has been observed in transcriptomic studies of tension wood, which is known to be rich in β-1,4-galactan (Andersson-Gunnerås et al., 2006). We therefore decided to investigate the function of GT92 proteins in Arabidopsis, which has three members of GT92. A phylogenetic tree is shown in Supplemental Figure 1 online (see Supplemental Data Set 1 and Supplemental References 1 online). The three Arabidopsis proteins fall in two clades, but only one of the clades is represented in rice (Oryza sativa), and in the lycophyte Selaginella moellendorffii, a single gene encodes a protein, which is about equally similar to proteins in either clade. A second GT92 protein encoded in S. moellendorffii may be more closely related to the animal β-1,4-galactosyltransferases.
Arabidopsis Loss-of-Function Mutants in GT92 Genes Are Deficient in β-1,4-Galactan
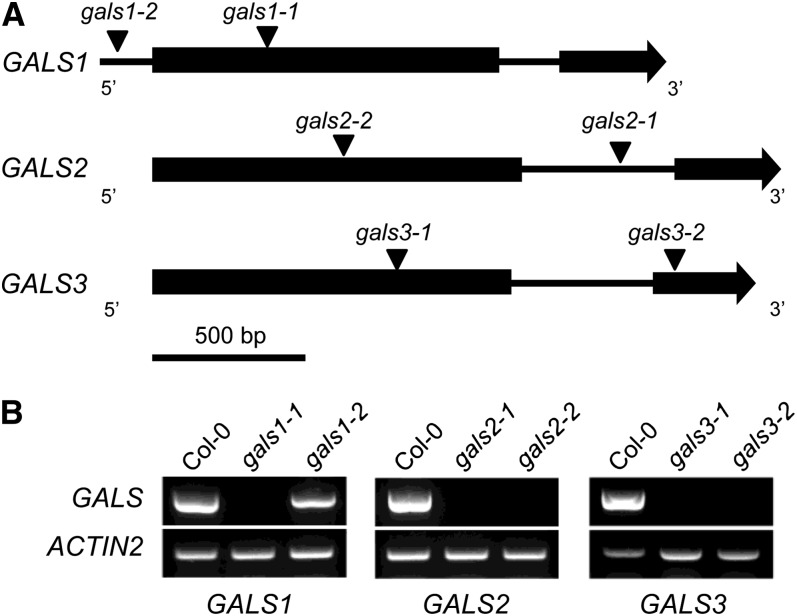
Two independent mutant lines with T-DNA insertions were identified for each of the three genes, and homozygous mutants were identified by PCR (Figure 1; see Supplemental Table 1 online). RT-PCR analysis showed that no transcript could be detected in five of the mutants, while one mutant with the T-DNA located in the promoter region (gals1-2) showed leaky expression (Figure 1). Based on the results shown below, the genes were designated GALACTAN SYNTHASE1 (GALS1), GALS2, and GALS3.
Figure 1.
Schematic Gene Models and Transcript Analysis of GALS1, GALS2, and GALS3 and T-DNA Mutants.
(A) Intron-exon structure of genes (exons shown as boxes and introns and 5′ regions as lines) and T-DNA insertions of mutants (triangles).
(B) RT-PCR analysis showing the lack of transcript in gals1-1 (Salk_016687), gals2-1 (Salk_121802), gals2-2 (GK-290D05), gals3-1 (WiscDsLox377-380G11), and gals3-2 (GK-133F03) mutants compared with Col-0 wild-type plants. The analyses were done with three biological replicates for each line. The gals1-2 mutant (WiscDsLox443D3) is leaky. ACTIN2 was amplified as the loading control (bottom panel). Mutant lines (Alonso et al., 2003; Woody et al., 2007; Kleinboelting et al., 2012) were obtained from the ABRC.
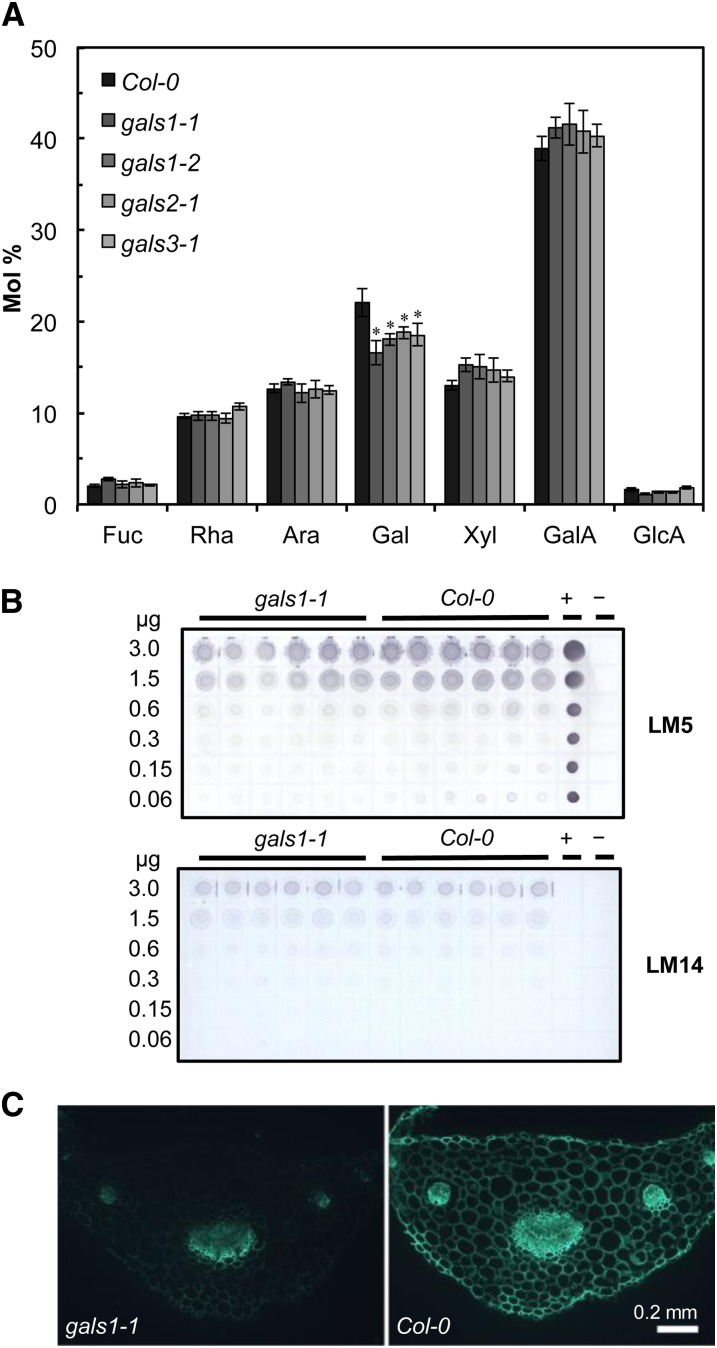
None of the mutants showed any obvious growth or developmental alterations compared with wild-type plants. Cell walls were prepared from rosette leaves and analyzed for monosaccharide composition. All six mutant lines showed a highly significant (analysis of variance [ANOVA] with Tukey test, P < 0.005) decrease of 14 to 25% in total cell wall Gal content, whereas the ratio between other sugars was not significantly changed (Figure 2A; see Supplemental Figure 2 online). Analysis of sugar composition in stems showed a significant 20 to 28% reduction in Gal in gals1-1 and gals3-1 (ANOVA with Tukey test, P < 0.001), whereas no significant differences were found in gals2-1 or in the other monosaccharides in gals1-1 and gals3-1 (see Supplemental Figure 3B online). Analysis of sugar composition in seeds showed a small but significant reduction in Gal in gals1 and gals2 mutant lines but not in gals3 (ANOVA with Tukey test, P < 0.02) (see Supplemental Figure 3 online). For the subsequent studies, we focused on GALS1 and the gals1-1 mutant, which had the largest reduction in Gal in leaf cell walls. The polysaccharide affected by the mutation was determined in immunodot assays using LM5, a monoclonal antibody that specifically recognizes pectic β-1,4-galactan (Jones et al., 1997). Indeed, when assessing its epitope recognition, the LM5 antibody showed less binding in the mutants than the wild type (Figure 2B). By contrast, LM14, a monoclonal antibody against arabinogalactan proteins (Moller et al., 2008), did not show any difference in binding (Figure 2B). The reduction in pectic galactan was further investigated by immunomicroscopy of transverse sections of petioles, a tissue chosen because it is conducive to sectioning and immunomicroscopy and because GALS1 is highly expressed (Figure 3). Immunofluorescence microscopy is not quantitative, but the detection of the LM5 galactan epitope was strongly decreased in gals1-1 when compared with the wild type (Figure 2C). These data provide evidence that GALS1 is specifically involved in the biosynthesis of β-1,4-galactan.
Figure 2.
Cell Wall Composition of gals Mutants.
(A) Monosaccharide composition of cell walls of the wild type and the gals1-1, gals1-2, gals2-1, and gals3-1 mutants. Cell walls were isolated from leaf tissue from 4-week-old plants, hydrolyzed with trifluoroacetic acid, and analyzed by HPAEC. The data show averages ± sd (n = 6 biological replicates). Monosaccharide composition for gals2-2 and gals3-2 is shown in Supplemental Figure 2 online. All six mutant lines showed a highly significant (ANOVA with Tukey test, P < 0.005, indicated with asterisks) decrease of 14 to 25% in total cell wall Gal content, whereas the ratio between other sugars was not significantly changed.
(B) Immunodot analysis of gals1-1 cell wall material. The blots were developed with the LM5 monoclonal antibody, which is specific for β-1,4-galactan, and LM14, which is specific for arabinogalactan proteins. Values on the left indicate the amount of cell wall material spotted on each line of dots. Potato galactan (+) and water (−) were used as controls.
(C) Immunodetection of the LM5 β-1,4-galactan epitope in transverse sections of petioles from the wild type and gals1-1.
[See online article for color version of this figure.]
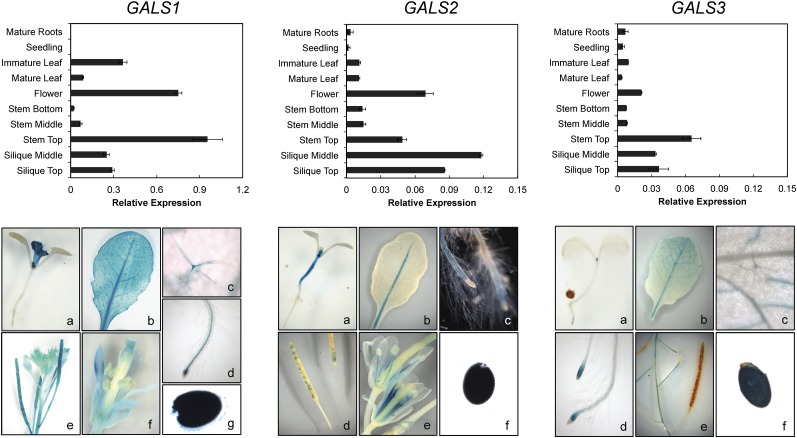
Figure 3.
Expression Pattern of the GALS1, GALS2, and GALS3 Genes.
Top panels: RT-PCR analysis of GALS1, GALS2, and GALS3 expression in major organs during development. The level of expression is calculated relative to the UBQ10 gene (mean ± sd of three biological replicates). Note the different scale for GALS1. Bottom panels: GUS activity patterns in transgenic Arabidopsis plants expressing the GUS reporter gene fused to the GALS1, GALS2, and GALS3 promoter, respectively. Expression analysis of GALS1 as revealed by promoter GUS analysis showing activity in a 9-d-old seedling (a), mature leaf (b), trichome (c), root vasculature (d), siliques (e), floral tissue (f), and seeds (g). GUS expression of GALS2pro:GUS plants revealed expression in the stem of 6-d-old seedlings (a), the midrib of a mature leaf (b), the root vasculature (c), siliques (d), flower filaments (e), and seeds (f). GUS activity pattern of GALS3pro:GUS plants showing staining in a 6-d-old seedling (a), mature leaf (b), root cap (d), top of the stem (e), and seed (f), but not in trichomes (c).
[See online article for color version of this figure.]
GALS1, GALS2, and GALS3 Have Overlapping but Distinct Expression Patterns
To analyze the spatial and developmental expression of the GALS genes, we isolated RNA from different tissues and determined the transcript levels of the GALS genes by quantitative RT-PCR (qRT-PCR) (Figure 3). In addition, we expressed the β-glucuronidase (GUS) gene under the control of the native promoters of the three genes (Figure 3). Taken together, our qRT-PCR and promoter-GUS data are substantially consistent with publicly available microarray data. The results show that while the GALS genes appeared to be almost ubiquitously expressed in Arabidopsis, the absolute expression values vary and distinct differences in the expression patterns can be found. In general, GALS1 showed an almost 10-fold higher expression than GALS2 and GALS3. GALS1 and GALS3 were highly expressed in the top of the stem, whereas GALS2 showed the highest expression in flowers and siliques (Figure 3). GUS activity was observed in leaves, stems, roots, and dry seeds for all three GALS genes (Figure 3). While GALS1pro:GUS and GALS3pro:GUS plants had GUS signals in all leaf veins, GALS2pro:GUS plants only showed staining in the midrib. GUS activity in trichomes was specifically found in GALS1pro:GUS transgenic plants. All GALS genes showed GUS activity in roots; in GALS1pro:GUS and GALS2pro:GUS plants, staining occurred in the root vasculature, while it was restricted to the root tip in GAL3pro:GUS plants. Immature GALS1pro:GUS and GALS3pro:GUS plants had GUS staining in primary leaves and roots. By contrast, GALS2pro:GUS seedlings had strong GUS activity in the hypocotyl. GALS1pro:GUS and GALS2pro:GUS, but not GAL3pro:GUS, revealed GUS expression in floral tissues and siliques.
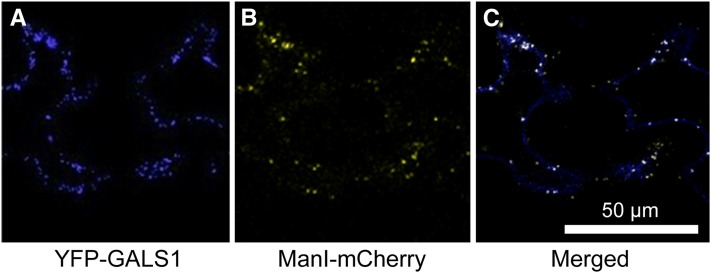
GALS1 Is Located in the Golgi Apparatus
The GALS proteins are all predicted to be Type II membrane proteins with a possible targeting to the secretory pathway (Krogh et al., 2001). The subcellular localization of GALS1 was investigated by heterologous expression of a yellow fluorescent protein (YFP) fusion in Nicotiana benthamiana and confocal laser scanning microscopy. This analysis showed that GALS1 was targeted to Golgi vesicles, consistent with a role in pectin biosynthesis and the reported location of β-1,4-galactan synthase activity (Geshi et al., 2004) (Figure 4). GALS2 and GALS3 have previously been identified as putative Golgi proteins in a proteomic study of purified Golgi vesicles from Arabidopsis cell cultures (Parsons et al., 2012).
Figure 4.
Subcellular Localization of GALS1.
(A) YFP-GALS1 fusion protein expressed under the control of the 35S promoter in N. benthamiana and imaged by confocal laser scanning microscopy.
(B) mCherry-tagged α-mannosidase, a Golgi marker, expressed in the same cells and imaged in the same way.
(C) Merged signals; colocalizations are visible as white dots.
GALS1 Is a β-1,4-Galactosyltransferase
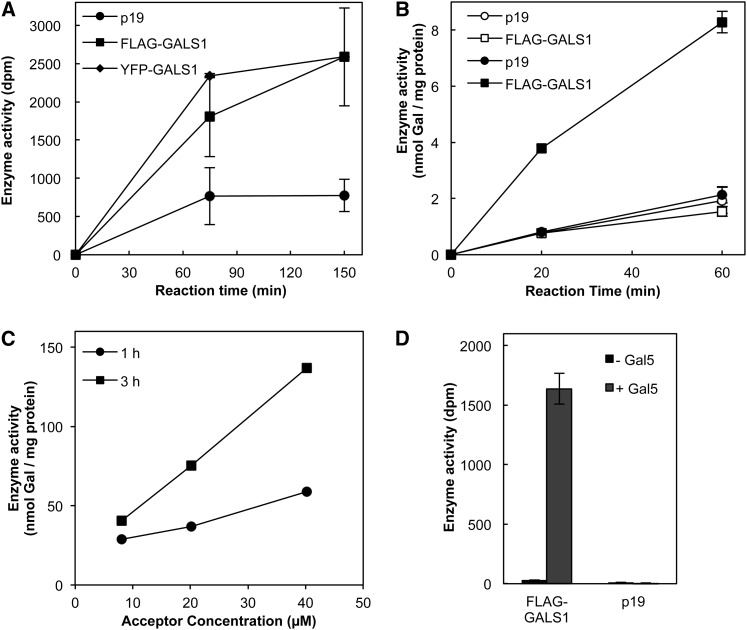
Because the animal GT92 enzymes include β-1,4-galactosyltransferases and the Arabidopsis mutants had a specific deficiency in β-1,4-galactan, we concluded that the Arabidopsis GT92 enzymes were strong candidates for β-1,4-galactan synthase. To characterize the biochemical function of GALS1, the protein was heterologously expressed in N. benthamiana as a fusion protein with N-terminal FLAG or YFP tags. Galactosyltransferase assays using microsomal proteins and endogenous acceptors showed a high incorporation of Gal from UDP-Gal into polymeric products (Figure 5A). The identity of a large proportion of the total product as β-1,4-galactan was confirmed by digestion with β-1,4-galactanase from Aspergillus niger, which released 90% of the polymeric product as Gal and a compound migrating as β-1,4-galactobiose on thin layer chromatography (TLC; Figures 6A and 6B). About 96% of the product remained soluble after treatment with KOH followed by neutralization with ammonium formate and 80% ± 1% of the soluble material was retained on a Q-Sepharose column as expected for β-1,4-galactan bound to a pectic backbone.
Figure 5.
Galactan Synthase Activity of GALS1.
(A) Galactosyltransferase activity is shown from 100 μg of microsomal protein incubated with ∼700 Bq UDP-3H-Gal. No exogenous acceptor was added. Data show average ± se (n = 3).
(B) Galactan synthase activity in solubilized microsomal proteins (10 μg) incubated with 740 Bq UDP-14C-Gal. Closed symbols indicate reactions in the presence of 2 µg of β-1,4-galactopentaose as acceptor, while open symbols indicate control reactions without acceptor. Data show average ± se (n = 3).
(C) Galactan synthase activity in solubilized microsomal proteins (5 μg) incubated with 0.34 mM UDP-14C-Gal (7.4 kBq) and varying concentrations of β-1,4-galactopentaose.
(D) Galactan synthase activity of affinity purified FLAG-GALS1. The purified protein was incubated with 740 Bq UDP-14C-Gal and 2 µg of β-1,4-galactopentaose as acceptor. Control reactions were mock-purified proteins from microsomes of p19-expressing plants and reactions without acceptor. Data show average ± se (n = 3 independently purified samples).
Microsomal membranes were isolated from N. benthamiana infiltrated with Agrobacterium for expression of GALS1 protein with an N-terminal FLAG or YFP fusion. The control plants were infiltrated only with p19 strain.
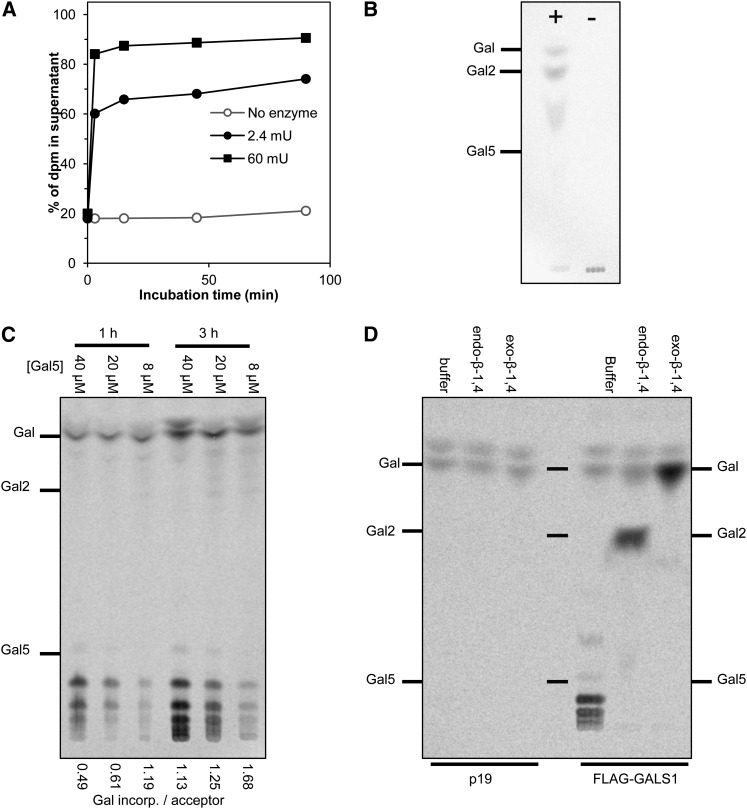
Figure 6.
Analysis of the GALS1 Reaction Product.
(A) Radiolabeled product generated with endogenous acceptors and 82 µM UDP-14C-Gal (6.3 kBq) was treated with 2.4 or 60 milliunits (mU) of endo-β-1,4-galactanase and separated into digested material and polymeric pellet. The graph shows the percentage of total radioactivity (dpm) that was soluble in 70% ethanol after the digestion. A major part of the product is β-1,4-galactan.
(B) Radiolabeled product as in (A) digested with endo-β-1,4-galactanase (+) for 4 h was analyzed by TLC and phosphorimaging. The control reaction was incubated with buffer (−). Galactose, β-1,4-galactobiose (Gal2) and β-1,4-galactopentaose (Gal5) were used as migration standards.
(C) Radiolabeled product was generated with exogenous β-1,4-galactopentaose by incubating for 1 or 3 h with 5 µg microsomal proteins and 0.34 mM UDP-14C-Gal (7.4 kBq). The products were chromatographed by TLC and phosphorimaging. The amount of Gal incorporated per acceptor molecule was calculated by scintillation counting of replicate samples and is written below each lane.
(D) Radiolabeled product was generated with exogenous β-1,4-galactopentaose by incubation for 1 h with 0.27 mM UDP-14C-Gal (14.8 kBq) and 2 µg galactopentaose acceptor. Aliquots were digested with endo-β-1,4-galactanase or exo-β-1,4-galactosidase, subjected to TLC, and analyzed by phosphorimaging.
To further characterize the galactosyltransferase activity, we chemically synthesized β-1,4-galactopentaose by stepwise coupling of monomeric building blocks (Davis, 2000) followed by global deprotection (Andersen, 2010) (see Supplemental Figure 4 online). After solubilization of microsomal membranes with Triton X-100 and incubation in the presence of the acceptor and UDP-[14C]Gal, the unincorporated UDP-[14C]Gal was removed by anion exchange chromatography and the product analyzed by liquid scintillation counting. The results showed a substantial activity of transfer of Gal onto the acceptor with microsomes from plants expressing FLAG-tagged GALS1 (Figure 5B). The galactan synthase activity was very stable as seen from the essentially linear time course. The activity obtained with FLAG-GALS1 corresponds to more than 50 nmol Gal h−1 mg protein−1 with 0.34 mM UDP-Gal (Figure 5C). This specific activity is substantially higher than what has been reported for other cell wall–related glycosyltransferases after heterologous expression, even though those proteins were generally more pure (Faik et al., 2002; Madson et al., 2003; Petersen et al., 2009).
While the data using microsomal protein strongly indicated that GALS1 is indeed β-1,4-galactan synthase, microsomes contain many proteins, including endogenous galactan synthase. We therefore isolated the FLAG-GALS1 fusion protein by affinity purification (see Supplemental Figure 5 online). We were not able to quantify the amount of purified protein and therefore could not calculate specific activity or turnover number for the isolated protein. The isolated protein retained acceptor-dependent activity, whereas the control reactions (mock purified protein from p19-expressing microsomes) had no detectable activity (Figure 5D).
To investigate if GALS1 could add more than one Gal to the acceptor molecule, we analyzed the products at high UDP-Gal/acceptor ratio by scintillation counting (Figure 5C) and TLC (Figure 6C). Standards for galactooligosaccharides larger than the β-1,4-galactopentaose acceptor were not available, but the TLC demonstrates a ladder of products, with longer incubation times and higher UDP-Gal/acceptor ratios leading to increasingly larger products (Figure 6C). Given that other oligosaccharides, including β-1,4-galactopentaose, migrate as a single band on the TLC, it seems reasonable to assume that the discrete radiolabeled bands correspond to galactohexaose, galactoheptaose, galactooctaose, and galactononaose. A calculation of the ratio of incorporated Gal per acceptor molecule confirmed that incorporation of more than one Gal was taking place (Figure 6C). Interestingly, the TLC suggests that the average product size was larger than calculated. For example, with 3 h incubation and 8 µM acceptor, the calculated incorporation was 1.7 Gal added per acceptor molecule; therefore, the predicted average product size would be 6.7 galactosyl residues if incorporation was random. However, the TLC suggested an average product size close to eight galactosyl residues. This may be due to a combination of the newly synthesized product being closer to the enzyme than the bulk acceptor molecules and to a higher acceptor efficiency of the longer oligosaccharide products than the Gal5 acceptor. We do not think the data suggest any real processivity of the enzyme.
The endo-β-1,4-galactanase digestion established that Gal was added to the endogenous acceptor with β-1,4-linkages. The product with exogenous acceptor was likewise investigated with endo-β-1,4-galactanase as well as with a β-1,4-specific exo-galactosidase from Streptococcus pneumonia. The specificity of the exogalactosidase was confirmed in reactions with β-1,4-galactobiose, β-1,3-galactobiose, β-methyl-β-1,3-galactobiose, and β-1,6-galactobiose (see Supplemental Figure 6 online). When the radiolabeled product was treated with endo-β-1,4-galactanase, it was fully digested to Gal and galactobiose, and when it was treated with exo-β-1,4-galactosidase, it was fully digested to Gal.
GALS1 Overexpression Leads to Increased β-1,4-Galactan Accumulation
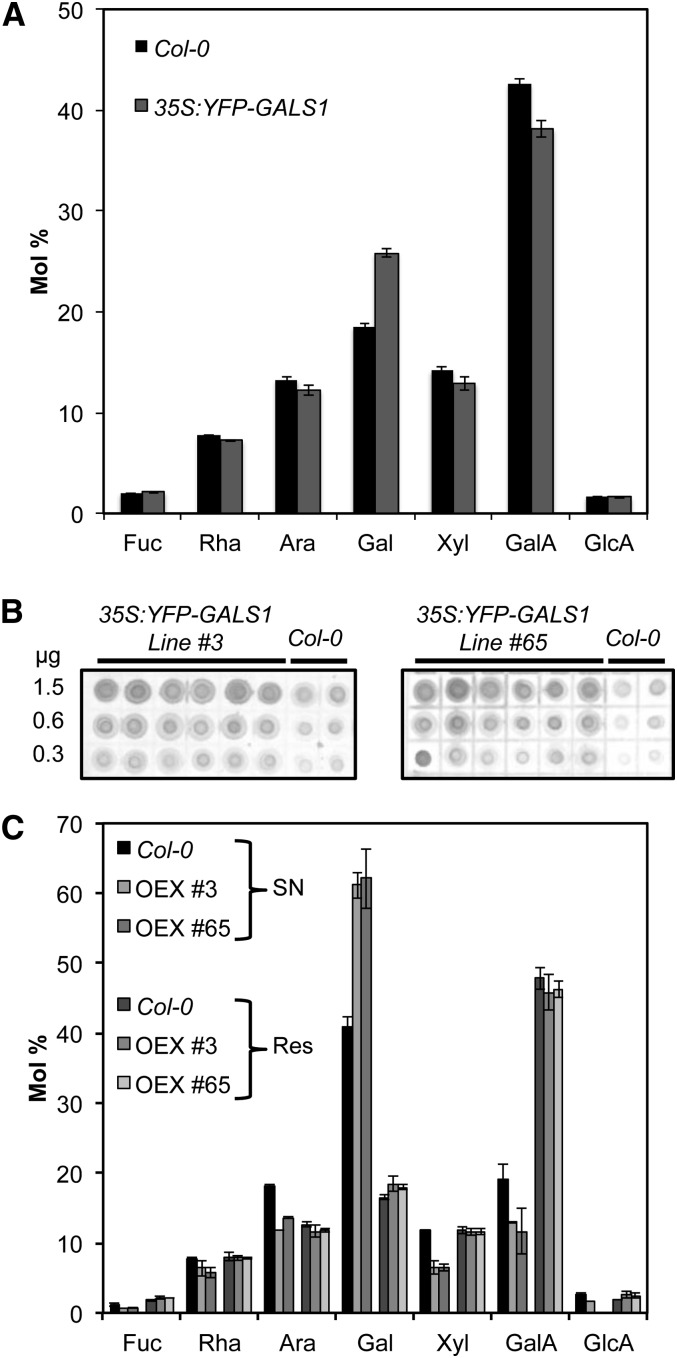
To investigate the function of GALS1 as galactan synthase in vivo, we expressed YFP-GALS1 in Arabidopsis under control of the 35S promoter. Several independent T1 lines with high expression level of the YFP-GALS1 fusion protein were selected based on immunofluorescence microscopy and immunoblotting of leaf extracts (see Supplemental Figure 7 online). Cell walls from leaves of seven independent lines were then analyzed, and six had ∼50% increased Gal content compared with the wild type (see Supplemental Figure 8 online). Two lines were selected for further analysis in T2, which showed that the content of cell wall Gal was significantly increased (ANOVA, P < 0.0001) by ∼50% (Figure 7A). The increase in galactan was accompanied by a relative decrease in the other monosaccharides, but there was no change in the relative amounts of the other monosaccharides between the wild type and overexpressors (ANOVA, P > 0.2). Immunodot blots with the LM5 antibody confirmed that the Gal increase was related to an increased accumulation of β-1,4-galactan (Figure 7B). Solubilization of the cell walls with KOH and subsequent digestion with endo-β-1,4-galactanase released 21.2% ± 1.8% of total Gal in the wild type and 36.8% ± 0.3% in the overexpressors (Figure 7C). No difference in monosaccharide composition between the wild type and overexpressors could be seen in the remaining pellet (Figure 7C).
Figure 7.
Characterization of Cell Walls in Plants Overexpressing GALS1.
(A) Cell wall monosaccharide composition of leaves from 4-week-old Arabidopsis Col-0 wild-type plants and overexpressing plants transformed with 35Spro:YFP-GALS1. Values are means ± sd obtained with six wild-type plants and seven T2 plants from lines #3 and #65. The cell wall Gal content in the overexpressors was significantly increased by ∼50% (ANOVA, P < 0.0001), whereas the ratio between other monosaccharides was not significantly changed.
(B) Immunodot blot analysis of leaf cell walls from 4-week-old T2 plants from two different T1 lines transformed with 35Spro:YFP-GALS1. The blots were developed with the LM5 monoclonal antibody and show increased content of β-1,4-galactan in the overexpressors. Values on the left indicate the amount of cell wall material spotted on each line of dots.
(C) Leaf cell walls from 4-week-old wild-type and YFP-GALS1 overexpressing plants were digested with endo-β-1,4-galactanase, separated into supernatant (SN) and residue (Res), and analyzed by HPAEC. The digestion released 21.2% ± 1.8% of total Gal in the wild type and 36.8% ± 0.3% in the overexpressors (OEX). Data show means ± sd (n = 4). Gal content was significantly different between overexpressors and the wild type in the supernatant (t test, P < 0.003) but not in the residue.
DISCUSSION
Pectin biosynthesis requires many different proteins for its synthesis, and identifying these proteins and determining their biochemical activity has been very challenging. Only one enzyme, GAUT1, has had its activity unambiguously shown previously (Sterling et al., 2006). Isolated RGXT proteins transfer Xyl onto Fuc in vitro and are involved in RGII biosynthesis even though the acceptor specificity has not been fully determined (Egelund et al., 2006). XGD1 appears to add Xyl to the backbone of xylogalacturonan, but the activity was only shown with crude membrane preparations and not with isolated protein (Jensen et al., 2008). Here, we provide clear biochemical evidence that glycosyltransferases of family GT92 in plants are β-1,4-galactan synthases. The GALS1 enzyme showed a high activity with galactooligosaccharide acceptor and is capable of elongating β-1,4-galactans. It is not clear if the same enzymes will add the first Gal residues onto the RGI backbone, but we find that unlikely due to the very different properties of the acceptor polysaccharide. Most likely, one or two as yet unidentified galactosyltransferases are required to initiate the β-1,4-galactan side chains. This activity has been described in an extract from potato that could initiate but not elongate β-1,4-galactan side chains in vitro (Geshi et al., 2000). Future studies should clarify this but will require a set of putative oligosaccharide substrates that are not easily obtained.
The GALS1 enzyme could add consecutive β-1,4-galactosyl residues to the β-1,4-galactopentaose acceptor, and the average product length suggested preferential addition of Gal to acceptors that had already been elongated. We think this pattern is more likely to show preference for acceptors longer than a pentaose rather than actual processivity of the enzyme. Since no acceptors longer than the pentaose were available, we could not test the specificity of GALS1 with acceptors of different length. However, Ishii et al. (2004) using 2-aminobenzamine (2AB)–labeled β-1,4-galactooligosaccharides and an enzyme preparation from mung bean (Vigna radiata) reported that initial transfer activity with 2AB-Gal6 and 2AB-Gal7 was ∼40 and 240% higher, respectively, than with 2AB-Gal5.
Overexpression of YFP-GALS1 in stable transformants resulted in a highly increased accumulation of β-1,4-galactan in the leaf cell walls, confirming that GALS1 functions as a β-1,4-galactan synthase in vivo. Previous experiments with potato and Arabidopsis have indicated that UDP-Gal rather than β-1,4-galactan synthase might be limiting for β-1,4-galactan synthesis (Seifert et al., 2002; Oomen et al., 2004; Rösti et al., 2007). However, our results indicate that Arabidopsis leaves contain sufficient UDP-Gal to support a substantial increase in galactan synthesis.
GT92 proteins are encoded in the genomes of all plants that have been analyzed and are found in a limited number of animal species (Hansen et al., 2012). They are not found in fungi or in green algae, such as Chlamydomonas reinhardtii, but are reported in apicomplexans of the Cryptosporidium genus. In animals, these proteins are known to add β-1,4-Gal to various N-glycans. However, this particular decoration is not well conserved, and the phylogeny indicates that GT92 members have been lost in many taxonomic groups. In plants, the GT92 proteins are also β-1,4-galactosyltransferases but have evolved specifically to synthesize β-1,4-galactans. The GT92 proteins contain Domain of Unknown Function 23, which is also found in a few bacteria. However, these bacterial proteins are quite divergent from the eukaryotic proteins, and none of them have been characterized.
The three Arabidopsis GALS genes show overlapping but not identical expression. This explains why a reduced content of cell wall Gal could be observed for mutants in all three genes, while all the mutants retained significant amounts of residual β-1,4-galactan. Mutants with combinations of two or three gene mutations will be needed to observe larger reductions in galactan. As previously observed in potato tubers (Oxenboll Sørensen et al., 2000; Martín et al., 2005) that exhibited a reduced amount of galactan, the gals mutants did not show any obvious phenotype. This absence of aberrant phenotype is in contrast with the proposed role of galactan in cell elongation that has been suggested based on the modulation of galactan occurrence by growth inhibitors in Arabidopsis roots (McCartney et al., 2003). In the potato tubers that were overexpressing a fungal galactanase, the decrease of galactan was accompanied by a more easily extractible pectin fraction, and the tubers showed slightly altered mechanical properties (Ulvskov et al., 2005). However, the potato tubers had a much more reduced galactan content (over 60% compared with control) than the gals mutants. Further studies will have to determine if any alteration of mechanical properties and cell wall extractability can be detected in the gals mutants.
β-1,4-Galactans have been implicated in the properties of flax fibers and reaction wood, but only few mutant studies have been performed. Galactanases and galactosidases that act on β-1,4-galactans are required for the development of flax fibers (Roach et al., 2011). Galactans and galactosidases are likely to play a similar function in tension wood, although xyloglucans are also abundant in tension wood and likely contribute significantly to its mechanical properties (Mellerowicz and Gorshkova, 2012). The specialized gelatinous fibers found in reaction wood and plants such as flax represent extreme forms of cell types that are present in most plants. However, the mutants described in this study had only a small decrease in stem Gal content; hence, the role of galactan in the formation and properties of secondary walls needs to be studied in double and triple mutants of the GALS genes.
β-1,4-Galactans are composed of hexoses, which in contrast with pentoses are easily used by fermenting microorganisms (e.g., for production of biofuels and other compounds). Hence, it would be advantageous to develop plants with increased galactan content instead of hemicelluloses consisting largely of pentoses. Plants overexpressing GALS1 already had a significantly increased C6/C5 sugar ratio in their cell walls and did not show any adverse phenotype. Given that UDP-Gal appeared to be limiting for galactan synthesis in some studies (Seifert et al., 2002; Oomen et al., 2004; Rösti et al., 2007), it is conceivable that an even higher level of galactan accumulation can be achieved by simultaneously overexpressing an appropriate isoform of UDP-Gal epimerase. Hence, the GALS genes may be important tools for developing bioenergy crops.
METHODS
Phylogenetic Analysis
Amino acid sequences of the CAZy GT92 family were obtained from the National Center for Biotechnology Information. The evolutionary history was inferred using the neighbor-joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set. There were a total of 320 positions in the final data set. Phylogenetic analyses were conducted in MEGA5 (Tamura et al., 2011).
Identification of Homozygous Plants with T-DNA Insertions
Arabidopsis thaliana T-DNA insertion mutants in At2g33570 (gals1-1, Salk_016687; gals1-2, WiscDsLox443D3), At5g44670 (gals2-1, Salk_121802; gals2-2, GK-290D05), and At4g20170 (gals3-1, WiscDsLox377-380G11; gals3-2, GK-133F03) (Alonso et al., 2003; Woody et al., 2007; Kleinboelting et al., 2012) were obtained from the ABRC and the European Arabidopsis Stock Centre. Homozygous mutants were identified by PCR using primer sequences obtained with the T-DNA Primer Design Tool provided by the Salk Institute Genomics Analysis Laboratory (http://signal.salk.edu/tdnaprimers.2.html) (see Supplemental Table 1 online). To confirm homozygous plants at the transcript level, RNA was extracted, reverse transcribed, and analyzed by PCR using primers covering the full-length sequence for the respective gene (see Supplemental Table 2 online).
Plants were germinated and grown on soil (PRO-MIX; Premier Horticulture) at 22°C and 60% relative humidity. Except as indicated, the plants were grown under long-day conditions (16 h photoperiod, 120 μmol m−2 s−1 of fluorescent light).
Cloning and Plant Transformation with 35S:GALS1
A full-length cDNA clone for GALS1 was obtained from the ABRC. Destination clones for plant transformation were made by LR reaction according to the manufacturer’s protocol (Invitrogen) with the vectors pEarleyGate 202 and 104 (Earley et al., 2006) for the N-terminal FLAG and YFP tag, respectively. The resulting 35Spro:YFP-GALS1 and 35Spro:FLAG-GALS1 constructs were transformed into Agrobacterium tumefaciens strains GV3101 and C58-1 pGV3850 and used for transient expression in Nicotiana benthamiana according to Voinnet et al. (2003) or for stable transformation of Arabidopsis Columbia-0 (Col-0) plants by floral dip (Clough and Bent, 1998). Arabidopsis transformants were identified by BASTA selection, and lines with a high level of YFP-GALS1 protein were further selected by visual inspection of fluorescence and by immunoblotting using an anti–green fluorescent protein antibody.
Subcellular Localization of GALS1
Infiltration of 4-week-old N. benthamiana leaves was done using Agrobacterium strain C58-1 pGV3850 by coinfiltration with a strain carrying the tomato bushy stunt virus p19 gene (OD600 = 0.5), following the method described by Sparkes et al. (2006). Plants transiently expressing the YFP-GALS1 fusion protein were analyzed by confocal laser scanning microscopy 2 d after the infiltration. The microscopy was performed using a Zeiss LSM 710 device equipped with an argon laser (514 nm for YFP excitation) and an In Tune laser (536 nm for propidium iodide excitation; Zeiss). Emission was collected at 510 to 545 nm (YFP) and 610 to 650 nm (propidium iodide). A construct for expressing α-mannosidase-mCherry was used as Golgi marker (Nelson et al., 2007).
Expression Analysis
Sequences upstream of the translation start codon from GALS1, GALS2, and GALS3 were amplified by PCR from genomic DNA. The primer sequences, restriction sites, and the length of the putative promoter region are summarized in Supplemental Table 3 online. The DNA fragments were cloned into pBI101.3 upstream of the GUS reporter gene (Jefferson et al., 1987). After Agrobacterium-mediated transfection of plants, histochemical staining of positive transformants was assessed with the indigogenic substrate X-Gluc (Gold Biotechnology) in GUS incubation solution (50 mM phosphate buffer, 10 mM EDTA, 330 mg/L K3FeCN6, 0.1% [v/v] Triton X-100, and 0.1% [v/v] Tween 20) for several hours overnight at 37°C.
For expression analysis by qRT-PCR, RNA was isolated from various tissues and used to generate cDNA. Real-time PCR was done with ABsolute SYBR Green ROX Mix (ABgene) on a StepOnePlus Real-Time PCR system (Applied Biosystems) according to conditions described earlier (Czechowski et al., 2005) using StepOne 2.0 software (Applied Biosystems). Primer sequences are listed in Supplemental Table 4 online.
Analysis of Cell Wall Monosaccharide Composition
Alcohol-insoluble residue (AIR) was prepared as described (Harholt et al., 2006). The lyophilized AIR was hydrolyzed in 2 M trifluoroacetic acid at 120°C for 1 h and analyzed by high-performance anion exchange chromatography (HPAEC) as previously described (ØBro et al., 2004; Yin et al., 2011). Glc was not determined due to the presence of residual starch. Statistical analysis of the data was done by three-way ANOVA using monosaccharide as repeated measurement and Tukey’s test for contrasts. This was done with the XLSTAT 212.6.02 software (Addinsoft).
Cell wall preparations from overexpressors were further analyzed by digestion with endo-β-1,4-galactanase from Aspergillus niger (Megazyme). AIR (2 mg) was dissolved in 0.1 mL of 1 M KOH and adjusted to pH 4.7 with 2 mL of 100 mM acetic acid. Galactanase (two units, one unit releases 1 µmol Gal per min from potato [Solanum tuberosum] galactan at 40°C) was added and the samples incubated for 1 h at 40°C. After incubation, cold 95% ethanol with 10 mM EDTA was added to a final concentration of 70% and the sample centrifuged at 14,000g for 5 min at 4°C. The supernatant and pellet were separated, dried, hydrolyzed with trifluoroacetic acid, and analyzed by HPAEC as described above.
Immunodot Assays
AIR was treated with phenol:acetic acid:water 2:1:1 (v/v/v) to remove soluble proteins according to Harholt et al. (2006). Subsequently, 3 mg of cell wall preparation was homogenized by ball milling and extracted with 4 M KOH containing 0.1% NaBH4 as described (Sørensen and Willats, 2011). Dilutions of the extracts were spotted onto nitrocellulose and allowed to dry overnight. Immunodot assays were performed as described (Vega-Sánchez et al., 2012). The primary antibodies LM5 and LM14 are rat monoclonal antibodies (from PlantProbes) and were used at 10-fold dilution in PBS with 5% milk powder (MP-PBS). The secondary antibody (goat anti-rat IgG coupled with horseradish peroxidase; Sigma-Aldrich) was used at 100-fold dilution in MP-PBS. Potato galactan (Megazyme) was used as positive control.
Indirect Immunofluorescence Microscopy
Five petioles of gals1-1 and Col-0 were embedded in wax as described (Hervé et al., 2011). Immunolabeling was done according to Verhertbruggen et al. (2009). LM5 was diluted 10-fold in MP-PBS and the secondary antibody (goat anti-rat IgG coupled with fluorescein isothiocyanate; Sigma-Aldrich) 100-fold. Sections incubated without primary antibody were used to assess the autofluorescence of the samples.
Chemical Synthesis of β-d-(1→4)-Galactopentaose
The pentasaccharide was synthesized using general methods for monosaccharide protection and coupling (Davis, 2000) and deprotected essentially as described (Andersen, 2010). The fully protected pentasaccharide (200 mg; 0.10 mmol) was dissolved in methanol (6 mL) and tetrahydrofuran (2 mL), and five drops of freshly prepared 1 M NaOMe in methanol was added. The reaction mixture was stirred at room temperature until full conversion was observed by TLC (72 h). The reaction was quenched with Amberlite IR-120 (H+), filtered, and concentrated. The crude isolate was dissolved in methanol (6 mL), tetrahydrofuran (2 mL), and AcOH (0.5 mL). Then, 10% Pd/C (83 mg; 0.078 mmol) was added and an atmosphere of H2 (1 atmosphere) was installed by bubbling through the solution. The reaction was stirred for 2 h under H2 and then water (2 mL) was added to solubilize the more polar intermediates. The reaction was stirred until TLC indicated full conversion (48 h), filtered through celite, and concentrated. The product was purified by chromatography on C18-modified silica (95:5 water/methanol) to give a white solid. The yield was 61 mg (75%). The structure was confirmed by NMR (one-dimensional 1H, double quantum filtered correlation spectroscopy, gradient heteronuclear single quantum coherence, and gradient-selected heteronuclear multiple-bond correlation spectroscopy), acquired in D2O on a 500-MHz Varian Unity Inova operating at 25°C using residual HDO (4.77 ppm) as internal reference and by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry using 2-hydroxybenzoic acid as matrix and data acquisition in positive ion mode using a 4800 matrix-assisted laser-desorption ionization–tandem time-of-flight device (Applied Biosystems). Purity was >95% based on the absence of detectable contaminants in the NMR and matrix-assisted laser-desorption ionization time-of-flight spectra.
Galactan Synthase Activity Assays
UDP-14C-Gal (10 GBq/mmol) and UDP-3H-Gal (2 TBq/mmol) were obtained from American Radiolabeled Chemicals.
Leaves of N. benthamiana transiently expressing YFP-GALS1 and FLAG-GALS1 were harvested 4 d after infiltration. Microsomes were prepared according to Konishi et al. (2006). Activity with endogenous acceptors was determined essentially according to Ishii et al. (2004) in reaction mixtures containing 30 mM MES-KOH, pH 6.0, 5 mM MnCl2, 0.1 mM UDP-Gal, 740 Bq of UDP-14C-Gal or UDP-3H-Gal, and microsomes corresponding to 50 to 100 µg protein in a reaction volume of 40 µL. After incubation at 30°C, the reactions were stopped by addition of 0.5 mL of a solution of 70% ethanol, 10 mM EDTA, and 100 µg dextran, followed by centrifugation at 14,000g for 5 min at 4°C. The pellet was resuspended in 300 μL of 10 mM EDTA and polymeric material repelleted by the addition of 700 μL of 100% ethanol followed by centrifugation. This step was repeated twice. The final pellet was resuspended in 200 μL of 20 mM EDTA and subjected to liquid scintillation counting. Control reactions contained microsomes from leaves expressing only the p19 protein.
Acceptor-dependent assays included 1 to 10 µg microsome protein, 40 mM MES-KOH, pH 6.5, 5 mM MnCl2, 160 mM Suc, 0.75% Triton X-100, 32 µM UDP-Gal, 740 Bq UDP-14C-Gal, and 2 µg β-1,4-galactopentaose in a reaction volume of 40 µL. In some experiments, higher concentrations of UDP-Gal and lower concentrations of acceptor were used as detailed in the figure legends. After the reaction, 0.5 mL of ice-cold water was added to the sample, which was passed over a 0.6-mL bed of Dowex 1×8 200-400 Mesh CL (Sigma-Aldrich) to capture unincorporated UDP-14C-Gal. The Dowex was washed twice with 0.5 mL of water and the eluents combined and analyzed by liquid scintillation counting.
Purification of GALS1
FLAG-GALS1 was purified as described (Rennie et al., 2012) by incubating microsomes corresponding to 0.6 mg protein with 1% Triton X-100, 50 mM PIPES-KOH, pH 7.0, 400 mM Suc, 1 mM phenylmethylsulfonyl fluoride, and Complete Mini Protease Inhibitor Cocktail (1 tablet per 10 mL; Roche) for 10 min and subsequently centrifuging at 100,000g for 30 min. The supernatant was incubated with EZview Red anti-FLAG M2 affinity gel (Sigma-Aldrich) for 3 h, washed three times with 1% Triton X-100, 400 mM Suc, 50 mM PIPES-KOH, pH 7.0, and 200 mM NaCl, and then three times with 400 mM Suc and 50 mM PIPES-KOH, pH 7.0. The gel with purified protein was used directly for activity assays as described above. Affinity-purified protein corresponding to 0.1 mg microsomal protein was used in each reaction and incubated for 3 h.
Characterization of Radiolabeled Product
The product generated with endogenous acceptors was analyzed by galactanase digestion. The precipitable product generated as described above was resuspended in 700 μL of 50 mM NaOAc, pH 4.5, boiled for 5 min, and incubated for 4 h at 40°C in the presence of 0.0024 and 0.06 units of endo-β-1,4-galactanase from A. niger (Megazyme). After the incubation, 700 μL of cold 70% ethanol and 10 mM EDTA was added, the sample centrifuged, and the pellet and supernatant analyzed by liquid scintillation counting. An aliquot of the digest with 60 milliunits was analyzed by TLC. The sample was lyophilized, extracted with pyridine, and applied to TLC using Silica gel 60 F254 plates (Merck) with development in 1-propanol:ethanol:water 7:1:2 (v/v/v). The TLC was analyzed by phosphorimaging using storage phosphor screens and a Typhoon 8600 Variable Mode Imager (Molecular Dynamics). Unlabeled standards (Gal, β-1,4-galactobiose and β-1,4-galactopentaose) were visualized by dipping the TLC plate in 10% sulfuric acid in ethanol and charring with a heat gun.
For anion exchange chromatography of the product with endogenous acceptors, the pellet was dissolved in 200 μL 1 M KOH, heated at 70°C for 15 min, diluted and neutralized with 6 mL of 40 mM ammonium formate and 10 mM EDTA, followed by centrifugation at 14,000g for 5 min at 25°C. The pellet was resuspended and analyzed by liquid scintillation counting. Aliquots of the supernatant were loaded onto a HiTrap Q FF column (GE Healthcare Bio-Sciences), the eluent was collected, and the column was washed with 2 mL of 40 mM ammonium formate and 10 mM EDTA and combined. The column was then eluted with 3 mL 0.1 HCL and 2 M NaCl.
To determine the average length of the product generated with the exogenous acceptor, microsomes (5 µg protein) from plants overexpressing FLAG-GALS1 were incubated as described above in the presence of galactopentaose acceptor. UDP-14C-Gal concentration was 0.34 mM (7.4 kBq), and galactopentaose concentration was 8 to 40 µM. After incubation for 1 or 3 h, the product was purified as described above, and a small aliquot analyzed by scintillation counting to determine the amount of Gal from UDP-Gal that was incorporated into product. The products were further analyzed by TLC with development four times in n-butanol:acetic acid:water 3:2:1 (v/v/v). The TLC was analyzed by phosphorimaging, and unlabeled standards (Gal, β-1,4-galactobiose, and β-1,4-galactopentaose) were visualized by dipping the TLC plate in 10% sulfuric acid in ethanol and charring with a heat gun.
To analyze the Gal linkage, products were generated using microsomes corresponding to 10 µg protein, 0.27 mM UDP-14C-Gal (14.8 kBq), and 2 µg galactopentaose acceptor and incubated for 1 h. Aliquots were lyophilized and dissolved in 50 mM NaOAc for digestion with endo-β-1,4-galactanase or in water for digestion with exo-β-1,4-galactosidase from Streptococcus pneumonia (ProZyme) using the buffer supplied with the enzyme. Digestion was done at 37°C with 0.4 units of galactanase for 2 h or 0.002 units of galactosidase for 16 h (one unit releases 1.0 µmol of p-nitrophenol from p-nitrophenol-β-galactoside per min at pH 6.0 and 37°C). After digestion, the sample was boiled for 5 min, centrifuged at 14,000g for 5 min, and subjected to TLC developed with n-propanol:ethanol:water 7:1:2 (v/v/v) and phosphorimaging as described above. The activity and specificity of the enzymes was tested in overnight incubations with 15 µg potato galactan (Megazyme), gum arabic (Sigma-Aldrich), and β-1,4-galactopentaose for the galactanase (0.4 units) and with β-1,3-galactobiose (Carbosynth), methyl-3-O-(β-d-galactopyranosyl)-β-d-galactopyranoside (Carbosynth), β-1,6-galactobiose (Carbosynth), β-1,4-galactobiose (Megazyme), and β-1,4-galactopentaose for the galactosidase (0.002 units). Incubations were done at least 10 times longer than required for complete digestion of substrates. The products were analyzed by TLC developed with n-propanol:ethanol:water 7:1:2 (v/v/v) and visualized by charring.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At2g33570/GALS1, At5g44670/GALS2, and At4g20170/GALS3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of GT92 Proteins.
Supplemental Figure 2. Monosaccharide Composition of Cell Walls of the Wild Type and the gals2-2 and gals3-2 Mutants.
Supplemental Figure 3. Monosaccharide Composition of Cell Walls from Wild-Type and Mutant Stems and Seeds.
Supplemental Figure 4. Fully Synthetic β-1,4-Galactopentaose Acceptor.
Supplemental Figure 5. Purification of GALS1.
Supplemental Figure 6. Confirmation of Substrate Specificity of Galactosyl Hydrolases.
Supplemental Figure 7. Immunoblot Analysis of YFP-GALS1 Overexpressors.
Supplemental Figure 8. Monosaccharide Composition Analysis of T1 Generation YFP-GALS1 Overexpressors.
Supplemental Table 1. Primers for Genotyping of T-DNA Mutants.
Supplemental Table 2. Primers Used for Amplification of the Full-Length Sequence for the GALS Genes.
Supplemental Table 3. Primers and Restriction Sites for Cloning of Promoters.
Supplemental Table 4. Primers Used for qRT-PCR Analysis.
Supplemental Data Set 1. Alignment of GT92 Proteins.
Acknowledgments
This work conducted by the Joint BioEnergy Institute was supported by the Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy and by the Danish Strategic Research Council. We thank Sherry Chan for assistance with plant growth.
AUTHOR CONTRIBUTIONS
H.V.S. directed the project. A.J.M.L, B.E., E.A.R, C.R., Y.V., A.O., M.C.F.A., M.H.C., and H.V.S. designed and performed research. A.J.M.L., B.E., and H.V.S. analyzed data. A.J.M.L., B.E., Y.V., and H.V.S. wrote the article. All authors have approved the final article.
Glossary
- ANOVA
analysis of variance
- qRT-PCR
quantitative RT-PCR
- YFP
yellow fluorescent protein
- TLC
thin layer chromatography
- 2AB
2-aminobenzamine
- Col-0
Columbia-0
- AIR
alcohol-insoluble residue
- HPAEC
high-performance anion exchange chromatography
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersen M.C.F. (2010). Synthesis of Oligosaccharides for Microarray Galectin Screening. Master’s thesis (Kongens Lyngby, Denmark: Technical University of Denmark).
- Andersson-Gunnerås S., Mellerowicz E.J., Love J., Segerman B., Ohmiya Y., Coutinho P.M., Nilsson P., Henrissat B., Moritz T., Sundberg B. (2006). Biosynthesis of cellulose-enriched tension wood in Populus: Global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 45: 144–165 [DOI] [PubMed] [Google Scholar]
- Arend M. (2008). Immunolocalization of (1,4)-beta-galactan in tension wood fibers of poplar. Tree Physiol. 28: 1263–1267 [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 37(Database issue): D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.G. (2000). Recent developments in oligosaccharide synthesis. J. Chem. Soc. Perkin Trans. 1 2000: 2137–2160 [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Egelund J., Damager I., Faber K., Olsen C.E., Ulvskov P., Petersen B.L. (2008). Functional characterisation of a putative rhamnogalacturonan II specific xylosyltransferase. FEBS Lett. 582: 3217–3222 [DOI] [PubMed] [Google Scholar]
- Egelund J., Petersen B.L., Motawia M.S., Damager I., Faik A., Olsen C.E., Ishii T., Clausen H., Ulvskov P., Geshi N. (2006). Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-alpha-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18: 2593–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A., Price N.J., Raikhel N.V., Keegstra K. (2002). An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl. Acad. Sci. USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshi N., Jørgensen B., Scheller H.V., Ulvskov P. (2000). In vitro biosynthesis of 1,4-beta-galactan attached to rhamnogalacturonan I. Planta 210: 622–629 [DOI] [PubMed] [Google Scholar]
- Geshi N., Jørgensen B., Ulvskov P. (2004). Subcellular localization and topology of beta(1→4)galactosyltransferase that elongates beta(1→4)galactan side chains in rhamnogalacturonan I in potato. Planta 218: 862–868 [DOI] [PubMed] [Google Scholar]
- Gorshkova T., Morvan C. (2006). Secondary cell-wall assembly in flax phloem fibres: Role of galactans. Planta 223: 149–158 [DOI] [PubMed] [Google Scholar]
- Ha M.A., Viëtor R.J., Jardine G.D., Apperley D.C., Jarvis M.C. (2005). Conformation and mobility of the arabinan and galactan side-chains of pectin. Phytochemistry 66: 1817–1824 [DOI] [PubMed] [Google Scholar]
- Hansen S.F., Harholt J., Oikawa A., Scheller H.V. (2012). Plant glycosyltransferases beyond CAZy: A perspective on DUF families. Front. Plant Sci. 3: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J., Jensen J.K., Sørensen S.O., Orfila C., Pauly M., Scheller H.V. (2006). ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J., Jensen J.K., Verhertbruggen Y., Søgaard C., Bernard S., Nafisi M., Poulsen C.P., Geshi N., Sakuragi Y., Driouich A., Knox J.P., Scheller H.V. (2012). ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236: 115–128 [DOI] [PubMed] [Google Scholar]
- Harholt J., Suttangkakul A., Vibe Scheller H. (2010). Biosynthesis of pectin. Plant Physiol. 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C., Marcus S.E., Knox J.P. (2011). Monoclonal antibodies, carbohydrate-binding modules, and the detection of polysaccharides in plant cell walls. Methods Mol. Biol. 715: 103–113 [DOI] [PubMed] [Google Scholar]
- Ishii T., Ohnishi-Kameyama M., Ono H. (2004). Identification of elongating beta-1,4-galactosyltransferase activity in mung bean (Vigna radiata) hypocotyls using 2-aminobenzaminated 1,4-linked beta-D-galactooligosaccharides as acceptor substrates. Planta 219: 310–318 [DOI] [PubMed] [Google Scholar]
- Jarvis M.C., Hall M.A., Threlfall D.R., Friend J. (1981). The polysaccharide structure of potato cell walls: Chemical fractionation. Planta 152: 93–100 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.K., et al. (2008). Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20: 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Seymour G.B., Knox J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1[->]4)-[beta]-D-galactan. Plant Physiol. 113: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B. (2012). GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40(Database issue): D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T., Ono H., Ohnishi-Kameyama M., Kaneko S., Ishii T. (2006). Identification of a mung bean arabinofuranosyltransferase that transfers arabinofuranosyl residues onto (1, 5)-linked alpha-L-arabino-oligosaccharides. Plant Physiol. 141: 1098–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Madson M., Dunand C., Li X.M., Verma R., Vanzin G.F., Caplan J., Shoue D.A., Carpita N.C., Reiter W.D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín I., Dopico B., Muñoz F.J., Esteban R., Oomen R.J., Driouich A., Vincken J.P., Visser R., Labrador E. (2005). In vivo expression of a Cicer arietinum beta-galactosidase in potato tubers leads to a reduction of the galactan side-chains in cell wall pectin. Plant Cell Physiol. 46: 1613–1622 [DOI] [PubMed] [Google Scholar]
- McCartney L., Ormerod A.P., Gidley M.J., Knox J.P. (2000). Temporal and spatial regulation of pectic (1→4)-beta-D-galactan in cell walls of developing pea cotyledons: Implications for mechanical properties. Plant J. 22: 105–113 [DOI] [PubMed] [Google Scholar]
- McCartney L., Steele-King C.G., Jordan E., Knox J.P. (2003). Cell wall pectic (1→4)-beta-d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 33: 447–454 [DOI] [PubMed] [Google Scholar]
- McNab J.M., Villemez C.L., Albersheim P. (1968). Biosynthesis of galactan by a particulate enzyme preparation from Phaseolus aureus seedlings. Biochem. J. 106: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz E.J., Gorshkova T.A. (2012). Tensional stress generation in gelatinous fibres: A review and possible mechanism based on cell-wall structure and composition. J. Exp. Bot. 63: 551–565 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Moller I., Marcus S.E., Haeger A., Verhertbruggen Y., Verhoef R., Schols H., Ulvskov P., Mikkelsen J.D., Knox J.P., Willats W. (2008). High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 25: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- ØBro J., Harholt J., Scheller H.V., Orfila C. (2004). Rhamnogalacturonan I in Solanum tuberosum tubers contains complex arabinogalactan structures. Phytochemistry 65: 1429–1438 [DOI] [PubMed] [Google Scholar]
- Obro J., Borkhardt B., Harholt J., Skjøt M., Willats W.G., Ulvskov P. (2009). Simultaneous in vivo truncation of pectic side chains. Transgenic Res. 18: 961–969 [DOI] [PubMed] [Google Scholar]
- Oomen R.J.F.J., Dao-Thi B., Tzitzikas E.N., Bakx E.J., Schols H.A., Visser R.G.F., Vincken J.P. (2004). Overexpression of two different potato UDP-Glc 4-epimerases can increase the galactose content of potato tuber cell walls. Plant Sci. 166: 1097–1104 [Google Scholar]
- Orfila C., Knox J.P. (2000). Spatial regulation of pectic polysaccharides in relation to pit fields in cell walls of tomato fruit pericarp. Plant Physiol. 122: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenboll Sørensen S., Pauly M., Bush M., Skjøt M., McCann M.C., Borkhardt B., Ulvskov P. (2000). Pectin engineering: modification of potato pectin by in vivo expression of an endo-1,4-beta-D-galactanase. Proc. Natl. Acad. Sci. USA 97: 7639–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons H.T., et al. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.L., Egelund J., Damager I., Faber K., Jensen J.K., Yang Z., Bennett E.P., Scheller H.V., Ulvskov P. (2009). Assay and heterologous expression in Pichia pastoris of plant cell wall type-II membrane anchored glycosyltransferases. Glycoconj. J. 26: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie E.A., Hansen S.F., Baidoo E.E., Hadi M.Z., Keasling J.D., Scheller H.V. (2012). Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 159: 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach M.J., Mokshina N.Y., Badhan A., Snegireva A.V., Hobson N., Deyholos M.K., Gorshkova T.A. (2011). Development of cellulosic secondary walls in flax fibers requires beta-galactosidase. Plant Physiol. 156: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösti J., Barton C.J., Albrecht S., Dupree P., Pauly M., Findlay K., Roberts K., Seifert G.J. (2007). UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19: 1565–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G.J., Barber C., Wells B., Dolan L., Roberts K. (2002). Galactose biosynthesis in Arabidopsis: Genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr. Biol. 12: 1840–1845 [DOI] [PubMed] [Google Scholar]
- Sørensen I., Willats W.G. (2011). Screening and characterization of plant cell walls using carbohydrate microarrays. Methods Mol. Biol. 715: 115–121 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sterling J.D., Atmodjo M.A., Inwood S.E., Kumar Kolli V.S., Quigley H.F., Hahn M.G., Mohnen D. (2006). Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Yamamoto K. (2010). Molecular cloning of pigeon UDP-galactose:beta-D-galactoside alpha1,4-galactosyltransferase and UDP-galactose:beta-D-galactoside beta1,4-galactosyltransferase, two novel enzymes catalyzing the formation of Gal alpha1-4Gal beta1-4Gal beta1-4GlcNAc sequence. J. Biol. Chem. 285: 5178–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Belton P.S., Ng A., Ryden P. (1999). 13C MAS NMR studies of the effects of hydration on the cell walls of potatoes and Chinese water chestnuts. J. Agric. Food Chem. 47: 510–517 [DOI] [PubMed] [Google Scholar]
- Titz A., Butschi A., Henrissat B., Fan Y.Y., Hennet T., Razzazi-Fazeli E., Hengartner M.O., Wilson I.B.H., Künzler M., Aebi M. (2009). Molecular basis for galactosylation of core fucose residues in invertebrates: Identification of Caenorhabditis elegans N-glycan core alpha1,6-fucoside beta1,4-galactosyltransferase GALT-1 as a member of a novel glycosyltransferase family. J. Biol. Chem. 284: 36223–36233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvskov P., Wium H., Bruce D., Jørgensen B., Qvist K.B., Skjøt M., Hepworth D., Borkhardt B., Sørensen S.O. (2005). Biophysical consequences of remodeling the neutral side chains of rhamnogalacturonan I in tubers of transgenic potatoes. Planta 220: 609–620 [DOI] [PubMed] [Google Scholar]
- Vega-Sánchez M.E., et al. (2012). Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159: 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y., Marcus S.E., Haeger A., Verhoef R., Schols H.A., McCleary B.V., McKee L., Gilbert H.J., Knox J.P. (2009). Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J. 59: 413–425 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: An Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Yin L., Verhertbruggen Y., Oikawa A., Manisseri C., Knierim B., Prak L., Jensen J.K., Knox J.P., Auer M., Willats W.G., Scheller H.V. (2011). The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 4: 1024–1037 [DOI] [PubMed] [Google Scholar]