A combination of crystallography, interaction analysis, and nuclear import assays demonstrates a distinct mode of autoinhibition in rice importin- α1a and the binding of plant-specific nuclear localization signals (NLSs) to its minor NLS binding site.
Abstract
In the classical nucleocytoplasmic import pathway, nuclear localization signals (NLSs) in cargo proteins are recognized by the import receptor importin-α. Importin-α has two separate NLS binding sites (the major and the minor site), both of which recognize positively charged amino acid clusters in NLSs. Little is known about the molecular basis of the unique features of the classical nuclear import pathway in plants. We determined the crystal structure of rice (Oryza sativa) importin-α1a at 2-Å resolution. The structure reveals that the autoinhibitory mechanism mediated by the importin-β binding domain of importin-α operates in plants, with NLS-mimicking sequences binding to both minor and major NLS binding sites. Consistent with yeast and mammalian proteins, rice importin-α binds the prototypical NLS from simian virus 40 large T-antigen preferentially at the major NLS binding site. We show that two NLSs, previously described as plant specific, bind to and are functional with plant, mammalian, and yeast importin-α proteins but interact with rice importin-α more strongly. The crystal structures of their complexes with rice importin-α show that they bind to the minor NLS binding site. By contrast, the crystal structures of their complexes with mouse (Mus musculus) importin-α show preferential binding to the major NLS binding site. Our results reveal the molecular basis of a number of features of the classical nuclear transport pathway specific to plants.
INTRODUCTION
The nuclear envelope is a characteristic feature of eukaryotic cells. It separates two key cellular activities, transcription and translation, necessitating that proteins and RNA must travel constantly between the nuclear and cytoplasmic compartments in a regulated manner to ensure the distinctive composition of each component and allow precise control of cellular activities. Nuclear pore complexes (NPCs) perforate the nuclear envelope and form the gateway for the bidirectional movement of molecules. Passive diffusion of ions and small proteins is allowed through the NPCs, but macromolecules (>40 kDa) require an active transport process to pass the NPCs. Consequently, sophisticated transport machinery is necessary to recognize macromolecular cargos specifically in one compartment to carry them through the nuclear pore and to release them in the other compartment (Tran et al., 2007).
The classical import pathway is the best-characterized system to translocate cargo proteins selectively into the nucleus. In this pathway, the carrier protein importin-β (Impβ; karyopherin-β1) uses importin-α (Impα; karyopherin-α) as an adaptor to recruit proteins containing classical nuclear localization signals (cNLSs) and form an import complex (Lange et al., 2007; Marfori et al., 2011). In the nucleus, RanGTP (for GTP-bound Ras-related;nuclear protein) binds to Impβ and causes the disassembly of the import complex. RanGEF (for guanine nucleotide-exchange factor; termed RCC1 in mammals) recharges RanGDP with GTP in the nucleus, and RanGAP (Ran GTPase activating protein) stimulates GTP hydrolysis to yield the GDP-bound form in the cytoplasm (Quimby and Dasso, 2003). The nucleotide states of Ran have asymmetric distribution and impart directionality to nuclear transport. The nucleoporins in the NPCs assist the passage of the import complex in coordination with Impβ and also participate in the release of cargo protein from Impα (Matsuura and Stewart, 2005).
A crucial step in the classical nuclear import pathway is the specific recognition by Impα of the NLSs. The structures of Impα proteins from human (Homo sapiens), mouse (Mus musculus; mImpα), and yeast (Saccharomyces cerevisiae; yImpα) show conserved architectures; the proteins are constructed from a tandem series of 10 armadillo (ARM) repeats that generate an elongated and gently curving shape representing the NLS binding domain (Conti et al., 1998; Kobe, 1999; Tarendeau et al., 2007; Dias et al., 2009). The additional flexible N-terminal sequence constitutes the Impβ binding (IBB) domain (Görlich et al., 1996; Cingolani et al., 1999; Lott and Cingolani, 2011) and also includes an autoinhibitory sequence that mimics a basic NLS (Kobe, 1999; Kobe and Kemp, 1999). The NLS binding domain provides two distinct NLS binding sites, both formed by an array of Trp, Asn, and negatively charged residues located within a shallow groove on the inner concave surface. The two NLS binding sites span ARM repeats 2 to 4 (major site) and ARM repeats 6 to 8 (minor site), respectively. The cNLSs are composed of one (monopartite) or two (bipartite) stretches of positively charged amino acids. The major site is considered the high-affinity binding site for typical monopartite cNLSs (Conti et al., 1998; Fontes et al., 2000; Hodel et al., 2001), while bipartite NLSs bind to both minor and major sites. Structural and interaction studies of Impα:cNLS complexes have provided an understanding of recognition determinants of cNLSs and have led to the identification of consensus sequences for monopartite (K[K/R]X[K/R], corresponding to positions P2 to P5; [K/R] represents Lys or Arg, and X represents any amino acid) and bipartite ([K/R][K/R]X10-12[K/R]3/5, corresponding to positions P1′ and P2′ for the N-terminal basic cluster) cNLSs (Conti and Kuriyan, 2000; Fontes et al., 2000, 2003; Lange et al., 2007; Marfori et al., 2011).
Relatively little is known about the classical nuclear import pathway in plants. While plant orthologs have been identified for most proteins involved in the nuclear trafficking process in yeast and mammals, the equivalent of RCC1 has to our knowledge not been identified. Some further unique features of the classical nuclear import pathway in plants have also been noted. In Arabidopsis thaliana, Impα has been reported to mediate nuclear transport independent of Impβ (Hübner et al., 1999). Impα proteins from Arabidopsis and rice (Oryza sativa) display broader specificity than their mammalian counterparts (Hicks and Raikhel, 1995; Smith et al., 1997; Jiang et al., 1998; Tzfira et al., 2000). For example, the unusual NLS from yeast protein Matα2 is recognized by yeast and plant (Arabidopsis) Impα but not by the mammalian proteins (Hicks and Raikhel, 1995; Smith et al., 1997), and the VirE2 virulence protein from the plant pathogen Agrobacterium tumefaciens contains two bipartite NLSs functional only in plants (Guralnick et al., 1996; Tzfira et al., 2000). Recently, a random peptide library approach applied to human, plant, and yeast Impα variants identified a plant-specific NLS consensus sequence (LGKR[K/R][W/F/Y]) as one of six classes of cNLSs (Kosugi et al., 2009).
In this study, we investigated the molecular basis of the classical nuclear import pathway in plants. We first determined the crystal structure of rice Impα1a (rImpα1a). The structure reveals that the autoinhibitory mechanism mediated by the IBB domain of Impα operates in plants, with NLS-mimicking sequences from the IBB domain binding to NLS binding sites. The binding of IBB domain at the minor NLS binding site is distinct from what is observed in mImpα structure (Kobe, 1999). We further characterized structurally the binding of the prototypical monopartite cNLS from simian virus 40 (SV40) large T-antigen (SV40TAgNLS) to rImpα1a lacking the IBB domain (rImpα1aΔIBB), showing preferential binding at the major NLS binding site consistent with mammalian and yeast proteins. Finally, we characterized the binding of two plant-specific NLSs (Kosugi et al., 2009) to rImpα1a using quantitative binding assays, nuclear import assays in permeabilized cells, and crystallography. While we confirm that these NLSs preferentially bind to rice Impα as compared with mouse (Mus musculus) and yeast (Saccharomyces cerevisiae) proteins, they are functional using the mouse and yeast proteins also. Unexpectedly, the structures show that the reason for favored binding to rImpα1aΔIBB is that they bind preferentially to the minor NLS binding site, while they bind preferentially to the major NLS binding site in mImpαΔIBB. These crystallographic results were confirmed by comparing binding of the NLSs to minor- and major-site mutants of rice and mouse Impα proteins. Our data jointly advance our understanding of the plant-specific features of the essential cellular process of nuclear import.
RESULTS
Crystal Structure of Rice Impα1a and the Mechanism of Autoinhibition
To study the specific features of plant Impα proteins, we determined the crystal structure of rImpα1a at 2-Å resolution (Table 1). The structure shows the expected arrangement of 10 ARM repeats constituting the NLS binding domain (Figure 1; after superposition, root mean square distances are 1.63 Å for mouse Impα [for 377 Cα atoms; Kobe, 1999] and 1.18 Å for yeast Impα [for 375 Cα atoms; Matsuura and Stewart, 2004]) (Figure 2). Mapping of evolutionary conservation of plant Impα orthologs onto the rImpα1a surface (ConSurf; Ashkenazy et al., 2010) demonstrates that the NLS binding surface is highly conserved (Figure 2C).
Table 1. Crystallographic Data.
| Crystallographic data statistics | rImpα1a | rImpα1aΔIBB: SV40TAgNLS | rImpα1aΔIBB: A89NLS | rImpα1aΔIBB: B54NLS | mImpαΔIBB: A89NLS | mImpαΔIBB: B54NLS |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | C21 | C21 | P21 | P21 | P212121 | P212121 |
| Cell dimensions | ||||||
| a, b, c (Å) | 134.2, 72.7, 62.4 | 133.5, 73.8, 62.0 | 62.4, 141, 73.1 | 62.2, 141.4, 73.3 | 78.8, 90.1, 99.4 | 78.1, 90.4, 97.1 |
| α, β, γ (°) | 90, 91.20, 90 | 90, 91.80, 90 | 90, 89.98, 90 | 90, 90.20, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 19.84–2.00 (2.11–2.00) | 64.62–2.08 (2.19–2.08) | 19.82–2.30 (2.42–2.30) | 19.95–2.10 (2.21–2.10) | 19.85–2.10 (2.21–2.10) | 20.10–2.30 (2.42–2.30) |
| Rmergeb | 0.05 (0.22) | 0.08 (0.47) | 0.15 (0.48) | 0.11 (0.91) | 0.12 (1.06) | 0.13 (0.61) |
| <I/σ(I)> | 16.2 (5.4) | 15.1 (3.6) | 4.5 (2.0) | 10.8 (2.1) | 19.6 (2.9) | 7.1 (2.3) |
| Completeness (%) | 99.1 (95.7) | 97.1 (82.8) | 99.5 (98.7) | 99.8 (99.9) | 99.9 (100) | 99.2 (100) |
| Multiplicity | 3.7 (3.6) | 6.3 (5.6) | 3.3 (3.0) | 6.1 (5.7) | 12.2 (12.3) | 5.0 (5.1) |
| Wilson plot B (Å2) | 23.2 | 26.3 | 35.7 | 32.4 | 31.7 | 38.9 |
| Observations | 150,503 | 221,160 | 186,460 | 448,163 | 510,837 | 154,685 |
| Unique reflections | 40,273 | 35,070 | 55,804 | 73,652 | 41,996 | 30,886 |
| Refinement | ||||||
| Rwork/Rfreec | 14.96/17.78 | 14.92/17.36 | 16.8/19.34 | 17.48/19.74 | 15.4820.43 | 20.14/22.10 |
| Average B-factor | 33.7 | 30.9 | 41.9 | 50.3 | 40.5 | 44.4 |
| RMSDsd | ||||||
| Bond lengths (Å) | 0.012 | 0.011 | 0.009 | 0.014 | 0.014 | 0.013 |
| Bond angles (°) | 1.202 | 1.201 | 1.060 | 1.680 | 1.288 | 1.400 |
| Ramachandran plot (%)e | ||||||
| Favored | 98.8 | 99.5 | 94.6 | 98.7 | 98.8 | 98.1 |
| Allowed | 0.9 | 0.5 | 4.5 | 0.8 | 1.2 | 1.9 |
| Forbidden | 0.2 | 0 | 0.9 | 0.5 | 0 | 0 |
Numbers in parentheses refer to the statistics for the highest resolution shell.
Rmerge = ∑hkl(∑i(|I hkl,I − <I hkl >|))/∑hkl,i <I hkl>, where I hkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and <Ihkl> is the mean intensity of that reflection. Calculated for I > −3σ(I).
Rwork = Σhkl(||Fobshkl| − |Fcalchkl||)/|Fobshkl|, where |Fobshkl| and |Fcalchkl| represent the observed and calculated structure factor amplitudes. Rfree is equivalent to Rwork but calculated using 5% of the reflections.
Root mean square deviations.
Calculated using Molprobity (Davis et al., 2007).
Figure 1.
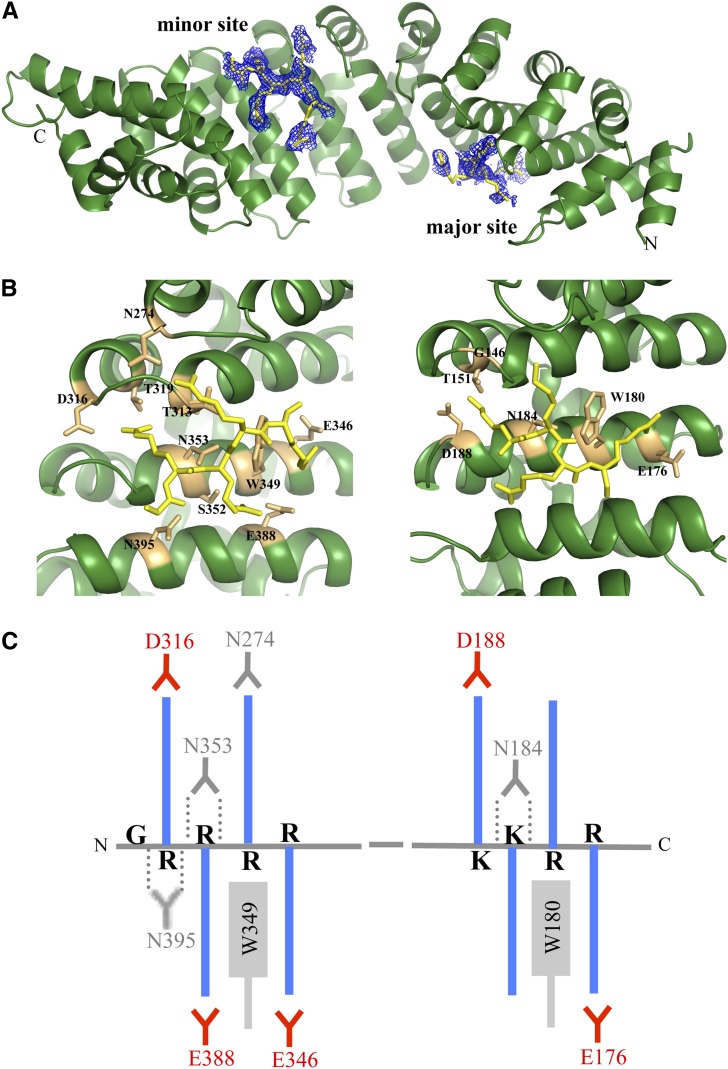
The Structure of Full-Length rImpα1a.
(A) The structure of rImpα1a comprises 10 ARM repeats (green, cartoon representation) and two NLS-like sequences from the N-terminal IBB domain (shown in yellow stick representation, superimposed with simulated annealing omit electron density map contoured at 2σ).
(B) The NLS-like sequences from the IBB domain (G25RRRR29 and K47KRR50 in yellow stick representation) interact with the labeled residues (in orange stick representation) from the minor (left) and major NLS binding sites (right), respectively, of rImpα1a.
(C) Schematic illustration of the main interactions between the IBB domain and rImp1a. The basic side chains (blue color) of Lys or Arg from NLS-like segments form salt bridges and electrostatic interactions with acid residues (red). The aliphatic portions of the basic chains interact with the Trp and Asn (in gray). The hydrogen bonds (dotted lines) are formed between Asn residues (in gray color) and the main-chain amides.
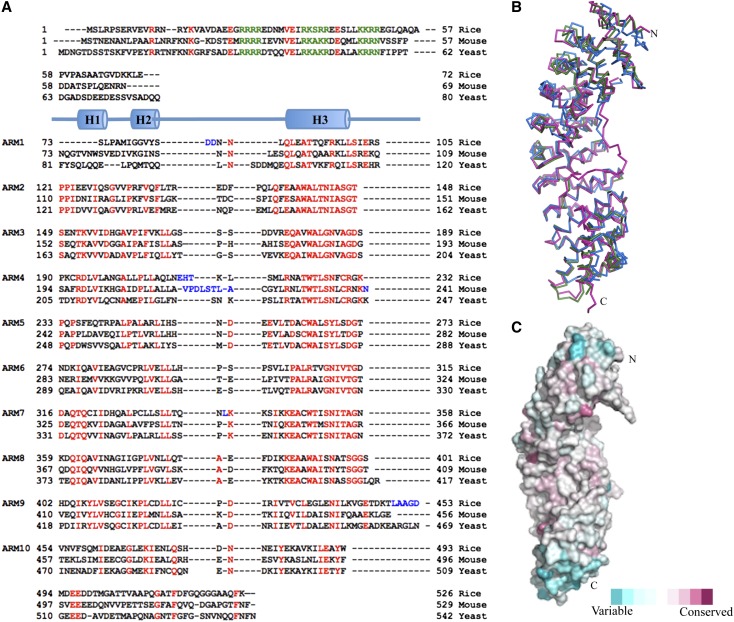
Figure 2.
Structure and Sequence Conservation in Impα Proteins.
(A) Structure-based alignment of mImpα, yImpα, and rImpα1a sequences (including the alignment of ARM repeats). Red highlights conserved residues; blue indicates insertions and deletions; green indicates the NLS-like segments.
(B) Superposition of full-length rice (green), mouse (blue; Kobe 1999), and yeast (magenta; from export complex structure; Matsuura and Stewart 2004) Impα proteins (ribbon representation).
(C) Sequence conservation of plant Impα proteins mapped onto the surface of rImpα1a (Consurf; Ashkenazy et al., 2010), based on the alignment of Impα proteins from Nicotiana benthamiana, Arabidopsis lyrata ssp lyrata, O. sativa ssp japonica, Trifolium pratense, Capsicum annuum, A. thaliana, and Z. mays. Red indicates conserved, while blue indicates variable.
The IBB domain was shown, in mammalian and yeast Impα, to contain an autoinhibitory region regulating NLS binding (Kobe, 1999; Fanara et al., 2000; Catimel et al., 2001; Harreman et al., 2003). The three NLS-like segments (R26RRR, R37KSRR, and K47KRR) in the IBB domain (Figure 2A) of rImpα1a are highly conserved in mouse and yeast proteins. The segment K47KRR resembles the autoinhibitory segment (K49RRN) identified in the mImpα structure (Kobe, 1999; Catimel et al., 2001). Most of the IBB domain in rImpα1 is disordered, but we were able to identify two small segments from this domain (G25RRRR and K47KRR) bound to the minor and major NLS binding sites, respectively (Figure 1A). As expected, the segment K47KRR (positions P2 to P5) forms hydrogen bonds and salt bridges with residues Asp-188, Gly-146, Thr-151, Asn-184, Trp-180, and Glu-176 in the major NLS binding site (Figures 1B and 1C, Table 2). However, the segment G25RRRR forms even more contacts with the ARM repeat domain in the minor NLS binding site (Figures 1B and 1C, Table 2). The average crystallographic B-factor (a parameter related to the mean square displacement of an atom and, therefore, atomic flexibility) of this segment (50.9 Å2) is also lower than of the major site binding segment (62.2 Å2). The observed pattern is unique to plant Impα because autoinhibitory contacts in mouse Impα were observed in the major site only (Kobe, 1999).
Table 2. NLS:Impα Interactions Based on the Corresponding Crystal Structures.
| Impα:NLS | NHB | NSB | Buried Surface Area (Å2) | Average B-Factor (Å2) ImpαΔIBB | Average B-Factor (Å2) NLS |
|---|---|---|---|---|---|
| mImpαΔIBB: A89 | 8 | 1 | 1163.6 | 44.15 | 67.28 |
| mImpαΔIBB: B54 | 10 (7) | 1 (3) | 1315.4 (935.6) | 49.30 | 68.80 (73.91) |
| rImpα1aΔIBB:A89 | (7) | (4) | (1139.9) | 44.01 | (35.13) |
| rImpα1aΔIBB:B54 | (11) | (6) | (1534.7) | 43.18 | (39.56) |
| rImpα1aΔIBB:SV40TAgNLS | 18 (13) | 1 (3) | 1521.1 (940.7) | 29.87 | 35.03 (46.41) |
| rImpα1a:IBB | 5 (15) | 2 (7) | 904.2 (1124.4) | 29.86 | 62.2 (50.94) |
Numbers of hydrogen bonds (NHB), salt bridges (NSB), and buried surface area of the interface between Impα and NLS peptides were calculated using PISA (Krissinel and Henrick, 2007). Values in parenthesis correspond to the minor site. Average B-factors for Impα and NLS peptides, indicating relative atomic flexibilities, were calculated using Baverage (Collaborative Computational Project, Number 4, 1994).
SV40 Large T-Antigen NLS Binds Preferentially to the Major NLS Binding Site in Rice Impα
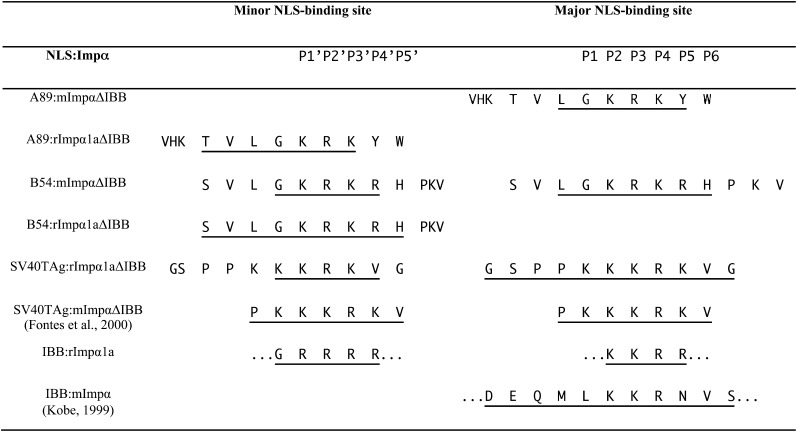
To determine how the prototypical monopartite cNLS from the SV40 large TAg binds to rImpα1a, we cocrystallized the corresponding peptide with rImpα1aΔIBB (the IBB domain was removed to avoid competition for binding between the autoinhibitory region and the NLS). Rice Impα and At-Impα were shown previously to bind to this NLS (Smith et al., 1997; Jiang et al., 1998). The structures are available for mImpα and yImpα in complex with SV40TAgNLS (Conti et al., 1998; Fontes et al., 2000). Consistent with these structures, a separate SV40TAgNLS peptide was found to interact each with both the major and minor NLS binding sites of rImpα1aΔIBB (see Supplemental Figure 1 online) but with more extensive interactions at the major site, which has generally been considered the biologically relevant binding site for most monopartite NLSs. Interestingly, the peptide binds in the minor site in a different register in rImpα1aΔIBB (with K129R in P1′-P2′ positions) than in yeast and mouse proteins (with K128K in P1′-P2′ positions).
Plant-Specific NLSs Are Functional Using Impα Proteins from Different Organisms, Although They Preferentially Bind to Rice Impα
Kosugi et al. (2009) used the in vitro virus method to screen random peptide libraries with Impα variants (rImpα1a and human Impα3, both lacking the IBB domain) as bait. We selected two peptides identified in this screen (A89 and B54) from the class 5 plant-specific NLSs (consensus sequence (LGKR[K/R][W/F/Y]) for further characterization. Kosugi et al. (2009) showed that class five NLSs were functional only in plant (tobacco [Nicotiana tabacum]) but not budding yeast and human (NIH3T3) cells and that they interacted only with rice Impα (and not yeast and human Impα proteins). We performed glutathione S-transferase (GST) pull-down assays to test if our two peptides bind to Impα proteins from different organisms (rImpα1aΔIBB, mImpαΔIBB, and yImpαΔIBB). In our hands, both peptides bound to all Impα variants, although they showed stronger binding to rImpα1aΔIBB (see Supplemental Figure 2 online). We quantified the interactions by microtiter plate binding assays using GST-SV40TAgNLS and GST as positive and negative controls, respectively (Table 3). GST-SV40TAgNLS bound to all three Impα variants with Kd values consistent with previous studies (Catimel et al., 2001; Leung et al., 2003). A89 and B54 NLSs bound to rImpα1aΔIBB with affinities in the nanomolar range (A89NLS, Kd = 140 nM; B54NLS, Kd = 25 nM), more than 10-fold higher than in the case of the other two Impα variants. Additionally, our binding assays showed that full-length rImpα1a interacted with A89 and B54 NLSs with lower affinity than rImpα1aΔIBB (Kd = 6 and 0.12 μM for A89NLS and B54NLS, respectively), consistent with the autoinhibitory function of the IBB domain.
Table 3. The Dissociation Constants (Kd, μM) for Impα:NLS Interactions.
| GST-NLS | mImpαΔIBB | yImpαΔIBB | rImpα1aΔIBB | mImpαΔIBB D192K | mImpαΔIBB E396R | rImpα1a | rImpα1aΔIBB D188K | rImpα1aΔIBB E388R |
|---|---|---|---|---|---|---|---|---|
| SV40TAg | 0.0070 ± 0.0019 | 0.0110 ± 0.0024 | 0.0040 ± 0.0002 | 3.91 ± 0.27 | 0.053 ± 0.009 | 1.73 ± 0.15 | 0.680 ± 0.094 | 0.0470 ± 0.008 |
| A89 | 2.90 ± 0.36 | 1.70 ± 0.20 | 0.140 ± 0.023 | Not detected | 7.65 ± 1.54 | 5.95 ± 1.63 | 0.410 ± 0.096 | 25.0 ± 8.5 |
| B54 | 1.70 ± 0.23 | 1.80 ± 0.20 | 0.025 ± 0.004 | 5.61 ± 2.00 | 4.95 ± 0.56 | 0.118 ± 0.017 | 0.0270 ± 0.0017 | 0.137 ± 0.019 |
The Kd values (in μM) were calculated using program GraphPad (Prism). Each assay was performed in triplicate, and the values with se correspond to the best fit to the one-site specific binding equation [Y = Bmax*X/(Kd + X), where Bmax is the maximum specific binding with the same unit as Y, Kd is the equilibrium binding constant, and X is ligand concentration]. The calculation assumed one-site binding to allow the comparison of overall binding affinities between different samples. The binding isotherms of representative measurements are provided in the Supplemental Figure 7 online.
We further examined the activity of A89 and B54 NLSs to import a nondiffusible cargo protein into the nucleus in digitonin-permeabilized HEp-2 cells using Impα variants from different organisms. The NLS peptides were tagged with GST–green fluorescent protein (GFP) (molecular mass > 50 kD; to prevent passive diffusion) to determine the cellular location of the cargo based on GFP fluorescence. The concentration of digitonin was optimized to ensure that the nuclear envelope of the cells was intact, as validated by the accumulation of Texas-red dextran (70 kD) only in the cytoplasm (Figure 3). The GST-GFP-SV40TAgNLS cargo was excluded from the nucleus when no ATP-generating system (creatine phosphate, creatine phosphate kinase, ATP, and GTP) or no transporter (Impα:mImpβ complex) was added and when the cells were treated with wheat germ agglutinin before reaction to block the nuclear pore and inhibit the nuclear import activity. Conversely, the GST-GFP-SV40TAgNLS cargo was observed in the nucleus when all reaction components were added using Impα proteins from different organisms, suggesting the reaction was based on Impα-dependent nuclear import. Using this assay, both GST-GFP-A89NLS and GST-GFP-B54NLS cargoes were functional using all three Impα proteins from different organisms.
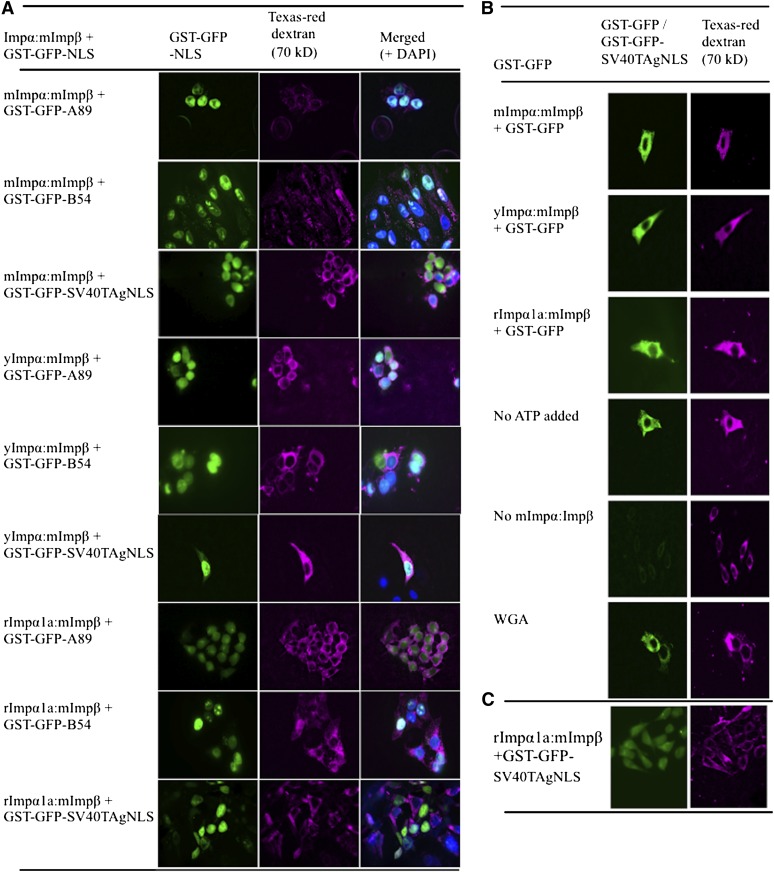
Figure 3.
A89 and B54 NLS Peptides Can Be Translocated into the Nucleus Using Impα Proteins from Different Organisms.
Nuclear import assays were performed in permeabilized human (HEp-2) cells. The cargo proteins (GST-GFP) fused to various NLS peptides (A89, B54, and SV40TAg) were transported into the nucleus by mouse Impβ and mImpα, yImpα, and rImpα1a.
(A) The left panel shows localization of GFP; the middle panel shows localization of Texas-red dextran (70 kD; magenta), showing the condition of the nuclear membranes; and the right panel is the merged image with 4′,6-diamidino-2-phenylindole staining indicating the location of the nuclei (blue).
(B) Import assay control experiments. The first three panels show that a cargo without NLS (GST-GFP) is not translocated into the nucleus by Impα:mImpβ. In the last three panels, GST-GFP-SV40TAgNLS import by mImpα1a:mImpβ failed due to the lack of an ATP generating system (no ATP added), lack of transporter proteins (no mImpα:Impβ), or addition of wheat germ agglutinin (WGA).
(C) The images show that the majority of the cells have intact nuclear membranes as indicated by the accumulation of Texas-red dextran (magenta) in the cytoplasm and GST-GFP-SV40TAgNLS (green) being translocated to the nucleus. We counted 2166 nuclei, and <4% of those were permeable to Texas-red dextran.
Crystal Structures Reveal Preferential Binding of A89 and B54 NLSs to the Minor NLS Binding Site of Rice Impα
To investigate the recognition of A89 and B54 NLSs by rImpα1a at the atomic level, we cocrystallized both NLSs with rImpα1aΔIBB. The rImpα1aΔIBB:A89NLS and rImpα1aΔIBB:B54NLS complex crystals diffracted to 2.1- and 2.3-Å resolution, respectively. Unexpectedly, both NLSs were found to bind specifically to the minor NLS binding site (Figure 4A). The main chain runs antiparallel to the direction of ARM repeats of rImpα1a, as observed for other Impα:NLS complexes (Marfori et al., 2011). While both NLSs bind in a similar conformation, B54NLS shows more contacts than A89NLS (Figure 4B). Both NLSs have Lys and Arg at P1′ and P2′ positions, respectively (Figure 5), as observed for most bipartite cNLS:Impα structures (Marfori et al., 2011, 2012); these residues interact with Glu-388, Asn-353, Val-312, and Ser-352 using hydrogen bonds and salt bridges. However, both NLSs make substantial interactions both N- and C-terminally to the conventional minor site binding positions. For example, the Leu residue at position P1′ from both NLSs binds to a highly complementary pocket on the rImpα1a surface, and residues Arg-8 and His-9 at the P4′ and P5′ positions of B54NLS interact extensively with Asp-271, Glu-346, and Glu-388 of rImpα1aΔIBB (Figure 4C).
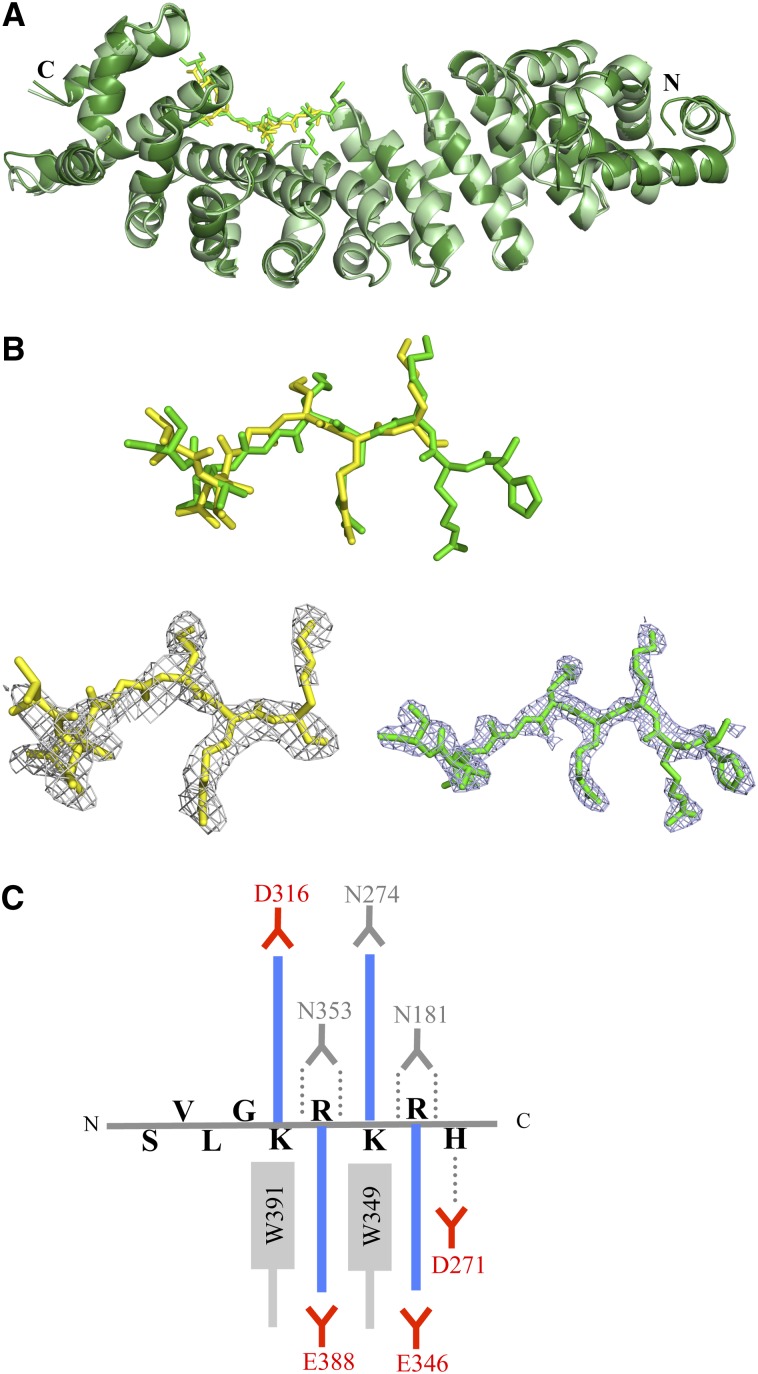
Figure 4.
A89 and B54 NLS Peptides Bind to the Minor NLS Binding Site of rImpα1aΔIBB.
(A) Superposition of rImpα1aΔIBB:A89 (light green/yellow) and rImpα1aΔIBB:B54 (dark green/green) complex structures (cartoon representation, NLS peptides in stick representation).
(B) Simulated annealing omit electron density maps (contoured at 2σ; gray mesh) superimposed onto the structure of A89 (top, yellow) and B54 (bottom, green) NLSs (stick representation).
(C) Schematic illustration of the interactions between B54NLS and rImp1a. The basic side chains (blue color) of Lys or Arg form salt bridges and electrostatic interactions with acid residues (red). The aliphatic portions of the basic chains interact with the Trp and Asn (in gray). The hydrogen bonds (dotted lines) are formed between Asn residues (in gray color) and the main-chain amides.
Figure 5.
NLS Binding to Specific Binding Pockets in rImpα1a and mImpα Proteins, Based on Structural Information.
The NLS-like sequences are aligned based on the interaction with Impα binding sites. The residues visible in the electron density map are underlined.
To investigate the reasons for the higher affinity binding to rice Impα than to the proteins from mouse and yeast, we cocrystallized A89 and B54 NLSs also with mImpαΔIBB. The structures of mImpαΔIBB:A89NLS and mImpαΔIBB:B54NLS were determined at 2.1- and 2.3-Å resolution, respectively (see Supplemental Figure 3 online). Unexpectedly, electron density maps showed binding of the NLSs at the major site, consistent with the binding of most monopartite NLSs (Marfori et al., 2011). The B54NLS additionally binds to the minor NLS binding site but is less well defined in the electron density and allowed us to model only the G4KRKR sequence. This interaction allows us to compare directly its binding to the minor site in rice and mouse proteins. Lys-5 and Arg-6 correspond to the P1′ and P2′ positions, respectively, binding to residues Thr-328, Asp-361, Glu-396, and Ser-360, while residues Lys-7 and Arg-8 occupy the P3′ and P4′ positions (Marfori et al., 2011). The P3′ Lys-7 participates in hydrogen binding interactions with Gly-281 and Asn-283, and the P4′ Arg-8 forms a salt bridge with Glu-396.
Mutational Analysis Confirms Plant-Specific NLS Binding Modes Observed Crystallographically
Point mutations were made in either the major or minor NLS binding sites of rImpα1aΔIBB and mImpαΔIBB to verify the NLS binding modes observed crystallographically. The rImpα1a residues Asp-188 and Glu-388 are conserved in different species (Asp-192 and Glu-396 in mImpα; Asp-203 and Glu-402 in yImpα) and are essential for NLS binding to the two corresponding binding sites (Robbins et al., 1991; Efthymiadis et al., 1997; Leung et al., 2003). SV40TAgNLS bound to the rImpα1aΔIBB minor-site mutant (E388R) with higher affinity than the major-site mutant (D188K; Table 3). An analogous result was obtained using mImpα1ΔIBB major-site (D192K) and minor-site (E396R) mutants. On the other hand, the mutation in the minor site of rImpα1aΔIBB (E388R) caused a 100-fold and sixfold reduction in binding compared with the wild-type protein for A89 and B54 NLS, respectively (Table 3), while the mutation in the major site (D188K) had much less impact on the binding affinity for these NLSs. The converse was the case for mImpα mutants; the binding was decreased more in the major-site than the minor-site mutants. All these results are consistent with the NLS binding modes observed in the crystal structure.
DISCUSSION
Structure of Full-Length Rice Impα1a Reveals a Previously Uncharacterized Mode of Autoinhibition
Currently, two structures of unliganded full-length Impα proteins are available: the structure of the mouse protein (Kobe, 1999) and the structure presented here for the rice protein. These two proteins share 46% sequence identity (Figure 2A) and display virtually identical folds, composed of 10 tandem ARM repeats. The N-terminal IBB domain is mostly disordered in rImpα1a, with the exception of two NLS-like sequences (K47KRR and G25RRRR) in the IBB domain that occupy the major and minor NLS binding sites, respectively. The IBB domain of virtually all Impα proteins contains three clusters of positively charged residues (R26RRR, R37KSRR, and K47KRR in rImpα1a) (see Supplemental Figure 4 online). The third cluster (K49RRN in mImpα) of the IBB domain occupies the major NLS binding site of mImpα (Kobe, 1999; Catimel et al., 2001). In yImpα, the C-terminal cluster has similarly been implicated in autoinhibition, based on mutational analyses and in vivo functional assays (Harreman et al., 2003). The C-terminal cluster K47KRR similarly occupies the major NLS binding site in rImpα1a, but in this case, the minor site is occupied by the N-terminal cluster G25RRRR. The structure of the export complex Cse1p:Kap60p:RanGTP (Matsuura and Stewart, 2004) similarly shows the equivalent cluster R33RRR bound at the minor NLS binding site of yImpα, although the interaction is likely reinforced by the presence of Cse1p. The additional interactions between the IBB domain and the minor site of rImpα1a could be an important feature in plant Impα proteins, reinforced by the observed binding mode of plant-specific NLSs to the minor NLS binding site. Further experiments are needed to establish the differential roles of the NLS-like segments from rImpα1a IBB, but our structural studies suggest that both N- and C-terminal NLS-like sequences in rImpα1a play a role in autoinhibition. In addition to the autoinhibitory function, the IBB domain plays a regulatory role in facilitating the formation of the import complex, translocation of the import complex, and recycling of Impα and Impβ (Lott and Cingolani, 2011); the observed differences may therefore have complex effects on the nucleocytoplasmic import cycle.
Distinct Features of NLS Binding to Rice Impα
The structure of the rImpα1aΔIBB bound to the well-characterized prototypical monopartite SV40TAgNLS reveals the expected binding mode in the major NLS binding site, analogous to yeast and mouse proteins (Conti et al., 1998; Fontes et al., 2000). To examine the specific features of plant Impα specificity, we analyzed the binding to Impα of plant-specific NLSs, identified based on peptide library experiments (Kosugi et al., 2009). GST pull-down assays showed that our two NLSs tested (A89 and B54) bind not only to rice Impα, but also to the yeast and mouse proteins. Both plant-specific NLSs showed stronger binding to the rice Impα, and we confirmed this by determining Kd values using microtiter plate-based assays. The affinities of both NLSs for rImpα1aΔIBB were in the nanomolar range, which is consistent with the values obtained for other functional NLSs (Hu and Jans, 1999; Fanara et al., 2000; Catimel et al., 2001). By contrast, these NLSs showed micromolar affinities for binding yImpα and mImpα. The NLS:Impα binding affinity has been shown to correlate with NLS import activity, and micromolar affinity falls at the lower limit for a functional NLS (Hu and Jans, 1999; Hodel et al., 2001). We therefore tested if these NLSs were functional using nuclear import assays in permeabilized cells and found that both NLSs were capable of facilitating the import of GST-GFP-NLS cargo using the Impα variants from all three organisms. The NLS from the human phospholipid scramblase 4 (hPLSCR4), binding Impα with Kd ∼48 μM, was similarly shown to be functional (Lott et al., 2011).
Consistent with the results of Kosugi et al. (2009), both A89 and B54 NLSs bound to rImpα1a more strongly than to yeast and mammalian proteins. Tracing the phylogeny of eukaryotic Impα proteins, they originate from an ancestral α1-like gene sequence, which gave rise to the animal α1 genes and fungal and plant α1-like genes (Goldfarb et al., 2004). The animal α1 gene family is more similar to plant and fungal α1-like genes than they are to the animal α2 or α3 groups (Mason et al., 2009). Animal α2 and α3 genes occur only in metazoan animals and are likely linked to cell and tissue development and differentiation (Goldfarb et al., 2004). The yImpα, rImpα1a, and mImpα proteins used in our study belong to fungal α1-like, plant α1-like, and animal α2 families, respectively (see Supplemental Figure 5 and Supplemental Data Set 1 online). These proteins differ more than Impα variants from the same species (e.g., rImpα1a and 1b share 83% sequence identity). It is therefore not unexpected that significant differences in NLS binding specificity exist among these Impα proteins. The crystal structures of A89 and B54 NLSs in complex with rice and mouse Impα proteins unexpectedly show that the basis for the differential binding affinities is that these NLSs bind preferentially to the minor NLS binding site of plant Impα, but to the major NLS binding site of the mammalian protein. We validated the binding modes observed crystallographically using Impα mutants; whereas a minor-site mutant of rImpα1a bound these NLSs with reduced affinity, it was the major-site mutant that had the analogous effect in mImpα. While most monopartite NLSs bind to the major NLS binding site (Marfori et al., 2011), minor site–specific NLSs have been identified previously. Minor site–specific consensus sequences (KRX[W/F/Y]XXAF and [R/P]XXKR[K/R][-D/E], where X represents any residue, residues in the square brackets indicate the favorable residues at that position, and “-” signifies residues that are not found at that position) have been identified by Kosugi et al. (2009) through screening random peptide libraries and mutational analysis; however, the binding modes of these NLSs have not been characterized structurally. The A89 and B54 sequences do not match either of these two consensus sequences. On the other hand, the binding of the NLSs from TPX2 (target protein for Xenopus laevis kinesin-like protein 2) (Giesecke and Stewart, 2010) and hPLSCR4 (Lott et al., 2011) to the minor NLS binding site has been observed structurally. A89 and B54 NLSs make use not only of the usual minor-site cavities (positions P1′ to P4′) but also have more extended contacts in the C-terminal region of rImpα1a. By contrast, the TPX2 NLS (K284RKH) and hPLSCR4 NLS (G273SIIRKWN) contain a short basic cluster and form only a few interactions at the minor binding site of the mImpα. These differential interactions observed in the structures are also reflected in the binding affinity. The Kd values for A89 and B54 binding to rImpα1a are in the nanomolar range, much higher than the micromolar Kd reported for hPLSCR4 NLS.
Structural Basis of Differential Binding of NLSs to Impα Proteins from Different Organisms
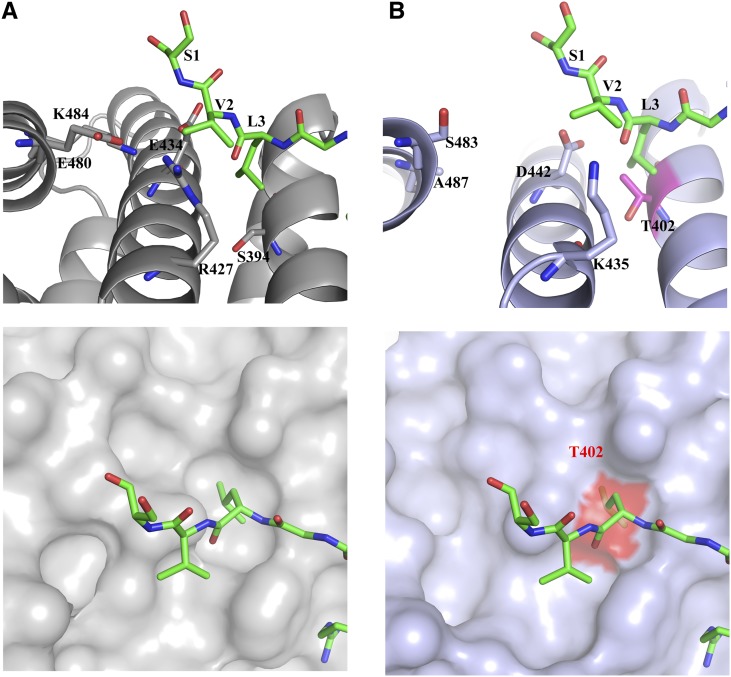
Structural analyses of NLS:Impα complexes suggest that the differences in the C-terminal region of rice and mouse Impα proteins are likely responsible for the distinct binding modes. Unique residues can be found in ARM repeats 8 and 9; for example, Thr-402, Lys-435, Asp-442, Ser-483, and Ala-487 in mImpα correspond to Ser-394, Arg-427, Glu-434, Glu-480, and Lys-484 in rImpα1a (Figure 6A). This creates a less favorable binding site for the residues SVL3 of B54 and TVL6 of A89 in mImpα. In particular, the shared Leu residue shows a considerable buried surface area in rImpα1a, but because rImpα1a Ser-394 is replaced by Thr-402 in mImpα, an equivalent binding mode is prevented through steric hindrance (Figure 6B). Sequence alignments (see Supplemental Figure 6 online) show high conservation of the minor-site residues toward the N terminus and lower identity toward the C terminus. Mapping the amino acids that differ among Impα proteins from α1, α2, and α3 groups (Mason et al., 2009) onto the surface uncovers several variations at the edges of the binding groove, which could result in differences in specificity among these proteins. The observed specificity could be further influenced by the aromatic (W/F/Y) residue that follows the basic cluster in the plant-specific NLS consensus sequence; a positively charged residue in this position could find higher complementarity in the major site, so by not having such a residue in this position, binding to the minor site may be promoted.
Figure 6.
Differential Binding of Plant-Specific NLSs to rImpα1a and mImpα.
(A) B54NLS:rImpα1aΔIBB complex. Top: The C-terminal residues of B54NLS (SVL3 in green stick representation) bound to rImpα1aΔIBB (gray cartoon representation, contact residues in gray stick representation). Bottom: Top view of the pocket (in gray surface representation) that accommodates Leu3 from the peptide.
(B) Superposition of the mImpαΔIBB:B54NLS and rImpα1aΔIBB:B54NLS structures. Top: mImpαΔIBB:B54NLS (light blue, cartoon representation, B54NLS omitted) and rImpα1aΔIBB:B54NLS (green, stick representation, rImpα1a omitted) in a view analogous to (A). Leu-3 from the peptide shows steric hindrance with Thr-402 (in magenta stick representation) from mImpαΔIBB. Bottom: Top view of the pocket (in light blue surface representation). Thr-402 is shown in red surface representation.
Figure 5 summarizes the binding of all NLSs studied here, relative to the binding pockets in the NLS binding groove of Impα. Positions P2 to P4 (in the major site) and P1′ to P3′ (in the minor site) are mainly occupied by positively charged residues (Lys and Arg). Thus, the residues flanking these basic clusters are responsible for the differential binding we observed. For instance, in the highly conserved major site, SV40TAgNLS binding to ARM repeats 1 to 5 forms more contacts than B54NLS; in particular, the residues at the N terminus of the SV40TAgNLS (SPP126) form hydrogen bonds with rImpα1a residues Arg-230 and Trp-223, whereas there is no close contact between N-terminal residues of the B54NLS (VLG4) and mImpα. Instead, the small hydrophobic residues such as Val and Leu at P1′ position in plant-specific NLSs use distinct interacting residues from the C-terminal region of rImpα1a to bind specifically to the minor NLS binding site.
Proteins with Plant-Specific NLSs
To investigate the prevalence of plant-specific NLSs resembling A89 or B54 in different species of plants, we constructed a position weight matrix to search protein sequences in the phylum Streptophyta. There are a number of proteins that contain the core basic segment (LGKR[K/R][W/F/Y]) within these NLSs (see Supplemental Data Set 2A online). Using Fisher’s exact test, the proportion of proteins containing the core basic segment in the Streptophyta phylum is noticeably greater than in the taxonomic class Mammalia (P = 0.23) and the genus Saccharomyces (P = 0.04). A similar, but more pronounced, outcome is observed if the proteomes of specific species are compared. O. sativa ssp japonica has significantly more matches than other proteomes (compared with Arabidopsis, P = 0.05; human, P = 1.42e-11; mouse, P = 2.75e-06; and S. cerevisiae, P = 0.03; see Supplemental Table 1 online). There is no statistically significant difference between any pair of nonplant proteomes. Notably, Gene Ontology terms (Ashburner et al., 2000) show that a number of proteins containing plant-specific NLS motifs are involved in nuclear processes such as DNA and RNA binding, DNA replication, and DNA integration (see Supplemental Data Set 2B online). For example, the chromatin remodeling factor PICKLE (PKL; UniProt ID Q9S775) contains the sequence L873GKRKR identical to all the key residues in B54NLS. In Arabidopsis, the PKL protein functions as a regulator to facilitate postgerminative growth by repressing the expression of embryonic trait genes (Ogas et al., 1999; Li et al., 2005). This protein has been shown to accumulate in the nucleus (Li et al., 2005) and was identified in nuclei-enriched fractions from Arabidopsis (Jones et al., 2009). The same sequence motif is found in the Arabidopsis ethylene-responsive transcription factor ERF104 (UniProt ID Q9FKG1), a nuclear protein implicated in coordinating stress responses and plant defense (Bethke et al., 2009). A similar sequence (L128GKRRR) is found in the DNA cross-link repair protein SNM1 (UniProt ID Q38961), required for the repair of DNA lesions caused by oxidative stress in Arabidopsis (Molinier et al., 2004).
Plant-Specific Features of Nuclear Import
The general characteristics of nuclear transport pathways are highly conserved between plants and other eukaryotes (Hicks and Raikhel, 1995; Tzfira et al., 2000; Merkle, 2011), although some plant-specific features have been noted, including broader specificity of NLS recognition and crosstalk between nuclear import and other plant-excusive biological activities, such as regulation of temperature stress, light signaling, and disease resistance (Yamamoto and Deng, 1999; Liu and Coaker, 2008; Merkle, 2011). The A89 and B54 NLSs we examined here show preference for binding plant Impα through superior binding at the minor NLS binding site. The minor site therefore does not merely play a supplementary role to the major site in bipartite NLS binding but can substitute in a primary role and enable nanomolar-range affinity for cargoes containing plant-specific NLSs resembling A89 and B54. The changed usage of the minor NLS binding site may relate to the binding of the extra autoinhibitory segment from the IBB domain to the minor NLS binding site, observed in the plant but not mammalian Impα structure.
In conclusion, we determined the crystal structure of rice Impα as a representative plant protein. Although the structure of this protein is very similar to the structures of yeast and mammalian Impα proteins determined previously, as expected based on the conservation of nuclear transport machinery between these organisms, the structure reveals plant protein-specific features. In particular, the structure shows autoinhibitory interactions in both minor and major NLS binding sites. This may have important implications for the regulation of the import pathway in the light of our finding that plant-specific NLSs bind to the minor NLS binding site in rice Impα, but instead to the major site in mouse Impα. Our work reveals the molecular basis for this differential binding. However, while plant-specific NLSs show preference for plant Impα, we show they are not exclusively functional with the plant receptor but can also function with yeast and mammalian Impαs.
METHODS
Generation of Recombinant DNA Constructs
The cDNAs corresponding to NLSs, containing BamHI and EcoRI restriction sites at the 5′ end and 3′ ends, respectively, were cloned into the pGEX2T vector (GE Healthcare) pretreated with restriction enzymes (BamHI-HF and EcoRI-HF, 20 units/µL; NEB). The complementary oligonucleotides were ligated into the pGEX2T vector using Quick T4 DNA Ligase (New England Biolabs) according to the manufacturer’s instructions.
The expression plasmid (pGEX-Osimpα1ΔIBB) coding for rImpα1aΔIBB (rice [Oryza sativa] Impα1a residues 73 to 526; NP_001042611.1; Kosugi et al., 2009) was subcloned from the pGEX6T-1 into the pET30a (Novagen) vector with BamHI and SalI restriction sites at 5′ and 3′ ends, respectively. The cDNA corresponding to the IBB domain of rImpα1a (residues 1 to 72) was obtained by designing multiple oligonucleotides with the help of DNAWorks (Hoover and Lubkowski, 2002), followed by PCR. The BamHI and EcoRI restriction sites were added to 5′ and 3′ ends, respectively, of the first and the last designed oligonucleotides. The PCR steps (gene assembly and gene amplification) were performed following Hoover and Lubkowski (2002). The cDNAs corresponding to the IBB domain and rImpα1aΔIBB were used individually as templates with 5′ and 3′ primers containing BamHI and EcoRI restriction sites to perform PCR-driven overlap extension (Heckman and Pease, 2007). The PCR product was then cloned into the pGEX2T vector. The pET30a-rImpα1a construct was subcloned from pGEX2T-rImpα1a using the BamHI and SalI restriction sites. The mutations (D188K and E388R) in NLS binding sites were introduced into the pET30a-rImpα1aΔIBB construct using site-directed mutagenesis with KOD hot start polymerase (Novagen). The pGEX2T-EGFP-(NLS) constructs were generated by amplifying the enhanced GFP DNA from pEGFP-N1 vector (Clontech) with the designed primers. The amplified enhanced GFP DNA was then cloned into pGEX2T and pGEX2T-NLS constructs. All constructs above were verified by sequencing (Australian Genome Research Facility). The oligonucleotides used are listed in Supplemental Table 2 online.
Recombinant Protein Expression and Purification
Mouse (Mus musculus) Impα lacking the IBB domain (mImpαΔIBB, residues 70 to 529; NP_034785) was expressed as previously described (Fontes et al., 2000) and purified using a HisTrap column (5 mL; GE Healthcare), followed by size exclusion chromatography (S-200; GE Healthcare) in 20 mM Tris, pH 7.8, and 125 mM NaCl. Saccharomyces cerevisiae ImpαΔIBB (yImpαΔIBB, residues 88 to 530 of Kap60p; Marfori et al., 2012) and rImpα1aΔIBB (residues 73 to 526) were expressed using the autoinduction method in BL21 (DE3) cells (Studier, 2005) and purified in the same way as mImpαΔIBB. The ImpαΔIBB:NLS complexes were purified as described previously (Marfori et al., 2012). The NLS peptides were overexpressed as GST fusion proteins (GST-NLSs) using the autoinduction method and immobilized on a GSTrap column (5 mL; GE Healthcare). The column was further washed with the binding buffer (50 mM Tris, pH 7.8, and 125 mM NaCl) and injected with purified ImpαΔIBB. The GST-NLS:ImpαΔIBB complex was eluted in the elution buffer containing 10 mM glutathione and digested with thrombin at 4°C overnight. The protein was further purified using S-200 and GSTrap columns. The pure ImpαΔIBB:NLS complexes were concentrated to 11 to ∼18 mg/mL (Amicon filter, MWCO 10 kD; Millipore). The concentrated proteins were snap-frozen in liquid nitrogen in small aliquots. The expression of GST-Ran, GST-Impα (full-length mImpα, yImpα, and rImpα1a), GST-mImpβ, GST-GFP, and GST-GFP-NLSs was achieved by the autoinduction method. These GST fusion proteins were purified using GSTrap and S-200 columns and finally eluted in 20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT, and 1 mM EDTA. To obtain purified Impα:mImpβ complexes, the pure GST-mImpβ was immobilized on the GSTrap column and injected with pure full-length Impα proteins, pretreated with thrombin to remove the GST tag. The complexes were further treated with thrombin at 4°C overnight and then purified using a S-200 column. Purified Ran was also obtained in the same way and further incubated with GDP (Sigma-Aldrich) in 10-fold molar ratio. All proteins were concentrated to desired concentrations and snap-frozen in liquid nitrogen for storage at –80°C.
In Vitro GST Pull-Down Assay
Bacterial pellets from 5 mL of cell culture expressing GST or GST-NLS fusion proteins were resuspended in 500 μL GST pull-down buffer (50 mM Tris-HCl and 125 mM NaCl at pH 7.8). The cells were lysed by three cycles of freezing/thawing, and the crude cell extract was clarified by centrifugation at 13,000g for 10 min. Ten microliters of Glutathione-Sepharose resin (GE Healthcare) was added to the clarified supernatant containing an equivalent amount of GST or GST-NLS protein, as determined by SDS-PAGE gels. Samples were incubated on ice for 10 min prior to centrifugation to remove unbound protein. The beads were washed further with 3× 500 μL pull-down buffer. After washing, 20 μg of purified ImpαΔIBB was incubated with the beads on ice for 10 min. The beads were then spun down and the supernatant was disposed of. The beads were washed further with 3× 500 μL pull-down buffer and the sample was analyzed on SDS-PAGE gels stained with Coomassie Brilliant Blue.
Microtiter Plate Binding Assay
The solid-phase binding assay was performed essentially as described previously (Matsuura et al., 2003; Lange et al., 2010; Takeda et al., 2011; Marfori et al., 2012). The assay was performed on Immuno MaxiSorp 96-well plate (Thermo Fisher Scientific). The plates were coated with 50 nM GST-NLS or GST per well for 16 h at 4°C in the coating buffer (PBS supplemented with 2 mM DTT and 0.2 mM PMSF). Then, the plates were washed three times with PBS and incubated overnight at 4°C in binding buffer (coating buffer supplemented with 3% BSA and 0.1% Tween to avoid nonspecific binding). Binding reactions were performed for 2 h at 4°C with 100 µL/well of S-tagged ImpαΔIBB in binding buffer. After binding, the plates were washed three times with binding buffer without BSA, and proteins were incubated in cross-linking buffer [1 mg/mL 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide in the same buffer] for 15 min. The plates were then washed for 20 min in PBS-T (PBS and 0.2% Tween 20), 10 min with PBS-T containing 100 mM ethanolamine, and finally incubated for 10 min in PBS-T containing 3% BSA. After washing, the plates were incubated in S-protein–horseradish peroxidase conjugate (Novagen) in coating buffer containing 1% BSA and 0.1% Tween 20 for 1 h at 4°C. After 1 h, the plates were washed three times by immersion in PBS. Horseradish peroxidase substrate (100 µg/mL 3,3′,5,5′-tetramethylbenzidine; Sigma-Aldrich) was added for 10 min at room temperature and then the reaction was stopped by adding an equal volume of 0.5 M H2SO4. The signal was determined at 450 nm with a Molecular Devices plate reader (Spectra Max 250). Binding data were analyzed with GraphPad (Prism) using nonlinear regression assuming one-site binding.
In Vitro Nuclear Import Assay
The nuclear import assays were performed as previously described (Wu et al., 2007; Yang et al., 2010). The HEp-2 (human larynx epidermoid carcinoma) cells were cultured on 12-mm cover slips in a 24-well plate at a starting cell density of 30,000 cells/mol in Dulbecco’s modified Eagle’s medium/10% fetal calf serum in a humidified 37°C, 5% CO2 incubator. The cell culture plate was placed on ice for 5 min and the medium aspirated, and then 1 mL ice-cold transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT, and 1 mM EDTA) was gently added. After 5 min, the transport buffer was aspirated and 1 mL ice-cold digitonin solution (5 µg/mL in transport buffer) was added to permeabilize the cells. Thirty microliters of the import reaction mixture (3 µM Impα:Impβ, 5 µM RanGDP, 1 mM ATP, 1 mM GTP, 5 mM creatine phosphate, 2 μg/μL creatine phosphate kinase, 10 μg/mL aprotintin, 0.5 μL 10 mg/mL Texas-red dextran [70 kD; Invitrogen], and 4 to 6 mg/mL import substrate [GST-GFP-NLS]) was placed on the parafilm laid evenly on the bench. The cover slip was gently inverted onto the drop of import reaction mixture. After incubating 20 min at room temperature and covered from light, 250 μL of ice-cold transport buffer was gently loaded underneath the cover slip to stop the reaction. The cover slip was then placed back onto the 24-well plate (cell side up). The cells were fixed with ice-cold 3% paraformaldehyde. Finally, the cover slip was removed and excess liquid wicked off and gently inverted (cell side down) on a small drop of mounting medium (ProLong Gold antifading regent supplemented with 4′,6-diamidino-2-phenylindole; Invitrogen). The cells were observed by Zeiss Axioplan 2 epifluorescence/light microscope using oil objectives. Photographs were imaged at intensities below saturation. Texas-red dextran (70 kD) was used to check if the nuclear membrane was intact. All experiments were performed in triplicate.
Protein Crystallization
Crystallization conditions for the mImpαΔIBB:NLS complexes were identified by screening in 0.4 to 0.9 M sodium citrate, 0.1 M HEPES buffer, pH 6.5 to 8.0, and 10 mM DTT, based on mImpα crystallization conditions described previously (Teh et al., 1999). Crystallization conditions for rImpα1a and rImpα1aΔIBB:NLS complexes were explored using commercial sparse matrix screens (JCSG, PACT, Proplex, and PEGIon; Hampton Research). The crystals were further optimized with additive screens (Hampton Research) and by varying crystallization kinetics to improve the crystal morphology. The optimized crystals of rImpα1aΔIBB:A89 and rImpα1aΔIBB:B54 were grown in 0.1 M bis-Trispropane, pH 7.0, 15 to 17% polyethylene glycol 3350, and 0.2 M NaF. Due to a twinning defect of these crystals, further optimization was performed for rImpα1a and rImpα1aΔIBB:SV40TAgNLS, and addition of 0.2 M NDSB-221 (Sigma-Aldrich) was found to lead to different crystal packing with no twinning. All crystallization screens were performed using hanging drop vapor diffusion using Linbro plates (Hampton Research) at 18°C.
X-Ray Diffraction Data Collection and Structure Determination
Single crystals were cryoprotected in the reservoir solution supplemented with 20 to 25% glycerol and flash-cooled in liquid nitrogen. X-ray diffraction data were collected and processed using MX1 and MX2 beamlines at the Australian Synchrotron, using a charge-coupled device detector (ADSC Quantum 210r) and Blu-Ice (McPhillips et al., 2002) and XDS software (Kabsch, 2010). The program Xtriage (Adams et al., 2010) was used to assess data quality. The mImpαΔIBB complex structures were determined by molecular replacement using MolRep (Collaborative Computational Project, Number 4, 1994) with structure of mImpα (Kobe 1999; residues 44 to 54 omitted) as the search model. The initial solution was refined by rigid body refinement using the program Refmac (Collaborative Computational Project, Number 4, 1994), and further rounds of refinement were performed using PHENIX and BUSTER (Adams et al., 2010; Smart et al., 2012). The NLS peptides were built manually using the program Coot (Emsley et al., 2010). Crystals of rImpα1aΔIBB:B54NLS complex had a twinning defect as assessed by Xtriage. One pseudo-merohedral twin operation was found with twin law (h, -k, -l) and twin fraction >0.40. The structure of yImpαΔIBB:c-myc complex (Conti and Kuriyan, 2000; with the c-myc peptide omitted) was used as the search model for molecular replacement (MolRep). These crystals had two molecules in the asymmetric unit and the symmetry of the P21 space group. The initial model was further refined as described above and additionally using twinning refinement with PHENIX. The B54NLS peptide was built manually using the program Coot. The final structure (with B54NLS omitted) was used as the search model to solve the structure of rImpα1aΔIBB:A89NLS complex, and the structure was refined as for rImpα1aΔIBB:B54NLS. The crystals of rImpα1aΔIBB:SV40TAgNLS and rImpα1a, grown in the presence of NDSB-221 as the additive, had no twinning defect and a different crystallographic symmetry (C21 space group). The structures were determined as described for other rImpα1aΔIBB complexes but without twinning refinement. Crystallographic data are summarized in Table 1.
Position Weight Matrix Method-Based Proteome Screen
Kosugi et al. (2009) provide an alignment containing n = 19 peptides, based on peptide library screening with rImpαΔIBB as bait. From the alignment, we first selected the K most conserved columns (we used K = 6). Next, we determined nu,a, the number of times an amino acid a is observed at column u in the N sequences. The column-specific probability of a is defined as qu,a = (nu,a + bpa)/(N + b), where pa is the prior probability of a, and b is a so-called pseudo-count set to N1/2. Prior probabilities are in their turn determined by counting the occurrences of amino acids in proteome-wide sequence data. Finally, for the alignment (with K columns selected), we defined a position weight matrix M = [mu,a]K×20, where mu,a = log(qu,a/pa). Each protein subsequence S = s1, s2, …, sK, was scored additively, by index to m, using the expression
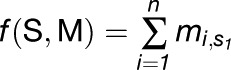 |
A greater score indicates that a functional localization signal is more probable. All possible positions in all protein sequences were scored, and the protein was assigned the maximum position-specific score. By imposing a threshold θ, we can distinguish between proteins that have at least one predicted functional site from those that do not. All protein sequence data were downloaded from UniProtKB (Streptophyta phylum, 444,613 sequences, numbers of sequences with a functional site [positives]: 262 [see Supplemental Data Set 2A online]; Mammalia class, 693,172 sequences, 384 positives; Saccharomyces genus, 80,054 sequences, 34 positives; O. sativa ssp japonica, 99,859 sequences, 108 positives; Arabidopsis thaliana, 54,435 sequences, 43 positives; Homo sapiens, 131,053 sequences, 46 positives; Mus musculus, 78,800 sequences, 37 positives; S. cerevisiae, 6652 sequences, two positives).
Gene Ontology and Species Enrichment Analyses
To explore whether proteins associated with plant-specific NLSs collectively exhibit some properties, we performed a Gene Ontology enrichment analysis (Ashburner et al., 2000). Specifically, all proteins annotated with the Streptophyta phylum in UniProtKB and predicted to have a plant-specific NLS form the foreground, and all proteins annotated with the Streptophyta phylum form the background. We used the complete Gene Ontology official release of annotations and term definitions (July, 2012). For each Gene Ontology term, we counted the number of proteins in the foreground set and the background set with this term.
A one-tailed Fisher’s exact test (Fisher, 1922) establishes the P value of the term: The probability of finding this split of protein counts or even greater proportion in favor of the foreground. The P value was corrected for multiple testing (Bonferroni correction; shown as E-value). A term is thus assigned a small E-value only if proteins annotated with that term occur in the foreground set with a higher prevalence than can be statistically explained by chance (when proteins are picked randomly from the background set). The same statistical methodology was also used to explore whether proteins with predicted NLSs are more prevalent in certain species. This time we ascertained a P value on the basis of the count of NLS-equipped proteins in a species-specific proteome, relative to that in another.
Accession Numbers
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (rImpα1a, 4B8J; rImpα1aΔIBB:SV40TAgNLS, 4B8O; rImpα1aΔIBB:A89NLS, 4B8P; rImpα1aΔIBB:B54NLS, 2YNS; mImpαΔIBB:A89NLS, 4BA3; mImpαΔIBB:B54NLS, 2YNR).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Crystal Structure of rImpα1aΔIBB:SV40TAgNLS Complex.
Supplemental Figure 2. A89 and B54 NLSs Bind to ImpαΔIBB Proteins from Different Organisms.
Supplemental Figure 3. A89 and B54 NLSs Bind to the Major NLS Binding Site of mImpαΔIBB.
Supplemental Figure 4. Sequence Alignment of IBB Domains from Various Impα Proteins.
Supplemental Figure 5. Phylogenetic Analysis of Impα Proteins from Different Organisms.
Supplemental Figure 6. Sequence Alignment of ARM Repeats 6 to 8 and the C-Terminal Region of Impα Proteins.
Supplemental Figure 7. Binding Isotherms from Representative Affinity Measurements.
Supplemental Table 1. Proportions of Proteins Containing Plant-Specific NLSs from Proteomes of Different Species.
Supplemental Table 2. List of Oligonucleotides Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Supplemental Figure 5.
Supplemental Data Set 2A. List of Plant Proteins Containing Plant-Specific NLS Motifs.
Supplemental Data Set 2B. Gene Ontology Term Analysis of Plant Proteins Containing Plant-Specific NLS Motifs.
Acknowledgments
We thank Shunichi Kosugi (Iwate Biotechnology Research Center, Kitakami, Japan) for the rImpα1aΔIBB expression plasmid; the Australian Synchrotron beamline scientists, Daniel J. Ericsson, Mary Marfori, Ahmed Mehdi, and Marc Leibundgut; and members of Kobe lab for help. We acknowledge the use of the University of Queensland Remote Operation Crystallisation and X-ray Diffraction Facility and the assistance of Karl Byriel and Gordon King. B.K. is a National Health and Medical Research Council Research Fellow.
AUTHOR CONTRIBUTIONS
C.-W.C., S.J.W., M.B., and B.K. designed research. C.-W.C. and R.L.M.C. performed research. M.B. contributed computational tools. All authors analyzed data and wrote the article.
Glossary
- NPC
nuclear pore complex
- cNLS
classical nuclear localization signal
- IBB
Impβ binding
- GST
glutathione S-transferase
- GFP
green fluorescent protein
References
- Adams P.D., et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. ; The Gene Ontology Consortium (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010). ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38(Web Server issue): W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catimel B., Teh T., Fontes M.R., Jennings I.G., Jans D.A., Howlett G.J., Nice E.C., Kobe B. (2001). Biophysical characterization of interactions involving importin-alpha during nuclear import. J. Biol. Chem. 276: 34189–34198 [DOI] [PubMed] [Google Scholar]
- Cingolani G., Petosa C., Weis K., Müller C.W. (1999). Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399: 221–229 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Conti E., Kuriyan J. (2000). Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure 8: 329–338 [DOI] [PubMed] [Google Scholar]
- Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998). Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94: 193–204 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., III, Snoeyink J., Richardson J.S., Richardson D.C. (2007). MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35(Web Server issue): W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S.M., Wilson K.F., Rojas K.S., Ambrosio A.L., Cerione R.A. (2009). The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 16: 930–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiadis A., Shao H., Hübner S., Jans D.A. (1997). Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. A comparison with the SV40 large T-antigen NLS. J. Biol. Chem. 272: 22134–22139 [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanara P., Hodel M.R., Corbett A.H., Hodel A.E. (2000). Quantitative analysis of nuclear localization signal (NLS)-importin alpha interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem. 275: 21218–21223 [DOI] [PubMed] [Google Scholar]
- Fisher R.A. (1922). On the interpretation of x(2) from contingency tables, and the calculation of P. J. R. Stat. Soc. 85: 87–94 [Google Scholar]
- Fontes M.R., Teh T., Jans D., Brinkworth R.I., Kobe B. (2003). Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 278: 27981–27987 [DOI] [PubMed] [Google Scholar]
- Fontes M.R., Teh T., Kobe B. (2000). Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Giesecke A., Stewart M. (2010). Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J. Biol. Chem. 285: 17628–17635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb D.S., Corbett A.H., Mason D.A., Harreman M.T., Adam S.A. (2004). Importin alpha: A multipurpose nuclear-transport receptor. Trends Cell Biol. 14: 505–514 [DOI] [PubMed] [Google Scholar]
- Görlich D., Henklein P., Laskey R.A., Hartmann E. (1996). A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15: 1810–1817 [PMC free article] [PubMed] [Google Scholar]
- Guralnick B., Thomsen G., Citovsky V. (1996). Transport of DNA into the nuclei of xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell 8: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harreman M.T., Hodel M.R., Fanara P., Hodel A.E., Corbett A.H. (2003). The auto-inhibitory function of importin alpha is essential in vivo. J. Biol. Chem. 278: 5854–5863 [DOI] [PubMed] [Google Scholar]
- Heckman K.L., Pease L.R. (2007). Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2: 924–932 [DOI] [PubMed] [Google Scholar]
- Hicks G.R., Raikhel N.V. (1995). Protein import into the nucleus: An integrated view. Annu. Rev. Cell Dev. Biol. 11: 155–188 [DOI] [PubMed] [Google Scholar]
- Hodel M.R., Corbett A.H., Hodel A.E. (2001). Dissection of a nuclear localization signal. J. Biol. Chem. 276: 1317–1325 [DOI] [PubMed] [Google Scholar]
- Hoover D.M., Lubkowski J. (2002). DNAWorks: An automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Jans D.A. (1999). Efficiency of importin alpha/beta-mediated nuclear localization sequence recognition and nuclear import. Differential role of NTF2. J. Biol. Chem. 274: 15820–15827 [DOI] [PubMed] [Google Scholar]
- Hübner S., Smith H.M., Hu W., Chan C.K., Rihs H.P., Paschal B.M., Raikhel N.V., Jans D.A. (1999). Plant importin alpha binds nuclear localization sequences with high affinity and can mediate nuclear import independent of importin beta. J. Biol. Chem. 274: 22610–22617 [DOI] [PubMed] [Google Scholar]
- Jiang C.J., Imamoto N., Matsuki R., Yoneda Y., Yamamoto N. (1998). Functional characterization of a plant importin alpha homologue. Nuclear localization signal (NLS)-selective binding and mediation of nuclear import of nls proteins in vitro. J. Biol. Chem. 273: 24083–24087 [DOI] [PubMed] [Google Scholar]
- Jones A.M., MacLean D., Studholme D.J., Serna-Sanz A., Andreasson E., Rathjen J.P., Peck S.C. (2009). Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J. Proteomics 72: 439–451 [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. (1999). Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6: 388–397 [DOI] [PubMed] [Google Scholar]
- Kobe B., Kemp B.E. (1999). Active site-directed protein regulation. Nature 402: 373–376 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009). Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284: 478–485 [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lange A., McLane L.M., Mills R.E., Devine S.E., Corbett A.H. (2010). Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11: 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. (2007). Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 282: 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung S.W., Harreman M.T., Hodel M.R., Hodel A.E., Corbett A.H. (2003). Dissection of the karyopherin alpha nuclear localization signal (NLS)-binding groove: Functional requirements for NLS binding. J. Biol. Chem. 278: 41947–41953 [DOI] [PubMed] [Google Scholar]
- Li H.C., Chuang K., Henderson J.T., Rider S.D., Jr, Bai Y., Zhang H., Fountain M., Gerber J., Ogas J. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Coaker G. (2008). Nuclear trafficking during plant innate immunity. Mol. Plant 1: 411–422 [DOI] [PubMed] [Google Scholar]
- Lott K., Bhardwaj A., Sims P.J., Cingolani G. (2011). A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin alpha1. J. Biol. Chem. 286: 28160–28169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K., Cingolani G. (2011). The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813: 1578–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M., Lonhienne T.G., Forwood J.K., Kobe B. (2012). Structural basis of high-affinity nuclear localization signal interactions with importin-α. Traffic 13: 532–548 [DOI] [PubMed] [Google Scholar]
- Marfori M., Mynott A., Ellis J.J., Mehdi A.M., Saunders N.F., Curmi P.M., Forwood J.K., Bodén M., Kobe B. (2011). Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813: 1562–1577 [DOI] [PubMed] [Google Scholar]
- Mason D.A., Stage D.E., Goldfarb D.S. (2009). Evolution of the metazoan-specific importin alpha gene family. J. Mol. Evol. 68: 351–365 [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Lange A., Harreman M.T., Corbett A.H., Stewart M. (2003). Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 22: 5358–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Stewart M. (2004). Structural basis for the assembly of a nuclear export complex. Nature 432: 872–877 [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Stewart M. (2005). Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 24: 3681–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips T.M., McPhillips S.E., Chiu H.J., Cohen A.E., Deacon A.M., Ellis P.J., Garman E., Gonzalez A., Sauter N.K., Phizackerley R.P., Soltis S.M., Kuhn P. (2002). Blu-Ice and the Distributed Control System: Software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9: 401–406 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2011). Nucleo-cytoplasmic transport of proteins and RNA in plants. Plant Cell Rep. 30: 153–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Stamm M.E., Hohn B. (2004). SNM-dependent recombinational repair of oxidatively induced DNA damage in Arabidopsis thaliana. EMBO Rep. 5: 994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby B.B., Dasso M. (2003). The small GTPase Ran: Interpreting the signs. Curr. Opin. Cell Biol. 15: 338–344 [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth S.M., Laskey R.A., Dingwall C. (1991). Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 64: 615–623 [DOI] [PubMed] [Google Scholar]
- Smart O.S., Womack T.O., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. (2012). Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.M., Hicks G.R., Raikhel N.V. (1997). Importin alpha from Arabidopsis thaliana is a nuclear import receptor that recognizes three classes of import signals. Plant Physiol. 114: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Takeda A.A., de Barros A.C., Chang C.W., Kobe B., Fontes M.R. (2011). Structural basis of importin-α-mediated nuclear transport for Ku70 and Ku80. J. Mol. Biol. 412: 226–234 [DOI] [PubMed] [Google Scholar]
- Tarendeau F., et al. (2007). Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14: 229–233 [DOI] [PubMed] [Google Scholar]
- Teh T., Tiganis T., Kobe B. (1999). Crystallization of importin alpha, the nuclear-import receptor. Acta Crystallogr. D Biol. Crystallogr. 55: 561–563 [DOI] [PubMed] [Google Scholar]
- Tran E.J., Bolger T.A., Wente S.R. (2007). SnapShot: Nuclear transport. Cell 131: 420. [DOI] [PubMed] [Google Scholar]
- Tzfira T., Rhee Y., Chen M.H., Kunik T., Citovsky V. (2000). Nucleic acid transport in plant-microbe interactions: The molecules that walk through the walls. Annu. Rev. Microbiol. 54: 187–219 [DOI] [PubMed] [Google Scholar]
- Wu W.W., Sun Y.H., Panté N. (2007). Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol. J. 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Deng X.W. (1999). Protein nucleocytoplasmic transport and its light regulation in plants. Genes Cells 4: 489–500 [DOI] [PubMed] [Google Scholar]
- Yang S.N., Takeda A.A., Fontes M.R., Harris J.M., Jans D.A., Kobe B. (2010). Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening. J. Biol. Chem. 285: 19935–19946 [DOI] [PMC free article] [PubMed] [Google Scholar]