This work examines effector recognition and signaling by NB-LRR resistance protein R3a, finding that recognition of AVR3A involves R3a relocalization to vesicular compartments in the endocytic pathway and attenuation of this relocalization suppressed the resultant hypersensitive cell death response.
Abstract
An important objective of plant–pathogen interactions research is to determine where resistance proteins detect pathogen effectors to mount an immune response. Many nucleotide binding–Leucine-rich repeat (NB-LRR) resistance proteins accumulate in the plant nucleus following effector recognition, where they initiate the hypersensitive response (HR). Here, we show that potato (Solanum tuberosum) resistance protein R3a relocates from the cytoplasm to endosomal compartments only when coexpressed with recognized Phytophthora infestans effector form AVR3aKI and not unrecognized form AVR3aEM. Moreover, AVR3aKI, but not AVR3aEM, is also relocalized to endosomes in the presence of R3a. Both R3a and AVR3aKI colocalized in close physical proximity at endosomes in planta. Treatment with brefeldin A (BFA) or wortmannin, inhibitors of the endocytic cycle, attenuated both the relocalization of R3a to endosomes and the R3a-mediated HR. No such effect of these inhibitors was observed on HRs triggered by the gene-for-gene pairs Rx1/PVX-CP and Sto1/IpiO1. An R3a(D501V) autoactive MHD mutant, which triggered HR in the absence of AVR3aKI, failed to localize to endosomes. Moreover, BFA and wortmannin did not alter cell death triggered by this mutant. We conclude that effector recognition and consequent HR signaling by NB-LRR resistance protein R3a require its relocalization to vesicles in the endocytic pathway.
INTRODUCTION
Plants have an efficient two-layer receptor system that detects potential pathogens and activates defense responses to prevent disease. Plasma membrane–localized pattern recognition receptors detect highly conserved microbial molecules known as microbe-associated molecular patterns (Zipfel, 2008). The consequent pattern-triggered immunity is suppressed by adapted pathogens, which deploy virulence factors known as effectors (Chisholm et al., 2006; Jones and Dangl, 2006). In recent years, our knowledge, especially of prokaryotic effectors, has demonstrated how microbes can use different strategies to manipulate host processes, creating a suitable environment for infection (Block et al., 2008).
In the second layer of the inducible immune system, effectors are perceived by direct or indirect interaction with intracellular nucleotide binding–Leucine-rich repeat (NB-LRR) resistance (R) receptors. NB-LRR genes are one of the largest gene families in plants. Approximately 150 such genes have been described in Arabidopsis thaliana (Meyers et al., 2003) and more than 430 in potato (Solanum tuberosum) (Jupe et al., 2012). Members of the NB-LRR family can be divided into TIR genes carrying an N-terminal domain with similarity to the Drosophila melanogaster Toll and human interleukin-1 receptor and those with a predicted coiled coil (CC) structure at the N terminus. Following perception of effectors (then termed avirulence, or AVR, proteins) R proteins activate effector-triggered immunity, which often includes a localized programmed cell death. Indirect recognition involves the R protein monitoring (or guarding) the biochemical state of a host protein that is targeted by an effector. Bacterial effectors are often recognized indirectly. For example, Pseudomonas syringae effectors AvrB and AvrRPM1 mediate phosphorylation of the host RPM1-INTERACTING PROTEIN4, RIN4, which is detected by the host immune receptor RESISTANCE TO PSEUDOMONAS SYRINGAE PV. MACULICOLA1, RPM1 (Chung et al., 2011), whereas RESISTANCE TO PSEUDOMONAS SYRINGAE2, RPS2, perceives the proteolytic cleavage of RIN4 by P. syringae effector AvrRpt2 (Belkhadir et al., 2004). In contrast with most bacterial effectors, recognition of filamentous pathogen AVR proteins by NB-LRR proteins is often direct (Jia et al., 2000; Dodds et al., 2006; Krasileva et al., 2010).
NB-LRR proteins in unchallenged plant cells have several subcellular localizations. Upon activation, resistance proteins often relocalize to the nucleus where defense responses are initiated (Caplan et al., 2008; Eitas and Dangl, 2010). The potato CC-NB-LRR protein Rx1, which confers extreme resistance to Potato virus X, is activated in the cytoplasm but both nuclear and cytoplasmic pools of Rx1 are required for full function, a partitioning which is regulated by Ran GTPase-activating protein 2 (Slootweg et al., 2010; Tameling et al., 2010). The nuclear localization of the tobacco (Nicotiana tabacum) TIR resistance protein N, which specifically recognizes the Tobacco mosaic virus p50 helicase domain, is essential for initiating defenses (Burch-Smith et al., 2007). Another example is the Arabidopsis RESISTANCE TO PSEUDOMONAS SYRINGAE 4 protein RPS4, for which accumulation in the nucleus is necessary to trigger immunity in the presence of the P. syringae effector AvrRps4 (Wirthmueller et al., 2007). Recent findings of Gao et al. (2011), however, argue against a universal rule regarding nuclear relocalization of NB-LRR proteins for defense induction. Nuclear localization of RPM1 protein is not required for efficient disease resistance upon activation caused by AvrB and AvrRpm1. Inactive and active RPM1 resides in the plant plasma membrane, an observation that is further supported by experiments using an autoactive RPM1 mutant (Gao et al., 2011). More recently, a number of NB-LRR resistance proteins have been shown to be constitutively localized to a variety of endomembranes, although the mechanistic basis for these localizations in terms of effector recognition and hypersensitive response (HR) signaling is unknown (Takemoto et al., 2012).
One of the best studied oomycete effectors is AVR3a from Phytophthora infestans. AVR3a occurs as two forms differing in only two amino acids: Avr3aE80M103 (Avr3aEM) and Avr3aK80I103 (Avr3aKI). The potato resistance protein R3a, a member of the CC-NB-LRR class, perceives Avr3aKI, whereas Avr3aEM evades recognition (Armstrong et al., 2005). Both forms of AVR3a suppress cell death caused by INF1, a microbe-associated molecular pattern from P. infestans (Bos et al., 2006, 2009), through their action on the host ubiquitin E3 ligase CMPG1 (Bos et al., 2010; Gilroy et al., 2011a). The C-terminal Tyr of Avr3aKI (Tyr-147) is dispensable for R3a perception but required for the suppression of CMPG1-mediated programmed cell death, allowing discrimination between these two effector properties (Bos et al., 2010; Gilroy et al., 2011a). CMPG1 is not required for R3a-mediated HR, suggesting that it is not a guardee for AVR3a perception.
In this study, we demonstrate that the potato resistance protein R3a is specifically localized to endosomes upon perception of P. infestans effectors that cause R3a-mediated HR. Moreover, in the presence of R3a, the recognized effector Avr3aKI, but not the unrecognized form Avr3aEM, is also localized to the same endosomal compartments prior to initiation of the HR. We provide experimental evidence that this relocalization is essential for R3a to induce the HR. Despite a lack of evidence for direct protein–protein interaction between R3a and AVR3a in yeast, both resistance protein and AVR3aKI colocalize at endosomes and are in close proximity in planta. Our data demonstrate that a resistance protein and its cognate avirulence protein can be relocalized to a component of the endocytic cycle, which acts as a site for subsequent activation of the immune response.
RESULTS
R3a Is Relocalized to Vesicles in the Presence of Recognized Effectors
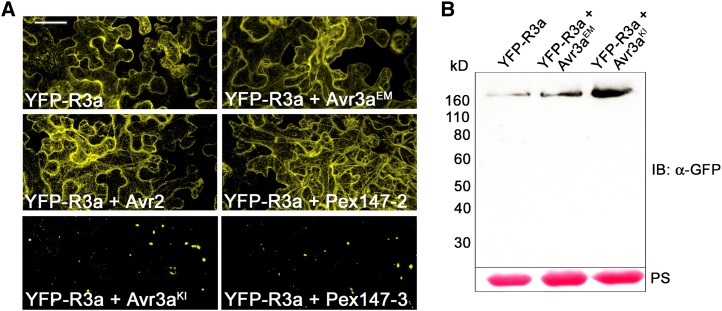
To investigate the localization of R3a upon activation, we fused yellow fluorescent protein (YFP) N-terminally to R3a and transiently expressed this construct in Nicotiana benthamiana in the presence or absence of P. infestans effectors. Fluorescence indicated a cytoplasmic localization of YFP-R3a in the absence of effector proteins (Figure 1A; see Supplemental Figure 1A online). R3a was excluded from the nucleus as fluorescence in the nucleoplasm was not detected. The cytoplasmic localization of R3a remained unaffected after coexpression with AVR3aEM, P. infestans avirulence effector AVR2 (Gilroy et al., 2011b), and Pex147-2, an AVR3a paralog (Armstrong et al., 2005) (Figure 1A), none of which trigger R3a-dependent HR (see Supplemental Figure 1B online). However, a dramatic relocalization of R3a from the cytoplasm to rapidly moving vesicle-like structures of various sizes occurred in the presence of AVR3aKI and Pex147-3 (Figure 1A), both of which are recognized by R3a (see Supplemental Figure 1B online; Armstrong et al., 2005; Oh et al., 2009). Immunoblot analyses (Figure 1B) confirm the stability of YFP-R3a fusion protein in the presence or absence of AVR3a, indicating that intact resistance protein fusion undergoes the observed relocalization. Interestingly, relocalization does not represent a symptom of dying cells caused by the onset of the HR, as YFP fused N-terminally to R3a effectively prevents the development of the HR in the presence of effectors known to cause R3a-mediated cell death (see Supplemental Figure 1B online). Since overexpression of CC domains alone of some NB-LRR proteins elicits an autoactive cell death phenotype (Collier et al., 2011), it is feasible that the N-terminal CC domain of R3a initiates the signaling process leading to cell death. In the presence of YFP fused to the CC domain of R3a, however, these signaling processes are likely to be disrupted, potentially due to steric hindrance.
Figure 1.
R3a Relocalizes to Vesicles in the Presence of Recognized Effectors.
(A) Confocal laser scanning microscopy following transient expression (by agroinfiltration) in N. benthamiana of YFP-R3a alone or transiently coexpressed with either AVR3aEM, AVR3aKI, AVR2, Pex147-2, or Pex147-3 as indicated in the panels at 2 d after inoculation. Experiments were repeated at least five times. Bar = 25 µm.
(B) Immunoblot probed with α-GFP following transient expression of YFP-R3a alone or coexpressed with AVR3aEM or AVR3aKI in N. benthamiana at 2 d after inoculation. Protein sizes are indicated (in kilodaltons), and protein loading is shown by Ponceau stain (PS).
To confirm that AVR3aKI causes a relocalization of YFP-R3a and to investigate the timing of this event, we coexpressed YFP-R3a with dexamethasone-inducible forms of AVR3aKI and AVR3aEM. Following dexamethasone treatment, YFP-R3a alone, or coexpressed with dex:AVR3aEM, remained cytoplasmic. However, when coexpressed with dex:AVR3aKI, relocalization of YFP-R3a was observed from cytoplasm to vesicles from 2 h post-treatment (hpt) with dexamethasone (Figure 2A). The visible symptoms of HR, following expression of untagged R3a with dex:AVR3aKI, were not detected at 4 hpt but were clearly visible by 24 hpt with dexamethasone (Figures 2B and 2C). However, weak HR symptoms were first detected in some R3a/dex:AVR3aKI coexpression sites by 8 hpt with dexamethasone (examples shown in Figure 2C). This suggests that relocalization of YFP-R3a occurs prior to the initiation of cell death.
Figure 2.
Relocalization of R3a Occurs before Development of HR Symptoms.
(A) Confocal laser scanning microscopy following transient expression (by agroinfiltration) in N. benthamiana of YFP-R3a alone or in the presence of conditionally coexpressed Avr3aEM and Avr3aKI at the indicated time points after dexamethasone treatment at 2 d after inoculation. Bar = 20 µm.
(B) Transient expression of R3a alone or in the presence of conditionally coexpressed Avr3aEM and Avr3aKI by agroinfiltration in N. benthamiana. Leaf photos were taken at indicated time points after dexamethasone treatment and are representative of three independent assays. Circles indicate the infiltrated area on the leaf panel.
(C) Close-up of leaf areas transiently expressing R3a with conditionally coexpressed Avr3aKI by agroinfiltration in N. benthamiana. Photos were taken at indicated time points after dexamethasone treatment.
R3a Is Relocalized to Late Endosomes
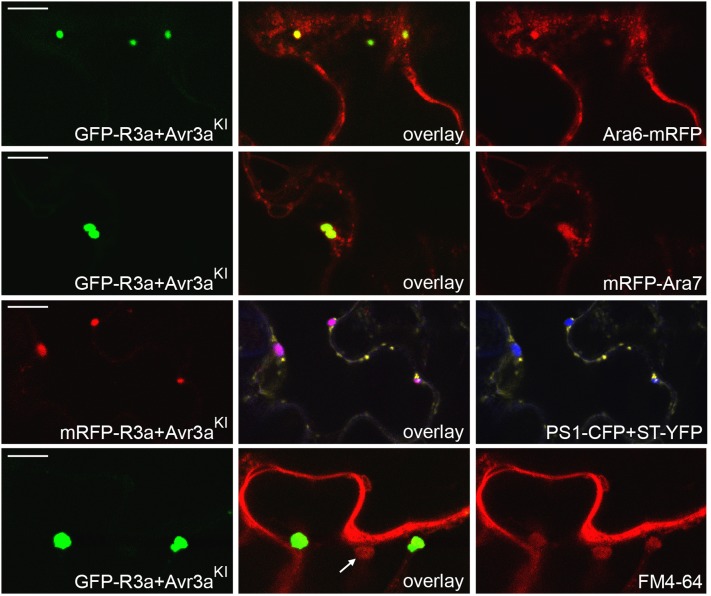
To identify the sites of R3a relocalization in the presence of AVR3aKI, we conducted a series of colocalization experiments with diverse subcellular markers. Supplemental Figure 2 online shows that fluorescent protein (FP)-R3a fusions do not relocalize to chloroplasts, nuclei, or peroxisomes. The vesicle-like structures exhibit a characteristic motility, which, together with their different sizes, prompted us to investigate components of the endocytic cycle. Using coexpression with a Golgi marker, we demonstrated that monomeric red fluorescent protein (mRFP)-R3a is not relocalized to these structures (see Supplemental Figure 2 online). We used two additional subcellular markers, Ara6 and Ara7, which belong to the RAB GTPase family. Ara6 and Ara7 are key regulators of endosomal trafficking, endocytosis, and vacuolar transport (Kotzer et al., 2004; Richter et al., 2009; Ebine et al., 2011). AVR3aKI-mediated relocalization of green fluorescent protein (GFP)-R3a partially coincided with the marker fluorescence of both Ara6-mRFP and mRFP-Ara7, indicating that, potentially, sorting endosomes are involved in the recognition process (Figure 3). Ara6-mRFP and mRFP-Ara7 have distinct but overlapping localization patterns (Ebine et al., 2011) and label a variety of endosomal compartments. We thus tried the specific prevacuolar compartment (PVC) marker PS1–cyan fluorescent protein (CFP) (Saint-Jean et al., 2010). We observed colocalization of mRFP-R3a with the PVC marker when coexpressed with AVR3aKI, demonstrating that late endosomes are a site of R3a relocalization (Figure 3).
Figure 3.
In the Presence of Recognized Effectors, R3a Relocalizes to Endosomal Compartments.
Colocalization studies using confocal laser scanning microscopy following transient coexpression (by agroinfiltration) in N. benthamiana of GFP-R3a or mRFP-R3a with 35S-driven AVR3aKI and different subcellular markers: Ara6-mRFP, mRFP-Ara7, PS1-CFP (blue PVC marker), ST-YFP (yellow Golgi-marker; same as in Supplemental Figure 2 online). Colocalization of GFP-R3a with FM4-64 (coexpressed with 35S:AVR3aKI) was examined 20 min after FM4-64 infiltration. Arrow indicates vesicle stained with FM4-64 but not GFP-R3a as described in the text. Left picture shows FP-R3a fluorescence, right picture shows marker fluorescence, and middle picture shows overlay of left and right pictures, all taken at 2 d after inoculation. Images showing colocalization of YFP-R3a with PS1-CFP, following coexpression with dex:AVR3aKI, were taken 2 h after dexamethasone treatment. Bars = 10 µm.
We extended our investigations with the endocytic tracer FM4-64, which labels early endosomes within a few minutes (Dettmer et al., 2006), followed by labeling of PVCs that constitute a site where endocytic and vacuolar trafficking merge (Tse et al., 2004). Confocal microscopy showed colocalization of FM4-64 with relocalized GFP-R3a (Figure 3), which could be observed from 10 up to 30 min after FM4-64 infiltration, supporting the observation that late endosomes are sites at which activated R3a is localized. Interestingly, not all endosomes labeled by FM4-64 were sites of GFP-R3a relocalization (Figure 3), indicating specificity in the endosomes to which R3a is associated.
In addition, we coexpressed YFP-R3a with dex:AVR3aKI and PS1-CFP. At 2 hpt with dexamethasone, the earliest point at which relocalization of YFP-R3a was observed, we saw colocalization between YFP-R3a and PS1-CFP fluorescence, indicating the presence of R3a at late endosomes (see Supplemental Figure 3 online). As with the FM4-64 tracer above, not all endosomes associated with the PS1-CFP marker were sites for relocalization of YFP-R3a, supporting specificity to certain endosomes in the relocalization process.
AVR3aKI, but Not AVR3aEM, Is Relocalized to Endosomes Prior to R3a-Mediated HR
The results shown in Figure 1 suggest that relocalization of R3a is associated with effector recognition, as unrecognized AVR3aEM and Pex147-2 do not provoke this response. Nevertheless, N-terminal FP tagging of wild-type R3a prevents effector-mediated cell death. To test whether relocalization of R3a is required for R3a-mediated cell death, we fused R3a C-terminally to GFP. However, the R3a-YFP fusion also did not trigger HR when coexpressed with AVR3aKI (see Supplemental Figure 4 online). As we had noted previously (Bos et al., 2010) that N-terminal FP tagging of AVR3aKI faithfully yielded R3a-dependent HR, we thus extended our analyses to the localization of recognized and unrecognized effector forms in the presence of untagged R3a.
We first performed colocalization studies following transient coexpression of AVR3aKI and R3a, both containing N-terminal fluorescent tags, in N. benthamiana. It was shown previously that AVR3aKI, overexpressed alone, is present in the cytoplasm and the nucleoplasm and does not localize to vesicles (Bos et al., 2010). After coexpression of a GFP-AVR3aKI fusion with mRFP-R3a, we observed the expected relocalization of the mRFP fluorescence to vesicles. In addition, however, we observed relocalization of GFP-AVR3aKI fluorescence to the same subcellular compartments (Figure 4A), indicating that recognition is associated with close proximity between R3a and AVR3aKI.
Figure 4.
Avr3aKI, but Not Avr3aEM, Relocalizes to Late Endosomes in the Presence of Untagged R3a Prior to HR Development.
(A) Confocal laser scanning microscopy following transient coexpression by agroinfiltration in N. benthamiana of GFP-AVR3aKI (left picture) and mRFP-R3a (right picture), with overlay (middle picture) indicating colocalization of GFP and mRFP fluorescence at 2 d after inoculation.
(B) GFP-Avr3aKI or GFP-Avr3aEM (left picture, as indicated), each coexpressed with PVC marker PS1-CFP (right picture) and untagged R3a, with overlay (middle picture), indicating colocalization of GFP and CFP fluorescence only in the case of GFP-Avr3aKI.
(C) GFP-Avr3aKI (left picture) coexpressed with untagged R3a. Localization of FM4-64 (right picture) was examined 20 min after its infiltration. Overlay (middle picture) indicates colocalization of GFP and FM4-64 fluorescence. For experiments in (B) and (C), agroinfiltration to express untagged R3a was performed 24 h after agroinfiltration to express GFP-Avr3a (and PS1-CFP, as indicated). Images in (B) and (C) were taken 2 d after inoculation of GFP-AVR3a (1 d after inoculation for R3a). Bars = 10 µm.
(D) Transient expression of untagged R3a and GFP-Avr3aKI alone or coexpressed (agroinfiltration to express R3a was 24 h after agroinfiltration to express GFP-Avr3a). Photos of HRs were taken at indicated time points (h p.i., hours post inoculation) and are representative of multiple independent assays. Circles indicate the infiltrated area on the leaf panel.
GFP-AVR3aKI and GFP-AVR3aEM were each coexpressed with untagged, functional R3a and with the PVC marker PS1-CFP. Agrobacterium tumefaciens expressing R3a was infiltrated into inoculation sites 24 h after infiltration of Agrobacterium strains expressing GFP-AVR3a alleles and PS1-CFP marker. Confocal images taken 24 h postinoculation of untagged R3a revealed that, whereas GFP-AVR3aKI clearly colocalized with the PS1-CFP marker, no such colocalization was seen between GFP-AVR3aEM and PS1-CFP (Figure 4B; see Supplemental Figure 5 online). Moreover, in the absence of R3a, AVR3aKI did not colocalize with the PS1 marker, indicating that its association with late endosomes is specific to coexpression with R3a (see Supplemental Figure 5 online). As seen for relocalization of FP-R3a in Figure 3, GFP-AVR3aKI was relocalized in the presence of untagged R3a to endosomes containing the endocytic tracer FM4-64 (Figure 4C). Critically, R3a-mediated HR was not inhibited in these assays, and macroscopic cell death was clearly visible by 48 h postinoculation of untagged R3a and only in the presence of GFP-AVR3aKI (Figure 4D). This indicates that relocalization of both R3a and AVR3aKI to endosomes occurs prior to HR symptoms.
To further investigate the close proximity between R3a and AVR3aKI, we used bimolecular fluorescence complementation (BiFC), or split-YFP, to analyze and localize protein–protein interactions in plant cells (Walter et al., 2004; Bos et al., 2010). We fused N and C terminus–encoding portions of YFP to the N terminus of R3a, AVR3aKI, and to the unrecognized effector form AVR3aEM. Constructs expressing fusions to complementary YFP halves were transiently expressed in N. benthamiana. As shown in Figure 5A, fluorescence was undetectable following coexpression of YN-R3a with YC-AVR3aEM. By contrast, YN-R3a/YC-AVR3aKI coexpression yielded strong fluorescence signals in a pattern reminiscent of relocalization to the endosomal vesicles, indicating close physical proximity between R3a and AVR3aKI sufficient to reconstitute YFP fluorescence. Immunoblot analyses confirmed the stability of all expressed fusion proteins in planta (Figure 5B). To confirm the identity of vesicles observed in these experiments, YN-R3a, YC-AVR3aKI, and the PS1-CFP marker were coexpressed and revealed colocalization of reconstituted YFP fluorescence with PS1-CFP at endosomal compartments (Figure 5C). These results support relocalization of both R3a and AVR3aKI into close proximity at compartments within the endocytic pathway.
Figure 5.
BiFC Indicates Close Proximity of R3a and AVR3aKI in Planta.
(A) and (C) Confocal laser scanning microscopy following transient coexpression in N. benthamiana by agroinfiltration of split-YFP constructs YN-R3a with either YC-AVR3aEM (left picture) or YC-AVR3aKI (right picture), as indicated, at 2 d after inoculation (A) and split-YFP constructs YN-R3a and YC-Avr3aKI, coexpressed with the PVC marker PS1-CFP, at 2 d after inoculation (C). Overlay (middle picture) indicates colocalization of YFP fluorescence (left picture) and CFP fluorescence (right picture). Bars = 20 µm.
(B) Immunoblot probed with α-cMyc or α-HA following transient expression of split-YFP constructs shown in (A) in N. benthamiana at 3 d after inoculation. Protein sizes are indicated (in kilodaltons) and protein loading is shown by Ponceau S (PS). Experiments in (A) and (C) were repeated on four occasions.
Direct Interaction between R3a and AVR3a Is Not Detectable Using Yeast Two-Hybrid or Coimmunoprecipitation Analyses
The close proximity observed between Avr3aKI and R3a in the colocalization and split-YFP experiment detailed above prompted us to investigate potential direct recognition of AVR3a by R3a. We conducted a series of yeast two-hybrid (Y2H) experiments using different R3a bait clones with three different AVR3a prey constructs: AVR3aEM, AVR3aKI, and AVR3aKIΔY147 (described in Bos et al., 2010). The AVR3aKIΔY147 mutant construct is unable to interact with the target protein in potato, CMPG1, but is nevertheless still recognized by R3a (Bos et al., 2010). No direct protein–protein interactions were observed between wild-type R3a with any AVR3a forms (see Supplemental Figure 6 online), since no colonies grew on triple-dropout medium (-L,-W,-Ade) and β-galactosidase activity was not detected. Moreover, no significant reporter gene activity was observed after coexpressing R3a subfragments with the AVR3a forms (see Supplemental Figure 6 online), indicating that, at least in the yeast system, AVR3a does not interact directly with R3a. This observation was not caused by fusion protein instability since all expressed R3a fusion proteins (see Supplemental Figure 6 online), and AVR3a fusion proteins (Bos et al., 2010) were detectable in yeast by immunoblot.
Although Y2H analysis is a facile, powerful, and high-throughput approach to analyze protein–protein interactions, it is prone to false-negative results, meaning that true interactions are sometimes not detected. Since yeast cytoplasmic and nucleoplasmic conditions do not mimic the plant cell environment in which effector recognition occurs naturally, misfolding of the fusion protein, steric hindrance, or nonspecific protein aggregation could prevent an R3a–AVR3a interaction, despite the proteins being stably expressed (see Supplemental Figure 6 online). To further investigate whether R3a interacts directly with AVR3a, we conducted in planta coexpression and coimmunoprecipitation studies.
We transiently coexpressed GFP-Avr3a fusions and FLAG-R3a fusions in N. benthamiana via agroinfiltration and immunoprecipitated GFP-Avr3a using magnetic GFP-Trap beads. Despite a detectable enrichment of GFP-Avr3a (see Supplemental Figure 7 online), we could not coimmunoprecipitate FLAG-R3a, indicating that a direct interaction between Avr3a and R3a possibly does not occur, that the interaction is too transient, or that it is potentially disrupted during the total protein extraction process. Unfortunately, it was not possible to conduct reciprocal coimmunoprecipitation experiments due to nonspecific binding of AVR3a to all beads tested.
Inhibitors of the Endocytic Cycle Attenuate R3a Relocalization to Late Endosomes
To investigate the contribution of late endosomes to either recognition of AVR3a and/or the signaling of R3a-dependent HR, we monitored the influence of two inhibitors of the endocytic cycle, brefeldin A (BFA) and wortmannin, on the relocalization of R3a in the presence of AVR3aKI. BFA is reported to inhibit the activation of ARF-GTPase that is involved in multiple trafficking pathways (Chardin and McCormick, 1999; Nielsen et al., 2008; Robinson et al., 2008). Golgi-localized proteins are redistributed to the endoplasmic reticulum (ER) upon BFA treatment. Furthermore, BFA causes aggregation of the trans-Golgi network and multivesicular endosomes (PVCs in plants) into BFA bodies (Uemura et al., 2004; Jaillais et al., 2008; Ebine et al., 2011). Wortmannin suppresses the formation of PVCs by inhibiting phosphatidylinositol 3-kinase and affects the later steps of clathrin-coated vesicle formation that eventually inhibits endocytosis (Emans et al., 2002; Ebine et al., 2011; Ito et al., 2011). Additionally, wortmannin causes late endosome/multivesicular endosome (PVC) deformation (Ebine et al., 2011).
The application of both compounds had a dramatic effect on FP-R3a relocalization. Only 20 to 30 min after treatment, both wortmannin and BFA (Figure 6A) caused a significant increase in cytoplasmic fluorescence compared with untreated samples. The cytoplasmic fluorescence in the presence of both BFA and wortmannin is not due to degradation of FP-R3a caused by mislocalization, as immunoblot analyses confirmed the stability of FP-R3a in planta following these treatments (Figure 6B). Due to the high cytoplasmic background of FP-AVR3a, it was not possible to perform reciprocal experiments to study the effects of the inhibitors on (preventing or reducing) FP-AVR3a relocalization in the presence of untagged R3a. However, we confirmed that GFP-AVR3a also remained stable in planta in the presence of mRFP-R3a following treatments with BFA and wortmannin (Figure 6B).
Figure 6.
Wortmannin and BFA Attenuate R3a Relocalization.
(A) Confocal laser scanning microscopy following transient coexpression in N. benthamiana by agroinfiltration of YFP-R3a with AVR3aKI in the absence (-wortmannin, -BFA, left pictures) and 30 min after infiltration of 33 µM Wortmannin or 20 min after infiltration of 20 μg/mL BFA (right picture). Bar = 25 (top images) and 20 (bottom images) µm. Pictures were taken 2 d after agroinfiltration, and experiments were repeated three times.
(B) Immunoblot probed with α-GFP following transient coexpression of YFP-R3a with AVR3aKI (left picture) and transient coexpression of GFP-Avr3aKI with mRFP-R3a (right picture) in N. benthamiana by agroinfiltration at 2 d after inoculation to show stability of R3a and Avr3a after application of 20 µg/mL BFA and 30 µM wortmannin for 2 h. Protein sizes are indicated (in kilodaltons), and protein loading is shown by Ponceau S (PS).
The R3a(D501V) Autoactive Mutant Is Cytoplasmic
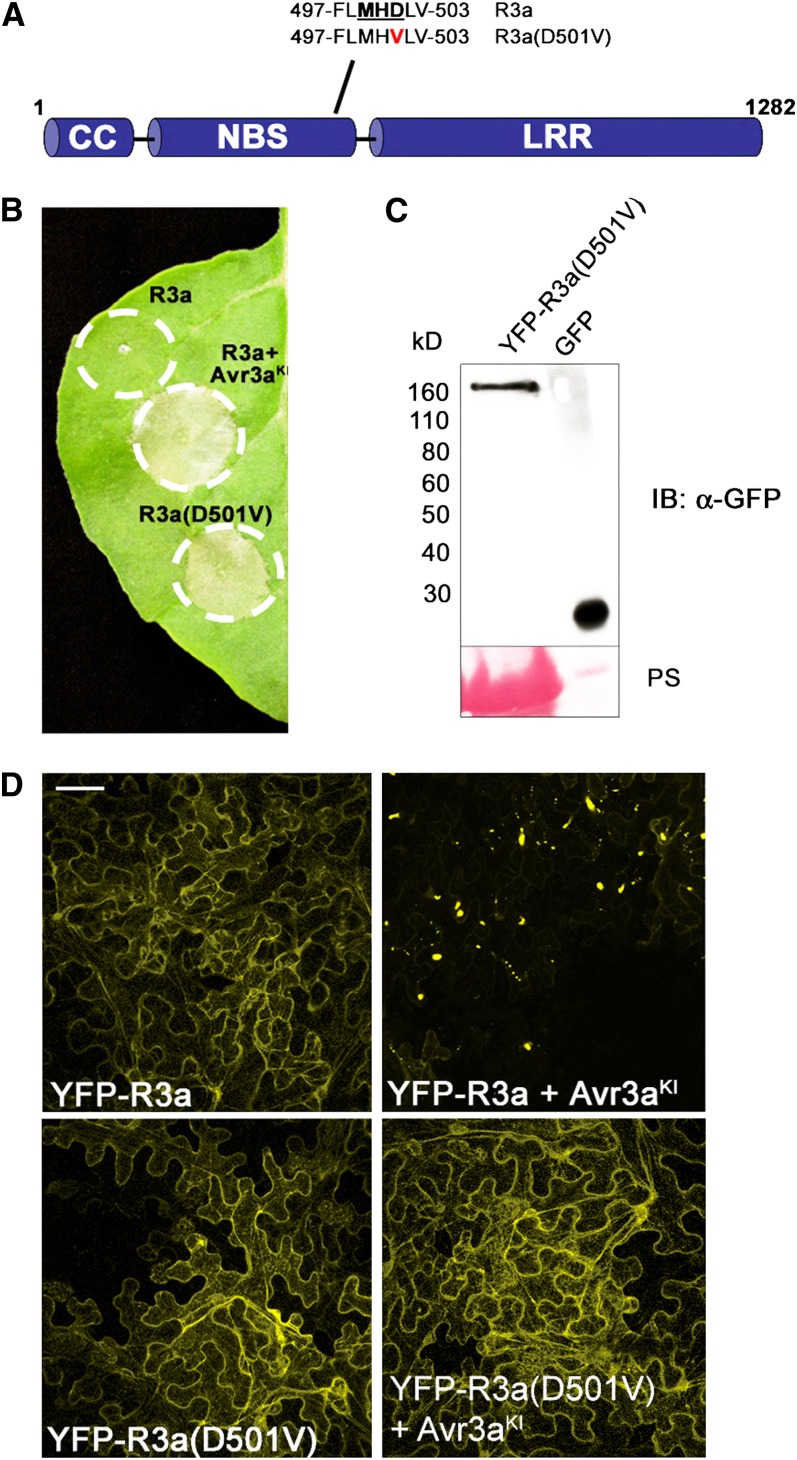
As some mutations in the NB-ARC domain in general, and in the Met-His-Asp (MHD) motif in particular, are known to cause autoactivation of resistance proteins (Tameling et al., 2006; Gao et al., 2011; Williams et al., 2011), we further investigated the significance of AVR3aKI-specific R3a relocalization by generating an autoactive R3a variant. We constructed R3a(D501V) (Figure 7A) by site-directed mutagenesis of the MHD motif and expressed this mutant with and without N-terminally fused YFP in N. benthamiana. In the absence of the effector, untagged R3a(D501V) is sufficient to induce HR cell death (Figure 7B). However, HR symptoms caused by R3a(D501V) alone developed a day later than HR triggered by R3a-mediated recognition of AVR3aKI (see Supplemental Figure 8 online). Delayed HR caused by mutations in the MHD motif of resistance proteins has been observed previously (Rairdan and Moffett, 2006).
Figure 7.
The Autoactive Mutant R3a(D501V) Is Cytoplasmic and Triggers HR in the Absence of Effectors.
(A) The MHD mutant of R3a used in this work. Wild-type sequence (top) and mutant sequence (bottom) is shown.
(B) Transient expression of R3a and R3a(D501V) alone and coexpression of R3a and AVR3aKI by agroinfiltration in N. benthamiana. Photo of HRs was taken 4 d after inoculation and is representative from multiple assays. Circles indicate the infiltrated area on the leaf panel.
(C) Immunoblot probed with α-GFP following transient expression of YFP-R3a(D501V) in N. benthamiana at 2 d after inoculation. Protein sizes are indicated (in kilodaltons), and protein loading is shown by Ponceau S (PS). A leaf sample constitutively expressing GFP was used as a positive control.
(D) Confocal laser scanning microscopy following transient expression in N. benthamiana of YFP-R3a alone (top left picture), YFP-R3a coexpressed with AVR3aKI (top right picture), YFP-R3a(D501V) alone (bottom left picture), and YFP-R3a(D501V) coexpressed with AVR3aKI (bottom right picture) at 2 d after inoculation. Bar = 20 µm.
The localization of the YFP-R3a(D501V) mutant was predominantly cytoplasmic, similar to YFP-R3a when expressed in the absence of recognized effectors. This cytoplasmic localization was maintained even when YFP-R3a(D501V) was coexpressed with AVR3aKI (Figure 7D). The observation that YFP-R3a(D501V) is largely cytoplasmic (Figure 7) suggests that HR signaling by this autoactive mutant has been uncoupled from relocalization.
As with the coexpression of wild-type YFP-R3a with AVR3aKI, YFP-R3a(D501V) does not trigger cell death symptoms (see Supplemental Figure 9 online), suggesting that downstream signaling is prevented by this fusion. Immunoblot analyses confirmed that YFP-R3a(D501V) was stable in planta, indicating that the fluorescence reports the location of intact fusion protein (Figure 7C).
Relocalization of R3a Is Required for the Immune Response
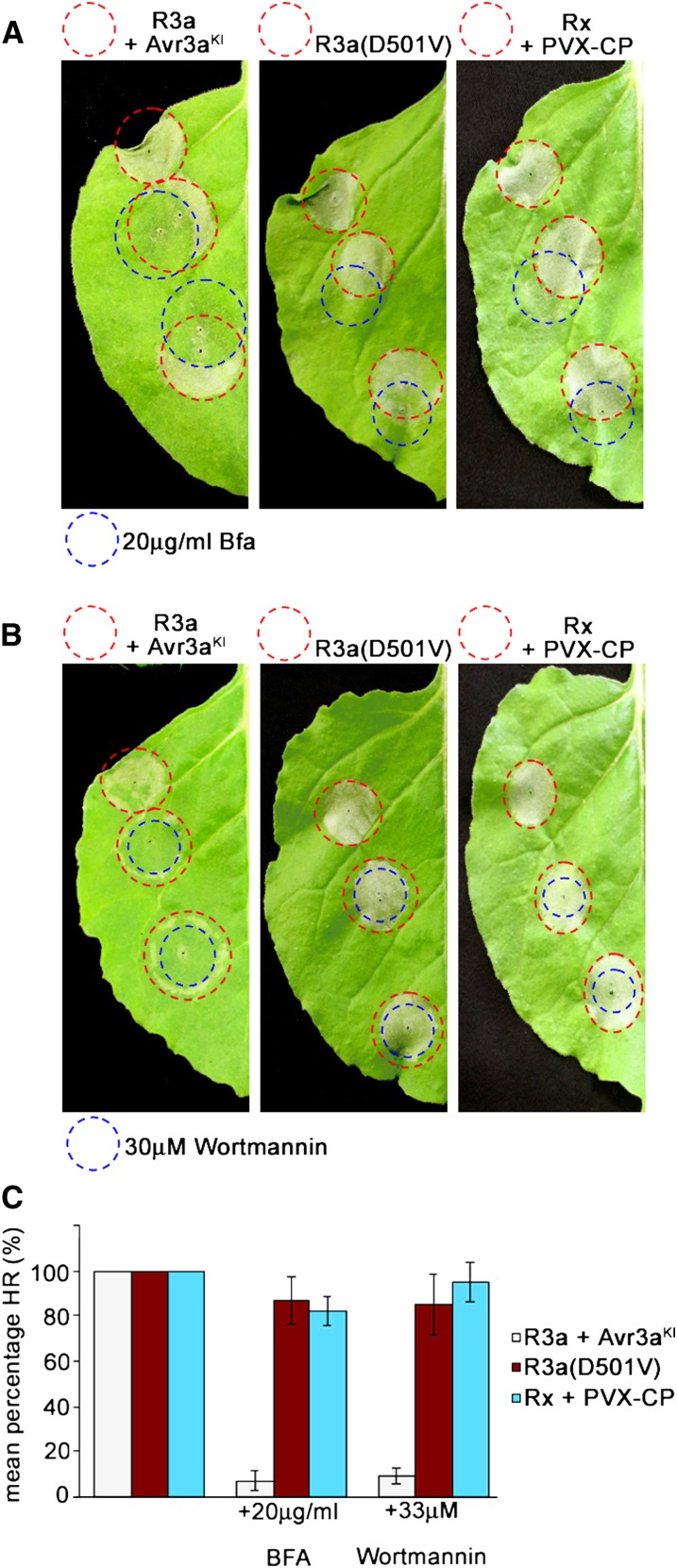
Since relocalization of R3a in the presence of AVR3aKI was attenuated by BFA and wortmannin treatment, we investigated the impact of these inhibitors on HR development. At 24 h after coagroinfiltration of N. benthamiana with Agrobacterium expressing R3a and AVR3aKI (24 h prior to the onset of visible HR symptoms), we infiltrated BFA and wortmannin into the agroinfiltrated leaf area and assessed HR development. As controls (showing similar timing of HR development), we transiently coexpressed the potato resistance protein Rx1 with the corresponding coat protein (CP) of Potato virus X (PVX) (Bendahmane et al., 1999), and Solanum stoloniferum resistance gene Sto1 with the corresponding P. infestans avirulence effector IpiO1 (Vleeshouwers et al., 2008). In the presence of PVX-CP, Rx1 triggers an HR that is not affected by BFA and wortmannin treatment (Figure 8; see Supplemental Figure 10 online). Likewise, we did not observe suppression of the Sto1-mediated HR by BFA and wortmannin in the presence of IpiO1 (see Supplemental Figure 10 online). The infiltration of either inhibitor alone did not cause any cell death symptoms within the time frame of assessing HR development (see Supplemental Figure 10 online). In contrast with the HR mediated by Rx1 and Sto1, both inhibitors considerably attenuated R3a-triggered HR (Figure 8; see Supplemental Figure 10B online). Within the entire inhibitor-infiltrated region, the HR normally caused by R3a/AVR3aKI did not develop.
Figure 8.
BFA and Wortmannin Significantly Attenuate HR Development Triggered by R3a in the Presence of AVR3aKI in Contrast with R3a(D501V)- and Rx1-Triggered HR.
(A) and (B) Transient coexpression of R3a with AVR3aKI (left picture) and Rx1(Rx) with PVX-CP (right picture) and expression of R3a(D501V) (middle picture) by agroinfiltration in N. benthamiana (red circles) was followed 24 h later [48 h later for R3a(D501V)-expressing leaves] by infiltration of 20 µg/mL BFA (A) and 33 µM wortmannin (B) (areas infiltrated by inhibitors indicated as blue circles). Photos of HRs were taken 48 h [72 h for R3a(D501V)-expressing leaves] after agroinfiltration.
(C) Graph shows the percentage of inhibitor (BFA and wortmannin) infiltration sites developing a clear HR at 2 d (for R3a and Rx1) and 3 d [for R3a(D501V)] after agroinfiltration. Experiments were repeated at least three times, each with no less than four leaves from four plants. Error bars indicate ± se.
We assessed whether BFA or wortmannin treatment would have a similar effect on the autoactive HR triggered by R3a(D501V). Inhibitors were applied from 24 h after inoculation of R3a(D501V) up to 48 h after inoculation (24 h prior to visible HR symptoms of this delayed cell death response, similar to application with wild-type R3a coexpressed with AVR3aKI). In contrast with wild-type R3a, the cell death triggered by the autoactive MHD mutant R3a(D501V) was unaffected by either BFA or wortmannin at any time point of inoculation (shown for 48 h after inoculation in Figure 8).
DISCUSSION
A vital objective of plant–pathogen interaction research is to determine how and where pathogen effectors are recognized by resistance proteins to mount a powerful immune response. General models of effector recognition by plant NB-LRR proteins are not yet well established (Caplan et al., 2008; Eitas and Dangl, 2010). Due to the different intrinsic virulence activities of pathogen effectors that, in turn, demand various recognition and signaling capabilities from R proteins, different model systems of effector recognition processes have been recently proposed (Bernoux et al., 2011). Our molecular and microscopy analyses of the potato resistance protein R3a and the oomycete effector protein AVR3a shed light on a particular activation process of a cytoplasmic NB-LRR protein by a pathogen effector. This study shows that R3a relocalizes to compartments of the endocytic pathway only in the presence of recognized effector forms prior to HR cell death. Interestingly, R3a relocalization is accompanied by relocalization also of the recognized effector form, AVR3aKI, and is required to mount a full immune response. We provide data demonstrating that both proteins colocalize to late endosomes in host plant cells prior to HR initiation.
Relocalization of R3a to Endosomes in the Presence of Recognized Effectors Is a Prerequisite for the Immune Response
In recent years, the observation that many resistance proteins accumulate in the nucleus in the presence of their corresponding effector proteins gave rise to a model where, upon pathogen perception, nuclear accumulation of R proteins is essential for activating the signaling process that leads to transcriptional reprogramming and ultimately the HR (Wiermer et al., 2007). In contrast with the nucleocytoplasmic trafficking that NB-LRR proteins like barley (Hordeum vulgare) MLA10, tobacco N, potato Rx, and Arabidopsis RPS4 and SNC1 undergo (Burch-Smith et al., 2007; Shen et al., 2007; Wirthmueller et al., 2007; Cheng et al., 2009; Slootweg et al., 2010; Tameling et al., 2010), Arabidopsis RPM1 has been shown recently to be constitutively plasma membrane localized, independent of the presence of AvrRpm1 (Gao et al., 2011), and is believed to initiate a cytosolic signaling pathway from this site. More recently, Takemoto et al. (2012) have demonstrated that a number of NB-LRR resistance proteins are constitutively associated with endomembranes, raising the likelihood that there are a number of possible subcellular locations from which the HR can be initiated. In addition, the importance of different subcellular localizations to downstream signaling events has been documented for animal NLR immune receptors, such as NOD2, which is only functional at the plasma membrane when activated (Lécine et al., 2007).
Our studies of potato R3a reveal a novel behavior: relocalization of this resistance protein from the cytoplasm to late endosomes only in the presence of recognized effector forms. The initial studies of this were based on FP-R3a fusion proteins, which are prevented from signaling the HR (Figures 1 to 3). Indeed, R3a-FP and FP-R3a(D501V) fusions were similarly prevented from eliciting HR. However, observations of the behavior of FP-AVR3a with untagged R3a, which leads to an active HR, revealed that the recognized FP-AVR3aKI, but not the unrecognized AVR3aEM, was similarly relocalized to late endosomes (Figures 4 and 5).
Studies of FP-R3a with dexamethasone-inducible AVR3aKI (Figure 2) and of FP-AVR3aKI with untagged R3a (Figure 4) both indicated that relocalization occurred prior to the initiation of HR, suggesting that such relocalization is an important step either in effector recognition or in activating R3a for subsequent HR signaling. Indeed, inhibitors of the endocytic cycle, wortmannin and BFA, both attenuated FP-R3a relocalization (Figure 6) and prevented R3a-mediated HR when applied 24 h prior to cell death symptoms (Figure 8), confirming the requirement for association with late endosomes for normal activation of wild-type R3a. Critically, these inhibitors had no effect on HR triggered by coexpression of Rx1 with PVX-CP or Sto1 with its cognate effector IpiO1. While we know little about the subcellular localization of Sto1, Rx1-mediated HR is dependent on nucleocytoplasmic shuttling of this resistance protein (Slootweg et al., 2010; Tameling et al., 2010). It is thus, perhaps, unsurprising that BFA and wortmannin treatments did not attenuate this HR.
The importance of the specific relocalization to late endosomes for the normal immune response of wild-type R3a was further emphasized by the behavior of the R3a(D501V) autoactive mutant form, which gave a delayed cell death response in the absence of effectors and failed to localize to late endosomes (Figure 7). This suggests that downstream signaling to trigger an HR by this form is uncoupled from effector-mediated relocalization. In keeping with this, wortmannin and BFA treatments failed to inhibit this HR (Figure 8).
Both BFA and wortmannin, which are inhibitors of different parts of the endocytic and secretory pathway (Geldner, 2004; Ebine et al., 2011; Ito et al., 2011), attenuate R3a association with late endosomes and have an effect on preexisting late endosomes with which R3a is already associated (Figure 7). Wortmannin causes deformations of late endosomes (Ebine et al., 2011) and homotypic fusions of late endosomes but also fusions between late endosomes and the trans-Golgi network to form small vacuoles (Wang et al., 2009). Treatment with BFA leads to reorganization of the ER–Golgi interaction, resulting in the formation of BFA bodies derived from modified ER cisternae (Orci et al., 1993; Uemura et al., 2004; Jaillais et al., 2008; Ebine et al., 2011). The fluorescing vesicles that we still detected after wortmannin and BFA treatments (Figure 6) may represent these late endosome fusions and BFA bodies, respectively. The assumed failure of further molecules of FP-R3a to relocalize in the presence of these inhibitors, leading to the observed cytoplasmic fluorescence, can be explained by the fact that the formation of late endosomes is inhibited by wortmannin and BFA (Geldner, 2004). Critically, the fact that we perturb normal R3a-mediated cell death with these inhibitors but not that of Rx1 or Sto1 (Figure 8; see Supplemental Figure 10 online) indicates that the accumulation of R3a at late endosomes is essential for HR initiation.
Influence of Mutations in the MHD Motif on R3a Function
Mutations in the highly conserved MHD motif in the C terminus of the ARC2 domain of NB-LRR resistance proteins are known to cause autoactivity (Bendahmane et al., 2002; Tameling et al., 2006; Van Oijen et al., 2008; Gao et al., 2011; Williams et al., 2011). As expected, mutation of the Asp in the MHD motif of R3a to Val gave rise to an autoactive cell death phenotype. Nevertheless, the cell death elicited by R3a(D501V) was significantly delayed compared with the effector-mediated cell death observed with wild-type R3a, a phenomenon shown also for an MHD mutant of Rx1 (Rairdan and Moffett, 2006).
Recent studies of an MHD-mutated flax NB-ARC-LRR protein, M (Williams et al., 2011), showed that the M(D555V) had significantly more ATP bound than ADP, which supports the molecular switch model, in which the ADP-bound R protein is inactive and an ATP-bound R protein is active (Tameling et al., 2006; Takken et al., 2006). Williams et al. (2011) speculate that mutations within the MHD motif may prevent ATP hydrolysis that, in turn, effectively prevents the R protein entering the inactive state. Accordingly, the mutation in R3a(D501V) should cause an increase binding of ATP, either due to increased ATP binding affinity as a result of a more open NB-ARC conformation or reduced hydrolysis of ATP (Van Oijen et al., 2008), leading to autoactivity.
One interesting observation is that R3a(D501V) is cytoplasmic. Accordingly, wortmannin and BFA treatments had no significant effect on the autoactive HR. One explanation of why the autoactivity is not compromised by BFA and wortmannin could be that the HR triggered by the autoactive R3a(D501V) mutant is mediated by a signaling pathway that is independent of regulatory factors at the late endosome. An alternative explanation, which is not mutually exclusive, is that wild-type R3a is only activated after relocalization to late endosomes and requires either direct or indirect interaction with AVR3a, or an AVR3a-target complex, at late endosomes to enter that active state. Following the reconformation of R3a into an active state, it is free to interact with signaling partners to trigger the HR. The site-specific activation prevents R3a from inappropriately becoming active in the absence of the effector and thus is a means to control the context of R3a activity.
R3a and AVR3aKI Association at Late Endosomes
Fluorescently tagged AVR3a in the absence of R3a is generally cytoplasmic and reveals no association with vesicles (Bos et al., 2010; Gilroy et al., 2011a). In this study, we show relocalization to late endosomes, prior to HR symptoms, specifically of AVR3aKI, but not AVR3aEM, in the presence of untagged (active) R3a (Figure 4). Both the colocalization study (Figure 4) and the BiFC experiment (Figure 5) indicate that R3a and AVR3aKI are in close proximity at the endosomes. However, direct, binary interaction between R3a and AVR3aKI was not supported by Y2H analyses (see Supplemental Figure 6 online), unlike notable examples, such as the flax (Linum usitatissimum) resistance L6 and flax rust (Melampsora lini) avirulence AvrL567 (Dodds et al., 2006). Moreover, coimmunoprecipitation experiments also failed to pull down an AVR3aKI-R3a complex (see Supplemental Figure 7 online). Nevertheless, Y2H is prone to false negative results, meaning that true interactions are sometimes not detected (Stellberger et al., 2010). In addition, the reconstitution of YFP during BiFC is a strong, irreversible bond. We cannot rule out a transient, direct interaction between R3a and AVR3aKI that was undetectable in either Y2H or coimmunoprecipitation experiments but effectively maintained by the BiFC analysis. Similarly, we cannot conclude whether the interaction is mediated by an additional host protein. Indeed, HR triggered by potato R2 and P. infestans AVR2 was recently shown to be mediated by the host protein BSL1, indicating that the involvement of additional protein partners in the recognition between resistance proteins and oomycete avirulence proteins can occur (Saunders et al., 2012).
The host ubiquitin E3 ligase, CMPG1, is an important virulence target of AVR3a and yet is not required for R3a HR (Bos et al., 2010; Gilroy et al., 2011a), demonstrating that it is not a guardee that mediates R3a recognition of AVR3a. However, a number of host proteins, as yet unverified in planta, have been shown to interact with AVR3a using Y2H, including two components of the exocyst, Sec3 and Sec5 (Bos et al., 2010). Sec3 and Sec5 interact with each other and are involved in endocytosis and exocytosis (Sommer et al., 2005; Hála et al., 2008; Zhang et al., 2010) and thus may be associated with compartments in the endocytic pathway. Further detailed studies, beyond the scope of this work, are needed to investigate the precise nature of the interaction between R3a and AVR3a, direct or indirect, at late endosomes.
Recently, a three-dimensional structure for AVR3a was elucidated, revealing key residues involved in binding the C-terminal effector domain to phosphoinositides (PIPs) (Yaeno et al., 2011). Mutation of these residues resulted in AVR3a failing to bind to PIPs. Moreover, the mutants failed to suppress INF1-triggered cell death or stabilize CMPG1 (Yaeno et al., 2011). However, they showed that none of the mutations attenuated R3a-mediated HR, showing that PIP binding is not required for AVR3a recognition by R3a. We thus conclude that such PIP binding is also not associated with the observed relocalization to late endosomes revealed in this study.
The Role Late Endosomes May Play in the R3a Immune Response
A recent report (Lu et al., 2012) indicated that development of the plant-haustorium interface in a compatible P. infestans–host interaction is accompanied by alterations to endosomal trafficking, emphasizing the potential importance of endocytic processes in establishing susceptibility or resistance. Our investigations led us to the conclusion that late endosomes are subcellular sites where R3a triggers a defense response upon AVR3a recognition.
Endosomal trafficking pathways are central regulators of plasma membrane protein homeostasis and also control multiple signaling pathways and developmental processes (Reyes et al., 2011). Apart from the literature that disagrees concerning the subcellular localization of the pentameric retromer complex (Reyes et al., 2011), indicating that proteins can be recycled from late endosomes, these compartments are nevertheless part of the degradative sorting pathway and the question unfolds: Why would R3a relocalize to an apparent dead end of the endosomal trafficking pathway?
Importantly, endocytosis of plant plasma membrane–localized receptors has been shown to be critical for their regulation and for their signaling following perception of associated ligands (Reyes et al., 2011). This has been described for pattern recognition receptors that perceive PAMPs. Upon flg22 stimulation, the Arabidopsis flagellin LRR–receptor-like kinase receptor FLAGELLIN-SENSING2 (FLS2) is ubiquitinated, internalized, and accumulates in late endosomes, prior to its degradation (Chinchilla et al., 2006; Robatzek et al., 2006; Salomon and Robatzek, 2006; Lu et al., 2011). Critically, endocytosis of FLS2 has been reported to be required for efficient pattern-triggered immunity (Robatzek et al., 2006), establishing a precedent in plants for signaling from endosomes. In animals, it has been well documented that receptor internalization into late endosomes is required, in a number of cases, for signaling to occur (Andersson, 2012). This establishes late endosomes as reported sites from which signaling can be initiated to activate an immune response.
Concluding Remarks
We provide evidence for novel behavior of an NB-LRR resistance protein: its relocalization, along with the recognized effector form AVR3aKI, to late endosomes. We show that this event occurs prior to the HR and is a prerequisite for the development of HR cell death. Our data contribute to a more general model of R protein activation, in which HR signaling can be triggered from diverse subcellular compartments and not exclusively within the nucleus. The major challenge will be to understand the chronological order of effector recognition and R3a activation in the context of relocalization and the putative involvement of additional factors required for signaling HR and for regulation of R3a activity.
METHODS
Microbial Strains and Growth Conditions
Agrobacterium tumefaciens strain AGL1 was used in molecular cloning and transient expression experiments and was routinely cultured at 28°C in yeast extract broth medium using appropriate antibiotics (Sambrook and Russell, 2001). DNA transformations of AGL1 were conducted by electroporation (Sambrook and Russell, 2001).
Plasmid Constructs
The constructs pENTR1A-Avr3aEM, pENTR1A-Avr3aKI, pENTR1A-Avr3aKIΔY, pENTR1A-R3a, pGRAB-Avr3aEM, pGRAB-Avr3aKI, pGRAB-Avr3aKIΔY, pGRAB-R3a, pGRAB-Avr2, pCL113-Avr3aEM, pCL113-Avr3aKI, Sto1-pK7WG2, and IpiO1-pGR106 were described previously (Armstrong et al., 2005; Vleeshouwers et al., 2008; Bos et al., 2010; Gilroy et al., 2011b). Rx (pB1:Rx-HA) and PVX CPs (pBin61:CP-TK) were described by Gilroy et al. (2011a).
The sequences for all the primers used in this study are shown in Supplemental Table 1 online in 5′ to 3′ orientation. After running the PCR products on an agarose gel, they were extracted with a Qiagen gel extraction kit, recombined into pDONR201 (Invitrogen) with Invitrogen BP clonase, and sequenced. The R3a(D501V) point mutation was introduced by site-directed mutagenesis using primers shown in Supplemental Table 1 online, followed by DpnI digestion. Fluorescent protein fusions were generated using the Gateway LR Clonase enzyme mix (Invitrogen) to recombine the entry vector clones of Avr3a and R3a with plant expression vectors. The vectors pCL112, pCL113, pB7WGY2, pB7WGF2, and pK7WGR2 were used to fuse YFP domains, YFP, GFP, and mRFP, respectively, to the N terminus of the protein of interest (Karimi et al., 2005; Bos et al., 2010).
For conditional expression, we recombined the entry vector clones of AVR3a with pBAV105 (Vinatzer et al., 2006) to express the effector from the DEX promoter.
The subcellular markers we used in this study were described previously: nuclear tag mRFP-H2B (Goodin et al., 2008; Martin et al., 2009), PVC tag CFP-PS1 (Saint-Jean et al., 2010), Golgi tag ST-YFP (Boevink et al., 1998; Brandizzi et al., 2002), peroxisome tag mRFP-SRL (Mathur et al., 2002; Sparkes et al., 2003), Ara6-mRFP (Ueda et al., 2001), mRFP-Ara7 (Kotzer et al., 2004; Ueda et al., 2004), and the ER marker tag RFP-KDEL (Tilsner et al., 2012).
We generated the entry vector pENTR1A-R3a(D501V) via site-directed mutagenesis of pENTR1A-R3a using the 5′ primer 5′-GAGAATTTATTCTTAATGCATGTCCTTGTCAATGATTTAGCGC-3′ in combination with the 3′ primer 5′-GCGCTAAATCATTGACAAGGACATGCATTAAGAATAAATTCTC-3′. After PCR, the template plasmid was digested with DpnI.
For Y2H experiments, DNA binding domain bait fusions and activation domain prey fusions were generated by recombination between entry vectors containing R3a, R3a subfragments, and Avr3a alleles with Clontech plasmids pGADT7 and pGBKT7 using Invitrogen LR recombinase. pGADT7 and pGBKT7 were rendered Gateway-compatible using the Invitrogen Gateway conversion kit.
Plant Material, Agroinfiltration, and Cell Death Assays
Nicotiana benthamiana was grown as described previously (Bos et al., 2010). All Agrobacterium cultures were grown overnight and, after pelleting, resuspended in sterile infiltration buffer (10 mM MES, 10 mM MgCl2, and 200 µM acetosyringone) to a final optical density at 600 nm (OD600) of 0.1 for confocal microscopy experiments (or as indicated otherwise) or 0.5 for cell death assays. For coexpression experiments, bacterial strains containing the constructs of interest, expressed from the 35S promoter, were mixed, adjusting each strain concentration to the required final OD600. After an incubation period of 2 to 4 h in darkness at room temperature, bacterial suspensions were infiltrated with a 1-mL blunt-end syringe through the abaxial leaf surface of 4- to 6-week-old plants, which were slightly wounded with a needle. On each plant, two to three leaves were used for each biological replicate; in total, at least five plants were used. FM 4-64 (3.33 μg/μL; in DMSO) was diluted 1:100 in water and infiltrated into leaf areas infected with constructs of interest 20 min before taking pictures. For cell death assays, BFA (20 µg/mL) and Wortmannin (33 µM) (Sigma-Aldrich) were infiltrated 24 to 72 h after agroinfiltration, and bright-field/UV pictures were taken 24, 48, 72, and 96 h after inhibitor infiltration. For conditional expression of AVR3a, dexamethasone (30 μM in a solution of 0.1% Tween 20 in water) was infiltrated into leaves infected with dexamethasone-inducible vectors 48 h after initial Agrobacterium infection. Confocal and cell death imaging was done at the indicated time points after dexamethasone treatment.
Y2H Assays
Both the bait protein-encoding vector pGBKT7 containing the R3a clones and the prey-encoding vector pGADT7 encoding the different Avr3a alleles were transformed into the yeast strain pJ69-4A (James et al., 1996) using the protocol described in Grefen et al. (2009). Initially, yeast transformants were plated onto complete supplement medium (CSM; Formedium) lacking Leu and Trp (-Leu, -Trp) and incubated at 30°C for 3 to 4 d. Colonies were picked and cultivated overnight in 5 mL of liquid CSM (-Leu, -Trp). Subsequently, a dilution series (10° to 10−3) was prepared of each suspension having an initial OD600 of 0.5. A total of 7.5 μL of each dilution was carefully dropped to CSM agar with (-Leu, -Trp) and CSM agar without adenine (-Leu, -Trp, -adenine). The plates were incubated at 30°C for 2 to 3 d prior to photographing. Positive interaction between the expressed proteins resulted in yeast growth and the activation of β-galactosidase independent of adenine. The β-galactosidase activity was measured via the 2-Nitrophenyl-β-D-galactopyranoside assay (Grefen et al., 2009). The interaction between murine p53 and SV40 large T-antigen served as the positive control (Li and Fields, 1993).
Coimmunoprecipitation Experiments and Immunoblot Analyses
Protein extraction from yeast cells was performed as described by Kushnirov (2000). Extraction of total protein from plant samples was done by grinding leaf tissue in liquid nitrogen followed by boiling for 5 min in SDS loading buffer supplemented with 1% β-mercaptoethanol. The presence of recombinant R3a, R3a(D501V), and Avr3a fusion proteins was determined by SDS-PAGE and protein gel blotting as described previously (Bos et al., 2010). To detect fusions containing cMyc, we used α-cMyc-horseradish peroxidase antibody (Invitrogen). For the detection of HA-containing fusions, we used α-HA from rabbit (Sigma-Aldrich). For the detection of GFP/YFP fusions, we used α-GFP from rabbit (Sigma-Aldrich, Santa Cruz Biotechnology); the latter were followed by α-rabbit-horseradish peroxidase from goat (Sigma-Aldrich). For the detection of FLAG-containing fusions, we used α-FLAG antibody from mouse (Santa Cruz Biotechnology) followed by α-mouse-horseradish peroxidase from goat (Sigma-Aldrich). Protein bands on the immunoblot were detected using ECL substrate (GE Healthcare).
For coimmunoprecipitation experiments, total proteins were extracted from N. benthamiana leaf discs 3 d after agroinfiltration by grinding in N2 and resuspending in GTEN buffer containing 0.5% Nonidet P-40, 1 mM PMSF, and 1× protease inhibitor cocktail (Sigma-Aldrich). To immunoprecipitate GFP fusion proteins, magnetic GFP-Trap (Chromotek) was used, following the manufacturer’s protocol.
Confocal Imaging
Imaging was conducted on a Leica TCS-SP2 AOBS (Leica Microsystems) using HCX APO L, ×40/0.8, and ×63x0.9 water dipping lenses. Images were at 512 × 512 resolution and taken using line-by-line sequential scanning. The optimal pinhole diameter and the same gain level for the photomultiplier tube was maintained at all times. PhotoshopCS software (Adobe Systems) was used for postacquisition image processing. The excitation wavelength for mRFP was 561 nm, and its emission was collected from 600 to 630 nm. GFP was imaged using 488 nm excitation, and its emission was collected from 500 to 530 nm. YFP was imaged using 514 nm excitation with an emission collected from 520 to 555 nm. CFP was imaged using 405-nm excitation, and its emission was collected from 455 to 490 nm. FM4-64 (Molecular Probes) was excited with the 561-nm line and emissions detected between 630 and 680nm. The images were taken no more than 10 min after infiltration with FM4-64. Coexpressed mRFP and YFP as well as GFP coexpressed with mRFP were imaged sequentially using a line-by-line mode. Chloroplasts were excited with the 488-nm line, and the emissions were detected between 660 and 700 nm.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: R3a (AY849382.1), Avr3a (PITG_14371), Avr2 (PITG_08943), Pex147-2 (PITG_14368), and Pex147-3 (PITG_14374).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FP-R3a Localizes to the Cytoplasm in the Absence of Effectors and Fails to Cause HR with AVR3aKI or pex147-3.
Supplemental Figure 2. In the Presence of AVR3aKI, R3a Does Not Relocalize to Chloroplasts, Nuclei, Peroxisomes, or Golgi.
Supplemental Figure 3. Cells Transiently coexpressing the YFP-R3a, CFP-PS1, and a Dexamethasone-Inducible Promoter-Driven Avr3aKI 2 h after Treatment with 30 µM Dexamethasone.
Supplemental Figure 4. YFP C-Terminally Fused to R3a and R3a(D501V) Prevents HR Development.
Supplemental Figure 5. GFP-Avr3aKI, but Not GFP-Avr3aEM, Relocalizes to Endosomal Compartments in the Presence of Untagged R3a.
Supplemental Figure 6. R3a and R3a Subfragments Do Not Interact with Avr3a Using Yeast Two-Hybrid Analysis.
Supplemental Figure 7. Immunoblots Showing R3a Fails to Coimmunoprecipitate with AVR3a in Planta.
Supplemental Figure 8. R3a(D501V) Causes a Delayed HR Development in Comparison to Wild-Type R3a.
Supplemental Figure 9. YFP N-Terminally Fused to R3a and R3a(D501V) Prevents HR Development.
Supplemental Figure 10. HR Triggered by Sto1 and Rx1 Is Not Affected by BFA or Wortmannin Treatment.
Supplemental Table 1. Primers Used in This Study.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council and the Scottish Government, UK, for financial support. S.E. was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft).
AUTHOR CONTRIBUTIONS
P.R.J.B., I.H., S.E., and P.C.B. designed the research. S.E., P.C.B., M.R.A., and M.B.R. performed the research. S.E., P.C.B., and P.R.J.B. analyzed the data. S.E., P.C.B., I.H., and P.R.J.B. wrote the article.
Glossary
- NB-LRR
nucleotide binding–Leu-rich repeat
- CC
coiled coil
- HR
hypersensitive response
- YFP
yellow fluorescent protein
- hpt
hours post-treatment
- GFP
green fluorescent protein
- PVC
prevacuolar compartment
- CFP
cyan fluorescent protein
- BiFC
bimolecular fluorescence complementation
- BFA
brefeldin A
- PVX
Potato virus X
- ER
endoplasmic reticulum
- MHD
Met-His-Asp
- Y2H
yeast two-hybrid
- PIP
phosphoinositide
- CSM
complete supplement medium
References
- Andersson E.R. (2012). The role of endocytosis in activating and regulating signal transduction. Cell. Mol. Life Sci. 69: 1755–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102: 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Nimchuk Z., Hubert D.A., Mackey D., Dangl J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ellis J.G., Dodds P.N. (2011). New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14: 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A., Li G., Fu Z.Q., Alfano J.R. (2008). Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11: 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Bos J.I.B., Chaparro-Garcia A., Quesada-Ocampo L.M, McSpadden Gardener B.B., Kamoun A. (2009). Distinct amino acids of the Phytophthora infestans effector AVR3a condition activation of R3a hypersensitivity and suppression of cell death. Mol. Plant Microbe Interact. 22: 269–281 [DOI] [PubMed] [Google Scholar]
- Bos J.I.B., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.I.B., Kanneganti T.-D., Young C., Cakir C., Huitema E., Win J., Armstrong M.R., Birch P.R.J., Kamoun S. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48: 165–176 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Snapp E.L., Roberts A.G., Lippincott-Schwartz J., Hawes C. (2002). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: Evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J., Padmanabhan M., Dinesh-Kumar S.P. (2008). Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135 [DOI] [PubMed] [Google Scholar]
- Chardin P., McCormick F. (1999). Brefeldin A: The advantage of being uncompetitive. Cell 97: 153–155 [DOI] [PubMed] [Google Scholar]
- Cheng Y.T., Germain H., Wiermer M., Bi D., Xu F., García A.V., Wirthmueller L., Després C., Parker J.E., Zhang Y., Li X. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Chung E.-H., da Cunha L., Wu A.-J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S.M., Hamel L.-P., Moffett P. (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.-M., Teh T., Wang C.-I.A., Ayliffe M.A., Kobe B., Ellis J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Eitas T.K., Dangl J.L. (2010). NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emans N., Zimmermann S., Fischer R. (2002). Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell 14: 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Chung E.H., Eitas T.K., Dangl J.L. (2011). Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 108: 7619–7624 Erratum. Proc. Natl. Acad. Sci. USA 108: 8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N. (2004). The plant endosomal system—Its structure and role in signal transduction and plant development. Planta 219: 547–560 [DOI] [PubMed] [Google Scholar]
- Gilroy E.M., et al. (2011b). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191: 763–776 [DOI] [PubMed] [Google Scholar]
- Gilroy E.M., Taylor R.M., Hein I., Boevink P., Sadanandom A., Birch P.R.J. (2011a). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190: 653–666 [DOI] [PubMed] [Google Scholar]
- Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. (2008). Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Grefen C., Obrdlik P., Harter K. (2009). The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 479: 217–233 [DOI] [PubMed] [Google Scholar]
- Hála M., Cole R., Synek L., Drdová E., Pecenková T., Nordheim A., Lamkemeyer T., Madlung J., Hochholdinger F., Fowler J.E., Zárský V. (2008). An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20: 1330–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E., Fujimoto M., Ebine K., Uemura T., Ueda T., Nakano A. (2011). Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69: 204–216 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Gaude T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 53: 237–247 [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jupe F., Pritchard L., Etherington G.J., Mackenzie K., Cock P.J., Wright F., Sharma S.K., Bolser D., Bryan G.J., Jones J.D., Hein I. (2012). Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics 13: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kotzer A.M., Brandizzi F., Neumann U., Paris N., Moore I., Hawes C. (2004). AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Lécine P., Esmiol S., Métais J.Y., Nicoletti C., Nourry C., McDonald C., Nunez G., Hugot J.P., Borg J.P., Ollendorff V. (2007). The NOD2-RICK complex signals from the plasma membrane. J. Biol. Chem. 282: 15197–15207 [DOI] [PubMed] [Google Scholar]
- Li B., Fields S. (1993). Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 7: 957–963 [DOI] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.-J., Schornack S., Spallek T., Geldner N., Chory J., Schellmann S., Schumacher K., Kamoun S., Robatzek S. (2012). Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell. Microbiol. 14: 682–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Hülskamp M. (2002). Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128: 1031–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Cheung A.Y., Ueda T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.K., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21: 2928–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Perrelet A., Ravazzola M., Wieland F.T., Schekman R., Rothman J.E. (1993). “BFA bodies”: A subcompartment of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 90: 11089–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan G.J., Moffett P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., And Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673 [DOI] [PubMed] [Google Scholar]
- Richter S., Voss U., Jürgens G. (2009). Post-Golgi traffic in plants. Traffic 10: 819–828 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Langhans M., Saint-Jore-Dupas C., Hawes C. (2008). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13: 405–408 [DOI] [PubMed] [Google Scholar]
- Saint-Jean B., Seveno-Carpentier E., Alcon C., Neuhaus J.M., Paris N. (2010). The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell 22: 2825–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S., Robatzek S. (2006). Induced endocytosis of the receptor kinase FLS2. Plant Signal. Behav. 1: 293–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Saunders D.G., Breen S., Win J., Schornak S., Hein I., Bozkurt T.O., Champouret N., Vleeshouwers V.G., Birch P.R.J., Gilroy E.M., Kamoun S. (2012). Host protein BSL1 associated with Phyyophthora infestans AVR2 and THE Solanum demissum R2 to mediate disease resistance. Plant Cell 24: 3420–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.-H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Slootweg E., et al. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Oprins A., Rabouille C., Munro S. (2005). The exocyst component Sec5 is present on endocytic vesicles in the oocyte of Drosophila melanogaster. J. Cell Biol. 169: 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Brandizzi F., Slocombe S.P., El-Shami M., Hawes C., Baker A. (2003). An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellberger T., Häuser R., Baiker A., Pothineni V.R., Haas J., Uetz P. (2010). Improving the yeast two-hybrid system with permutated fusions proteins: The Varicella Zoster Virus interactome. Proteome Sci. 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D., Rafiqi M., Hurley U., Lawrence G.J., Bernoux M., Hardham A.R., Ellis J.G., Dodds P.N., Jones D.A. (2012). N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol. Plant Microbe Interact. 25: 379–392 [DOI] [PubMed] [Google Scholar]
- Takken F.L.W., Albrecht M., Tameling W.I.L. (2006). Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Tameling W.I.L., Nooijen C., Ludwig N., Boter M., Slootweg E., Goverse A., Shirasu K., Joosten M.H.A.J. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Vossen J.H., Albrecht M., Lengauer T., Berden J.A., Haring M.A., Cornelissen B.J.C., Takken F.L.W. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Wright K.M., Bell K., Roberts A.G., Lacomme C., Cruz S.S., Oparka K.J. (2012). The TGB1 Movement Protein of Potato virus X Reorganizes Actin and Endomembranes into the X-Body, a Viral Replication Factory. Plant Physiol. 158: 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Van Oijen G., Mayr G., Kasiem M.M.A., Albrecht M., Cornelissen B.J.C., Takken F.L.W. (2008). Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 59: 1383–1397 [DOI] [PubMed] [Google Scholar]
- Vinatzer B.A., Teitzel G.M., Lee M.-W., Jelenska J., Hotton S., Fairfax K., Jenrette J., Greenberg J.T. (2006). The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62: 26–44 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V.G.A.A., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and phytophthora infestans avirulence genes. PLoS ONE 3: e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Palma K., Zhang Y., Li X. (2007). Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell. Microbiol. 9: 1880–1890 [DOI] [PubMed] [Google Scholar]
- Williams S.J., Sornaraj P., deCourcy-Ireland E., Menz R.I., Kobe B., Ellis J.G., Dodds P.N., Anderson P.A. (2011). An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 24: 897–906 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Yaeno T., Li H., Chaparro-Garcia A., Schornack S., Koshiba S., Watanabe S., Kigawa T., Kamoun S., Shirasu K. (2011). Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. USA 108: 14682–14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu C.-M., Emons A.-M.C., Ketelaar T. (2010). The plant exocyst. J. Integr. Plant Biol. 52: 138–146 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20: 10–16 [DOI] [PubMed] [Google Scholar]