This chemical genetics work reveals natural variation in a newly identified R protein homolog, named VICTR, that produces primary root growth arrest in response to the small molecule DFPM. DFPM perception and signal transduction require early components of the plant R gene resistance signaling network, and the R protein VICTR coresides in complexes not only with EDS1 but also PAD4.
Abstract
In a chemical genetics screen we identified the small-molecule [5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione (DFPM) that triggers rapid inhibition of early abscisic acid signal transduction via PHYTOALEXIN DEFICIENT4 (PAD4)- and ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1)-dependent immune signaling mechanisms. However, mechanisms upstream of EDS1 and PAD4 in DFPM-mediated signaling remain unknown. Here, we report that DFPM generates an Arabidopsis thaliana accession-specific root growth arrest in Columbia-0 (Col-0) plants. The genetic locus responsible for this natural variant, VICTR (VARIATION IN COMPOUND TRIGGERED ROOT growth response), encodes a TIR-NB-LRR (for Toll-Interleukin1 Receptor–nucleotide binding–Leucine-rich repeat) protein. Analyses of T-DNA insertion victr alleles showed that VICTR is necessary for DFPM-induced root growth arrest and inhibition of abscisic acid–induced stomatal closing. Transgenic expression of the Col-0 VICTR allele in DFPM-insensitive Arabidopsis accessions recapitulated the DFPM-induced root growth arrest. EDS1 and PAD4, both central regulators of basal resistance and effector-triggered immunity, as well as HSP90 chaperones and their cochaperones RAR1 and SGT1B, are required for the DFPM-induced root growth arrest. Salicylic acid and jasmonic acid signaling pathway components are dispensable. We further demonstrate that VICTR associates with EDS1 and PAD4 in a nuclear protein complex. These findings show a previously unexplored association between a TIR-NB-LRR protein and PAD4 and identify functions of plant immune signaling components in the regulation of root meristematic zone-targeted growth arrest.
INTRODUCTION
Chemical genetics provides a powerful tool to address redundancy and network robustness in signal transduction (Schreiber, 2000; Armstrong et al., 2004; Zouhar et al., 2004; Park et al., 2009). In a recent screen of a 9600-compound chemical library for inhibition of abscisic acid (ABA) signaling, a small molecule named [5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione (DFPM) was isolated that showed the capacity to inhibit several ABA responses, including rapid disruption of ABA-induced stomatal closing and ABA activation of guard cell anion channels (Kim et al., 2011). Detailed analyses revealed that DFPM stimulates an effector-triggered immune signaling pathway and thereby rapidly disrupts ABA signal transduction (Kim et al., 2011).
The central regulators of basal resistance and effector-triggered immunity, EDS1 and PAD4, as well as the cochaperones RAR1 (REQUIRED FOR Mla12 RESISTANCE) and SGT1b (SUPPRESSOR OF G-TWO ALLELE OF SKP1b) are required for DFPM-induced disruption of ABA signaling (Kim et al., 2011). Effector-triggered immunity relies on resistance (R) proteins as sensors that indirectly or directly recognize specific pathogen-derived effector molecules and trigger downstream disease resistance responses (Glazebrook, 2005; Jones and Dangl, 2006). Whether specific R proteins are required for the small-molecule DFPM-induced response remains unknown. Depending on their N-terminal domain, R proteins are generally divided into two groups, coiled-coil (CC)-nucleotide binding (NB)-leucine-rich repeat (LRR) and Toll-Interleukin1 Receptor (TIR)-NB-LRR (Meyers et al., 2003; Belkhadir et al., 2004; Chisholm et al., 2006; DeYoung and Innes, 2006; Shen and Schulze-Lefert, 2007; Caplan et al., 2008). The genome of Arabidopsis thaliana contains ∼150 NB-LRR genes, and up to 600 genes are found in rice (Oryza sativa; Kadota et al., 2010). The presence of many polymorphisms in NB-LRR genes has been proposed as a diversification process against rapidly evolving pathogens (Tian et al., 2003; Guo et al., 2011). In fact, NB-LRR genes represent the most variable plant gene family (Weigel, 2012).
Upon pathogen recognition, intramolecular structural changes of NB-LRR proteins induce ADP-ATP exchange in the NB domain and activate downstream responses. Effector-triggered activation of TIR-NB-LRR proteins requires the function of EDS1 and PAD4 to generate downstream disease resistance responses, including salicylic acid–mediated signal transduction (Wiermer et al., 2005). Recently, it was found that EDS1 resides in some complexes with TIR-NB-LRR proteins (Bhattacharjee et al., 2011; Heidrich et al., 2011).
Here, using a chemical genetics approach, we identified natural genetic variation in a primary root growth response induced by the small molecule DFPM. The Arabidopsis Columbia-0 (Col-0) accession responds specifically to DFPM by generating a primary root meristem arrest. Molecular identification of the responsible genetic locus and its functional characterization demonstrate that a natural genetic variant in the TIR-NB-LRR gene VICTR (for VARIATION IN COMPOUND TRIGGERED ROOT growth response) determines DFPM-induced root growth arrest. Involvement of additional components of effector-triggered immune signaling in this phenotype implicates pathogen-induced primary root growth arrest as a possible resistance response to soil-borne plant pathogens. Furthermore, we show an association of VICTR protein with EDS1 and with PAD4 within a nuclear complex. The finding of PAD4 within a TIR-NB-LRR–containing protein complex expands recent findings that TIR-NB-LRR R proteins can associate within complexes with the EDS1 protein (Bhattacharjee et al., 2011; Heidrich et al., 2011). The DFPM-induced PAD4- and EDS1-dependent root growth arrest is caused by inhibition of primary root meristem activity. The DFPM-induced and easily scored root growth arrest provides a powerful tool for further dissection of R protein/EDS1/PAD4-dependent signaling mechanisms, as genetic and cell biological components of immunity-induced cell death pathways can be identified, as shown here for VICTR.
RESULTS
Accession-Specific Primary Root Growth Response to the Small Compound DFPM
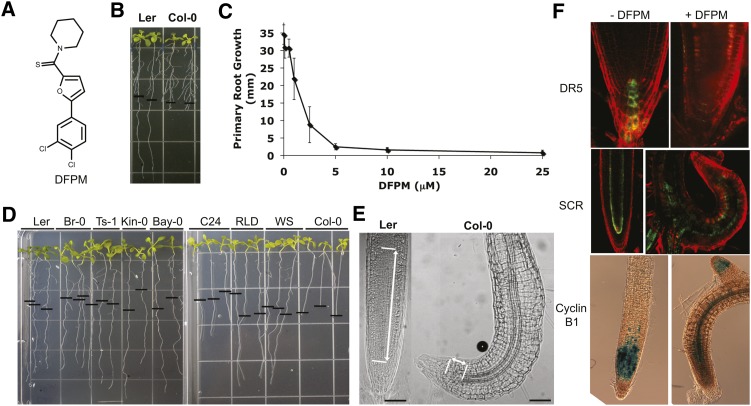
The chemical compound DFPM (Figure 1A) was isolated from a chemical library of 9600 compounds by screening for physiologically active small molecules that inhibit ABA-induced gene expression and signal transduction (Kim et al., 2011). An in-depth screen for additional visible phenotypic responses caused by DFPM application identified a rapid root growth inhibition after DFPM treatment. Intriguingly, this arrest was found to be specific for the Col-0 accession. DFPM did not affect root growth of the Arabidopsis Landsberg erecta (Ler) accession (Figure 1B). At a concentration of 5 μM DFPM, root growth of Col-0 was blocked (EC50 = 1.5 μM; Figure 1C). The seedling root growth of 50 other Arabidopsis accessions, including Ler, Kindalville-0 (Kin-0), Bayreuth-0 (Bay-0), and C24, were not affected by DFPM treatment (Figure 1D; see Supplemental Table 1 online). We concluded that the Col-0 accession contains unusual natural genetic variation that causes DFPM-induced root growth arrest. Based on the phenotype, we refer to this class of genes as VICTR. In response to DFPM, primary root growth of Col-0 was abrogated, but secondary lateral root growth was not (Figure 1B).
Figure 1.
Natural Genetic Variation in Compound-Specific Root Growth Arrest Specifically Targets the Primary Root Meristem in Col-0.
(A) Chemical structure of root growth arrest-causing DFPM.
(B) DFPM causes a primary root growth arrest phenotype in Col-0.
(C) DFPM-induced root growth inhibition of Col-0 (EC50 ∼1.5 μM). Error bars mean ± sd.
(D) While Col-0 roots are sensitive to DFPM, roots of Ler, Br-0 (Brunn-0), Ts-1 (Tossa de Mar-1), Kin-0 (Kindalville-0), Bay-0 (Bayreuth-0), C24, RLD (Rschew), WS (Wassilewskija), and 42 other Arabidopsis accessions (see Supplemental Table 1 online) produced no significant growth arrest response to DFPM. The black horizontal bars mark the starting position of roots when DFPM was applied.
(E) Microscopy examination of DFPM-treated roots reveals that while root differentiation is normal, meristematic and elongation zones of the primary root are reduced in size in response to DFPM in Col-0. Note that Ler produced normal root morphology in response to DFPM exposure (left). Meristematic and elongation zones are indicated by lines and arrows. Bars = 50 μm.
(F) Deregulated DR5 promoter expression by DFPM indicates that auxin accumulation is abrogated in the root meristem after 4 d of DFPM treatment. Expression of the endodermal marker gene SCR is also altered by DFPM. Expression of the CyclinB1 promoter suggests that cell cycle activity in the DFPM-treated primary root tip is reduced. All marker lines tested here are in the Col-0 background.
Chemical compounds structurally similar to DFPM (see Supplemental Figure 1A online) were analyzed to define critical structural motifs required for the triggering of the VICTR phenotype. These analyses showed that modifications in the structure of several DFPM-related compounds can reduce the potency of the DFPM response. Although the slightly modified molecules DFPM-1 and DFPM-2 exhibited biological activity similar to the original DFPM, a chlorine in the four position seemed to be critical for eliciting the VICTR phenotype (see Supplemental Figure 1 online). Loss of activity was also caused by modifications of side groups as shown in DFPM-3. Whether DFPM or a breakdown product thereof is the active compound remains to be determined. The root growth response caused by DFPM was found to be time dependent. Only 8 h of DFPM treatment was sufficient to initiate the VICTR phenotype in an irreversible manner (see Supplemental Figure 2C online). Primary root growth did not recover even 6 d after seedlings were retransferred to standard growth media.
DFPM Targets the Primary Root Meristematic Zone in Col-0
Microscopy examination of the morphological changes of Col-0 roots treated with DFPM revealed a large reduction of the meristematic and elongation zones, whereas root differentiation, such as vascular and lateral root formation, did not show any observable differences, indicating that DFPM may target the primary root meristematic zone (Figure 1E). To investigate molecular changes in DFPM-treated primary root tips, the pattern of local auxin accumulation was examined using DR5 promoter-driven green fluorescent protein (GFP) lines (Friml et al., 2003) in the Col-0 background (Figure 1F). DFPM treatment for 1 d did not change the pDR5:GFP activity at the columella at the root tip (see Supplemental Figure 2A online). After 4 d of DFPM treatment, no pDR5:GFP activity was visible at the columella of Col-0 (Figure 1F), consistent with the model that DFPM targets the primary root meristem. However, the pDR5:GFP construct has been reported to have certain limitations as an auxin reporter and to not necessarily correlate with other auxin-sensitive genes (Moreno-Risueno et al., 2010). Therefore, to measure the effect of DFPM on auxin transporters in the primary root, pPIN1:PIN1-GFP and pPIN2:PIN2-GFP Col-0 lines were monitored as a function of time after DFPM exposure (see Supplemental Figure 2B online). After 18 h of DFPM treatment, both PIN1:GFP and PIN2:GFP signals became weaker. At 24 h, when DFPM-mediated root growth arrest becomes visible, both PIN1:GFP and PIN2:GFP levels decreased substantially. These results indicate that altered PIN1 or PIN2 activities and resulting changes in the auxin gradient, which is important to maintain meristematic cell divisions (Blilou et al., 2005; Dello Ioio et al., 2007; Fernández-Marcos et al., 2011), might contribute to DFPM-mediated root growth inhibition.
SCARECROW (SCR) expression was analyzed as an endodermal and quiescent center marker gene (Di Laurenzio et al., 1996). SCR expression was disrupted after 4 d of DFPM treatment when DFPM had already produced obvious morphological differences (Figure 1F). However, SCR expression exhibited a normal pattern of expression after 1 d of DFPM treatment (see Supplemental Figure 2A online). Therefore, the disrupted SCR expression in Col-0 might be a secondary effect of the altered root morphology after DFPM treatment.
Consistent with the deregulated DR5 and SCR expression patterns induced after 4 d by DFPM, cell cycle activity monitored by the Cyclin B1 promoter (Colón-Carmona et al., 1999) was disrupted by DFPM treatment (Figure 1F). Retention of the normal expression pattern of Cyclin B1 in secondary root meristems demonstrates the tissue specificity of the DFPM effect (Figure 1F). These marker line analyses suggesting that DFPM targets the primary root meristematic zone led us to further explore how DFPM mediates root growth arrest (described further below).
The VICTR Gene Encodes a TIR-NB-LRR Protein
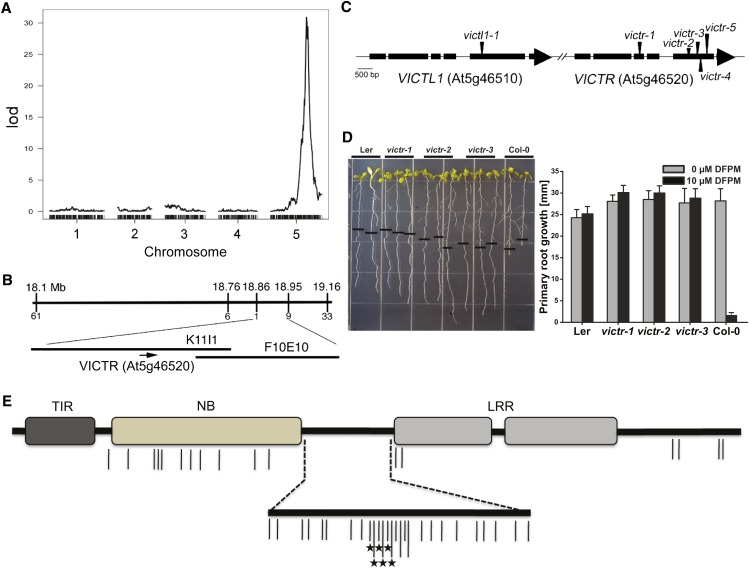
To identify the genetic element(s) in Col-0 producing the VICTR phenotype, 100 Col × Ler recombinant inbred lines (RILs) (Clarke et al., 1995) that have been genotyped using microarrays (Singer et al., 2006) were screened for the VICTR phenotype (see Supplemental Table 2 online). Quantitative trait locus (QTL) analyses of the phenotypic scores and available genetic map information for these RILs suggested that a single locus in Col-0 positioned on the lower arm of chromosome 5 might determine DFPM-induced root arrest (Figure 2A).
Figure 2.
Identification of the Natural Variation VICTR Locus.
(A) QTL analyses using the 100 Col × Ler RILs identified a single locus on chromosome 5 as a main element that triggers the VICTR phenotype in Col-0. lod, logarithm of odds.
(B) Map-based cloning using F2 segregants of Col-0 crosses to Ler narrowed down the natural mutation VICTRCol within a 100-kb region.
(C) Gene structure of VICTRCol and VICTL1 and the locations of T-DNA insertion mutants.
(D) T-DNA insertion mutants of VICTRCol showed complete resistance to DFPM in root growth assays. The black horizontal bars mark the starting position of roots when DFPM was applied. Error bars mean ± sd (n = 3 with 10 plants each).
(E) Protein motif structure of VICTR. Amino acid sequence comparison deduced from the cDNAs of Col-0, Bay-0, and Kin-0 identified multiple Col-0–specific amino acid polymorphisms within the coding region of VICTRCol. Vertical lines indicate positions where the Col-0 sequences differ from Bay-0 and Kin-0. It is important to note that Bay-0 and Kin-0 sequences are identical in these positions. Protein sequences other than the TIR, NB, and LRR domains are depicted by black horizontal bars. Asterisks mark amino acid deletions. Most amino acid polymorphisms are found in the NB domain (13 polymorphisms/44 total) and the linker regions between the NB and LRR domains (24 polymorphisms/44 total).
[See online article for color version of this figure.]
F1 seedlings of Col-0 crosses to Ler produced the Col-0 VICTR phenotype upon DFPM treatment, indicating that heterozygous expression of the VICTRCol allele is sufficient to confer the DFPM response in the Ler background. Consistent with QTL analyses of the Col-0 × Ler RILs, F2 seedlings of a Col-0 cross to Ler segregated at a ratio of 3:1 (P value of χ2 test is 0.665, n = 178), indicating that a single locus mediates the VICTR phenotype. Using these segregating VICTR F2 plants, map-based cloning narrowed down the candidate VICTR locus to a 100-kb region (Figures 2A and 2B). Since VICTR is a natural variant present in Col-0, any polymorphic gene within the target region could encode VICTR. Because VICTRCol was necessary and sufficient for the DFPM response and was hypothesized to represent an active allele, T-DNA insertion mutants of genes located within the target region in Col-0 were examined. Among the T-DNA lines tested, only T-DNA insertion lines in gene At5g46520 displayed complete insensitivity in DFPM-mediated root growth arrest (Figures 2C and 2D). Five independent T-DNA insertion mutants in the Col-0 At5g46520 gene showed loss of DFPM-triggered root arrest.
DFPM inhibits primary root growth in Col-0 but not in the victr mutants. To determine how DFPM mediates this effect, confocal images of Col-0, victr-1, and victr-2 primary roots were analyzed (see Supplemental Figure 3 online). While the quiescent center cells and cortical stem cells appeared unaffected by 24 h of DFPM treatment in all three genotypes, the division zone of the DFPM-treated Col-0 primary roots was smaller than that of the victr-1 and victr-2 primary roots. To further analyze this phenotype, we quantified the number of meristematic cells in the division zones of primary roots (see Supplemental Figure 3 online). Our data indicate that when treated with DFPM for 24 h, Col-0 primary roots show a reduced number of meristem cells in the division zone of the primary root compared with victr-1 and victr-2. This shorter division zone and the reduced number of meristematic cells are consistent with the inhibition of primary root growth first observable after 24 h of DFPM treatment in Col-0 wild type and may represent the initial phase of DFPM-induced growth inhibition.
The most similar gene to VICTR, VICTR Like1 (VICTL1; At5g46510), is located in tandem upstream of VICTR (Figure 2C; see Supplemental Figure 4B and Supplemental Data Set 1 online). A T-DNA mutant victl1-1 was not resistant to DFPM treatment in the root growth arrest response (see Supplemental Figure 4C online). This suggests that the VICTRCol (At5g46520) gene is required for the chemical DFPM response targeting primary root meristem activity. Chemical genetics can provide an approach for identifying phenotypes in potentially redundant signal transduction genes via small molecule targeting of specific proteins (Park et al., 2009).
VICTR is predicted to encode a previously uncharacterized member of the NB-LRR family with a TIR motif (TIR-NB-LRR) at the N terminus (Figure 2E). TIR-NB-LRR family members can function as immune signaling proteins encoded by R genes responding to specific pathogen effectors in plants (Belkhadir et al., 2004; Chisholm et al., 2006; Jones and Dangl, 2006; Shen and Schulze-Lefert, 2007; Caplan et al., 2008). Compared with characterized R proteins, the predicted VICTRCol protein shows high sequence similarity to that of At1g31540 (64.9% amino acid identity) (Katoh et al., 2002; Thompson et al., 2003; Raghava and Barton, 2006), the Col-0 allele of RAC1 from accession Ksk-1, which mediates PAD4-independent resistance to Albugo candida (Borhan et al., 2004). VICTRCol is in the same cluster of R genes as RPS6 (At5g46470; see Supplemental Figure 4B online) that specifies resistance to the Pseudomonas syringae effector HopA1 (Kim et al., 2009).
To determine whether VICTRCol contains unique polymorphism(s) that define the Col-0 sensitivity to DFPM, VICTR cDNA sequences from other accessions were reverse transcribed and compared with VICTRCol. The VICTR gene is highly diverged and possibly absent in Ler (Ziolkowski et al., 2009) and also in Wassilewskija-0 (Ws-0) based on PCR analysis (see Supplemental Figure 4E online). Comparison of the deduced amino acid sequences in the VICTR-containing ecotypes, Bay-0 and Kin-0, shows that VICTRCol has multiple unique amino acid polymorphisms centered in the NB domain and the linker region between the NB and LRR domains (Figure 2E; see Supplemental Figure 5A and Supplemental Data Set 2 online). The promoter sequences of the VICTR genes are highly conserved (see Supplemental Figure 5B and Supplemental Data Set 2 online).
VICTR Expression in DFPM-Insensitive Accessions Generates Col DFPM Responses
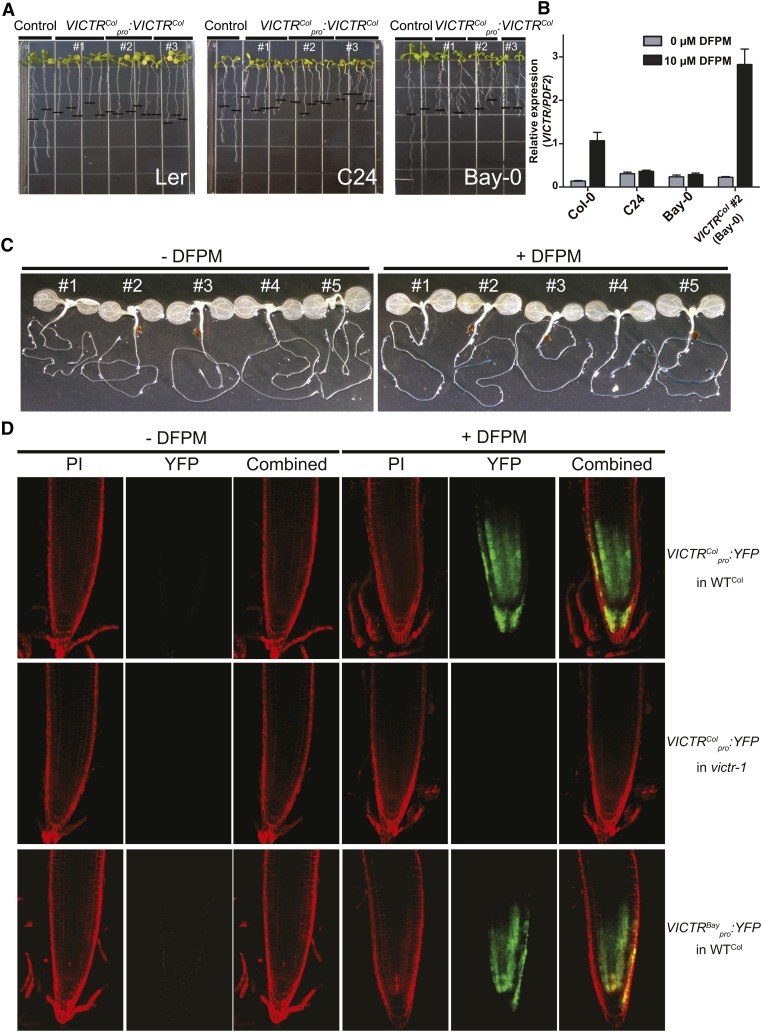
To further establish whether DFPM-induced primary root growth inhibition in Col-0 is dependent on polymorphisms present in the coding region of VICTRCol, the VICTRCol gene expressed under the control of its natural Col-0 promoter was introduced into accessions insensitive to DFPM: Ler, C24, and Bay-0. The introduction of VICTRCol was sufficient to generate the DFPM response in primary roots of Ler, C24, and Bay-0, demonstrating that VICTRCol is the only missing component required for the DFPM-induced primary root growth arrest in these accessions (Figure 3A). As reported for other R proteins, ectopic 35S:VICTRCol expression in the Col-0 background caused stunted plant growth (see Supplemental Figure 1D online).
Figure 3.
VICTRCol Expression Is Sufficient for Generation of the DFPM-Induced Root Meristem Arrest in the DFPM-Resistant Ecotypes Ler, C24, and Bay-0.
(A) Expression of the VICTRCol gene under the control of the VICTRCol promoter in Ler (left panel), C24 (middle), and Bay-0 (right) reproduced the Col-0 phenotype in response to DFPM. Three independent transgenic lines are shown together with a control transgenic line transformed with empty vector. The black horizontal bars mark the starting position of roots when DFPM was applied.
(B) DFPM-sensitive Arabidopsis lines show induction of VICTR expression by DFPM treatment: five biological replicates for Col-0 and Bay-0 and three for C24 and VICTRCol #2 (Bay-0).
(C) VICTRCol promoter-driven GUS reporter lines in Col-0 background showed tissue-specific and DFPM-dependent induction of VICTR gene expression. Five independent transgenic reporter lines are presented.
(D) VICTRCol promoter-driven YFP reporter line confirms the root tip–specific expression of VICTR after DFPM treatment. No induction of the VICTRCol promoter in victr-1 indicated that VICTRCol is required for the transcriptional DFPM induction of the VICTRCol promoter. The VICTRBay promoter drove YFP reporter expression similarly to the VICTRCol promoter. WT, the wild type. PI, propidium iodide.
VICTR Expression Is Increased in the Root Meristematic Zone in a DFPM- and VICTR-Dependent Manner
Resistance genes are generally expressed ubiquitously and at very low levels in unchallenged plants. Electronic Arabidopsis gene expression resources, including AtGenExpress and eFP Browser indicate that VICTR is expressed at a low level in many different tissue types (Schmid et al., 2005; Winter et al., 2007). Interestingly, when treated with DFPM for 24 h, VICTR was highly induced (eightfold) in the Col-0 background (Figure 3B). Bay-0 and C24, which carry a polymorphic VICTR gene relative to VICTRCol but are not sensitive to DFPM, did not exhibit enhancement of VICTR expression in response to DFPM treatment (Figure 3B). More detailed analyses of VICTRCol promoter driven-GUS (for β-glucuronidase) reporter and yellow fluorescent protein (YFP) reporter lines showed dramatic DFPM-enhanced expression of the VICTR gene in the root meristematic zone (Figures 3C and 3D). These results suggest that the DFPM-induced root growth arrest may be caused by differential spatial upregulation of VICTR expression in the primary root tip region.
Notably, induction of the VICTR gene by DFPM treatment requires functional VICTRCol because DFPM could not induce the VICTRCol promoter in the victr-1 mutant (Figure 3D, middle panels). On the other hand, the promoter of VICTRBay, whose sequence is highly similar to that of VICTRCol (see Supplemental Figure 5B and Supplemental Data Set 2 online), activated YFP expression by DFPM treatment when introduced into the Col-0 wild type (Figure 3D, bottom panels). Altogether, these results suggest that functional VICTR mediates DFPM-induced positive feedback regulation of VICTR expression and that amino acid polymorphism(s) in VICTRCol are responsible for the VICTR phenotype. Consistent with this, Bay-0 expressing VICTRCol under the VICTRCol promoter exhibited DFPM-mediated root growth arrest (Figure 3A) and also increased VICTR expression upon DFPM treatment (Figure 3B). Such positive feedback regulation has been observed for R genes and is part of the resistance response induced by corresponding avirulent pathogens (Zhang and Gassmann, 2007).
VICTR Requires Effector-Triggered Immune Signaling Components for the DFPM Response
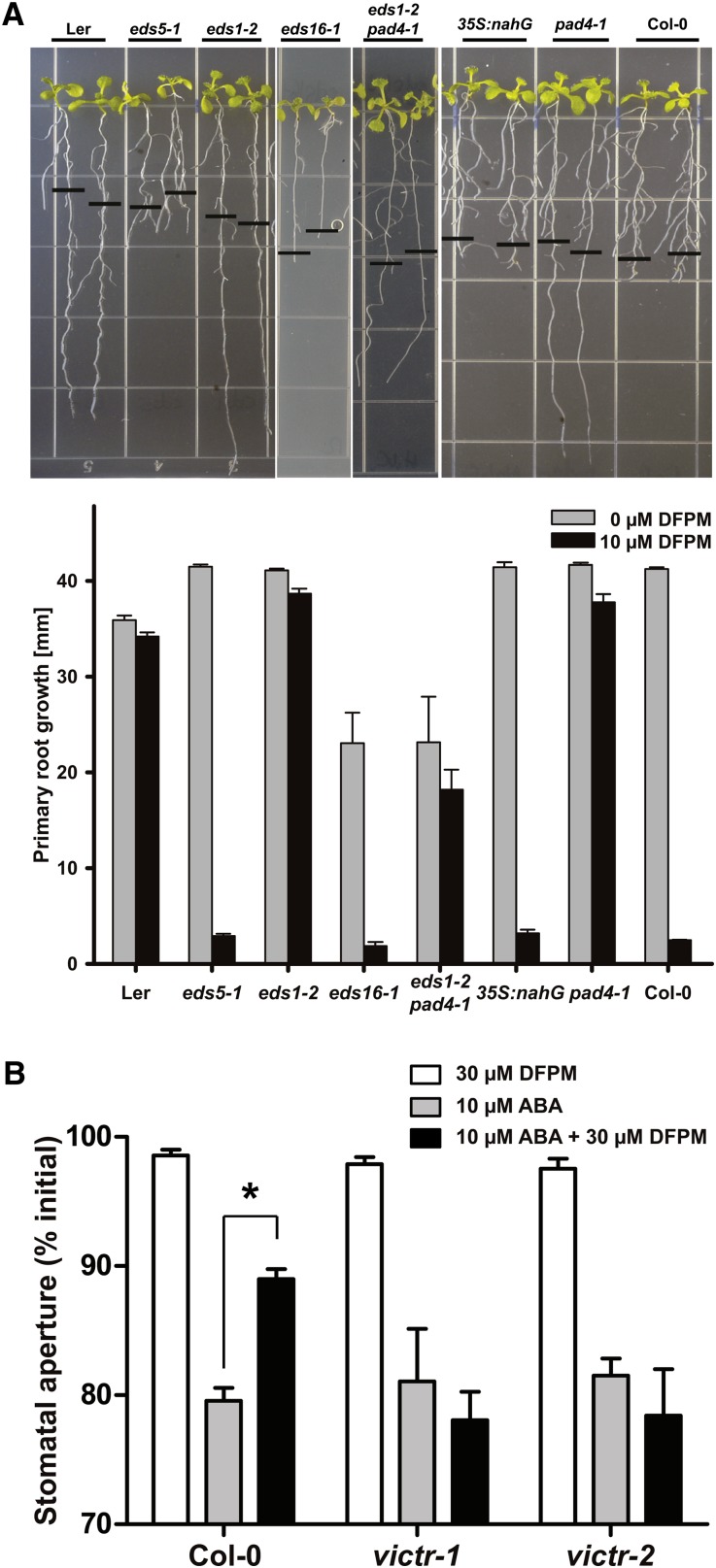
The finding that VICTR encodes a TIR-NB-LRR family R-like protein and previous indications that DFPM induces defense-related gene expression (Kim et al., 2011) raise the hypothesis that pathogen-triggered biotic stress signaling and/or salicylic acid (SA) signaling is responsible for generation of the VICTR root growth arrest phenotype. To test this hypothesis and to identify additional genetic components of the DFPM-mediated signaling pathway, pathogen mutants in Col-0 were analyzed to determine whether they exhibit altered root growth responses to DFPM. Among the mutants analyzed, eds1-2 (Bartsch et al., 2006), eds1-23, and pad4-1 (Glazebrook et al., 1997) mutations disrupted the DFPM-induced root growth arrest, as did the eds1-2 pad4-1 double mutant (Figure 4A; see Supplemental Figure 4A online). No DFPM resistance was observed in eds5-1 and eds16-1.
Figure 4.
DFPM-Induced Signal Transduction Requires Pathogen Signaling Components.
(A) Chemical signaling by VICTRCol is dependent on EDS1 and PAD4; thus, eds1-2 and pad4-1 mutants were insensitive to DFPM, in contrast with eds5-1 and 35S:nahG. The black horizontal bars mark the starting position of roots when DFPM was applied. Error bars show ± sd (n = 3 with 10 plants each).
(B) VICTR functions in DFPM inhibition of ABA-induced stomatal closure. Guard cells of victr-1 and victr-2 mutants remained sensitive to ABA even in the presence of DFPM. n = 3 experiments with 42 to 50 stomata analyzed per condition. Error bars are se. *P < 0.01.
[See online article for color version of this figure.]
EDS1 and PAD4 are also required for DFPM inhibition of ABA-induced stomatal closure (Kim et al., 2011). Therefore, victr-1 and victr-2 mutants were analyzed for the ability of DFPM to impair ABA-induced stomatal closing in genotype blind assays. Both victr-1 and victr-2 plants showed impairment in DFPM inhibition of ABA-induced stomatal closing (Figure 4B).
VICTR Subcellular Localization in Transiently Transformed Cells
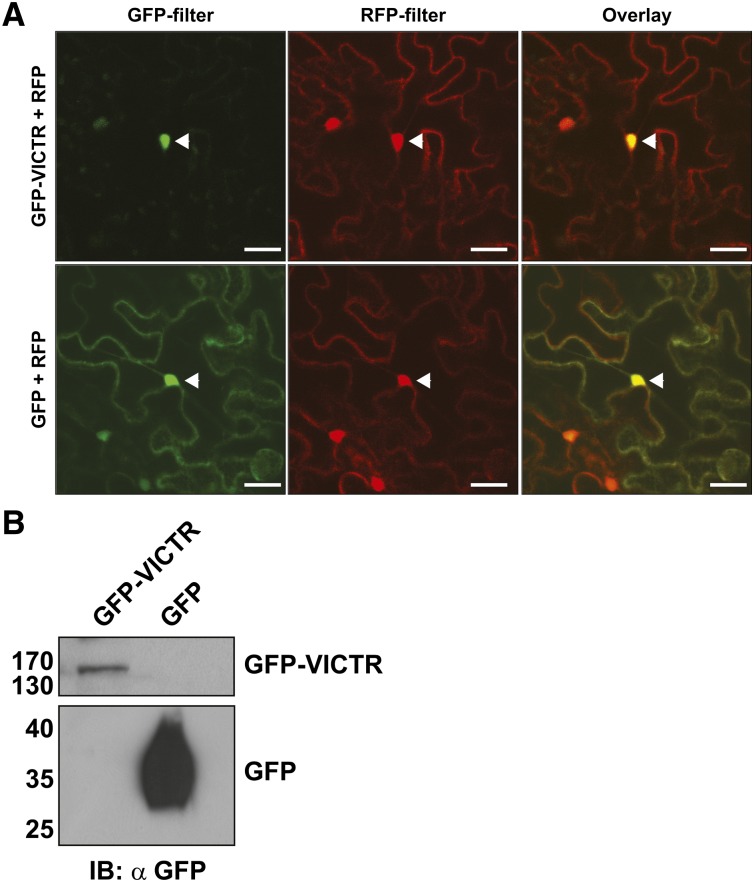
To investigate the subcellular localization of VICTR, YFP or GFP fluorophores were fused to either its N or C terminus. All constructs showed that VICTRCol localized to the nucleus with localization also in the cytoplasm as indicated by faint fluorescence in transient onion (Allium cepa) epidermis expression assays (see Supplemental Figure 4D online). Arabidopsis TIR-NB-LRR proteins, such as RPS4 and RPS6, also display nucleocytoplasmic distributions (Wirthmueller et al., 2007; Kim et al., 2009). In transient Agrobacterium tumefaciens–mediated overexpression and immunological detection of the VICTR constructs in Nicotiana benthamiana plants, VICTR fusion protein levels were low but detectable based on GFP fluorescence and immunoblots (Figure 5). In N. benthamiana expression analyses, we coexpressed red fluorescent protein (RFP) constructs, allowing us to clearly identify transformed cells, visualize nuclei, and perform colocalization analyses. The GFP-VICTR fusion protein was preferentially nuclear localized, and GFP-VICTR fluorescence merged with nuclear RFP (Figure 5A). Although >70% of cells showed RFP expression, GFP-VICTR was detected in only 15 to 20% of transformed cells. Immunoblot analyses identified GFP-VICTR fusions at the expected size (∼160 kD), suggesting that the detected fluorescence is not due to either GFP alone or GFP fused to truncated VICTR (Figure 5B).
Figure 5.
VICTR Localizes to the Nucleus in Plant Cells.
(A) Agrobacterium strains expressing GFP-VICTR fusions or GFP alone were coinfiltrated with RFP-expressing strains in N. benthamiana leaves. Two days after infiltration, leaf sections were analyzed with a confocal microscope. The left and the middle panels show GFP and RFP fluorescence, respectively. The right panels show an overlap of GFP and RFP fluorescence. Arrowheads point out the nucleus. The assay was repeated with similar localizations. Bars = 20 μm.
(B) Immunoblots (IB) detecting GFP-VICTR in N. benthamiana extracts and showing absence of free GFP or degradation products in GFP-VICTR sample.
VICTR Resides in Protein Complexes with Nuclear EDS1 and PAD4
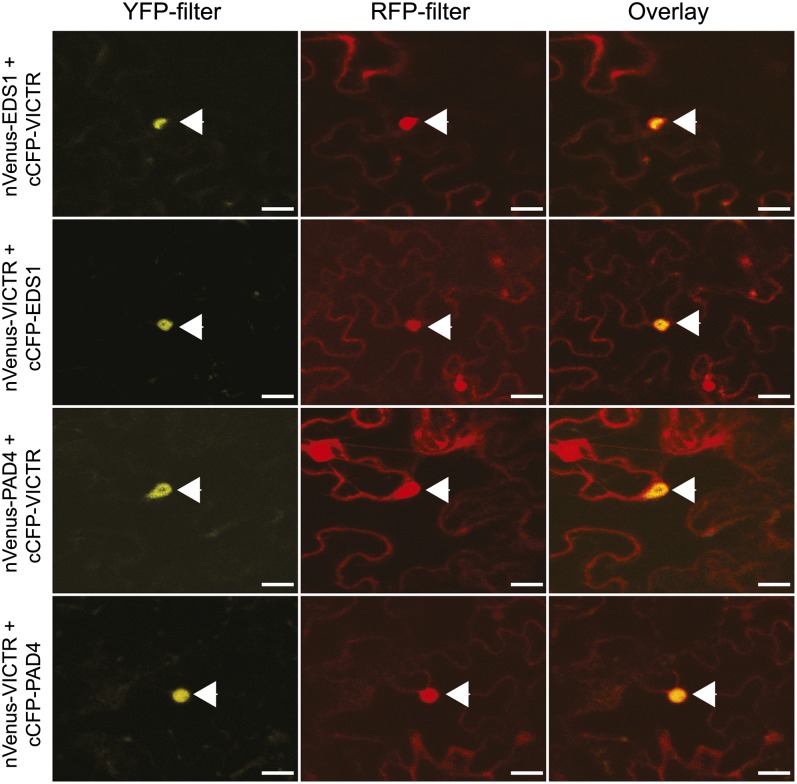
The requirement for EDS1 and PAD4 in DFPM-dependent root inhibition prompted us to investigate in vivo protein complex associations of VICTR with these proteins. Recent studies have shown that the TIR-NB-LRR resistance proteins RPS4 and RPS6 coreside in protein complexes with EDS1 (Bhattacharjee et al., 2011; Heidrich et al., 2011). Pathogen effectors that might signal VICTR-dependent primary root growth arrest are presently not known and effectors that cause root growth arrest have not yet been determined. Such putative effectors may also redundantly use the tandem repeat homolog VICTL1. Identification of molecular interactors of VICTR is relevant toward understanding VICTR protein functions. Bimolecular fluorescence complementation (BiFC), an efficient tool to study in vivo protein–protein interactions, can be used to identify molecular complexes of key innate immunity components (Bhattacharjee et al., 2011). An inherent feature of this assay is its irreversibility, allowing capture of transient interactions. Confocal microscopy analysis of transiently expressed BiFC fusion combinations of VICTR with PAD4 or EDS1 in N. benthamiana cells suggested nuclear complexes of VICTR with these two proteins (Figure 6). VICTR did not show associations with the reciprocal BiFC vectors expressing GUS that also accumulated inside nuclei (see Supplemental Figure 6A online). Heterodimers of EDS1-PAD4 display nucleocytoplasmic distributions (Feys et al., 2005). However, our BiFC results cannot distinguish whether VICTR associates with EDS1-PAD4 heterodimer-including complexes or within protein complexes individually with EDS1 or PAD4 proteins, which warrants further investigations. Our results suggest cocomplex formation of a TIR-NB-LRR protein with the EDS1 signaling partner PAD4. This is in marked contrast with the TIR-NB-LRR R proteins RPS4 and RPS6, which did not show an interaction with PAD4 in transient expression assays (Bhattacharjee et al., 2011).
Figure 6.
VICTR Associates with Nuclear Pool of EDS1 and PAD4.
Confocal analysis of N. benthamiana leaves agroinfiltrated with BiFC vector combinations of VICTR with EDS1 or PAD4. These vector combinations use the N-terminal fragment of an enhanced version of YFP (nVenus) and the C-terminal fragment of CFP (cCFP), which when reconstituted gives yellow fluorescence (Hu and Kerppola, 2003). Localization of indicated molecular complexes is shown by yellow fluorescence. Red fluorescence indicates free RFP. Panels show images with separate YFP, RFP, and an overlap of YFP + RFP fluorescence. Arrowheads indicate the nucleus. The experiment was repeated with identical results (n = 3). Bars = 20 μm.
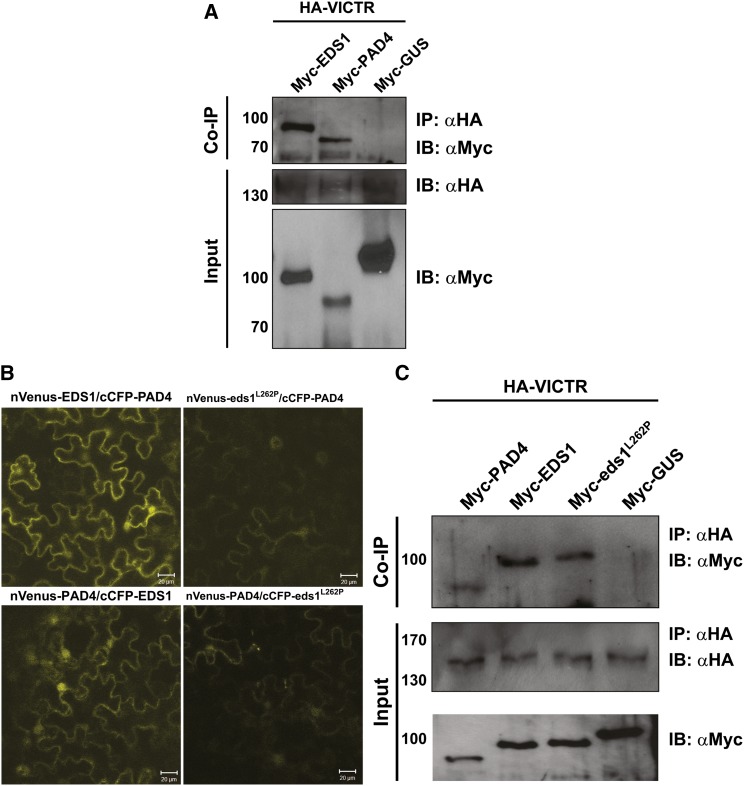
We independently investigated the above results by performing coimmunoprecipitation (co-IP) assays on transiently transformed N. benthamiana cells (Figure 7A). N-terminally hemagglutinin (HA) epitope-tagged VICTR (HA-VICTR) was coexpressed either with cMyc-tagged-EDS1, -PAD4 or -GUS. Because VICTR expression was enriched in the nucleus, we isolated purified nuclei from the infiltrated leaf tissues and performed nuclear co-IP assays. Both EDS1 and PAD4, but not GUS, coimmunoprecipitated with VICTR (Figure 7A and 7C). These results validated the associations of VICTR with EDS1 and PAD4 proteins in nuclei detected by BiFC (Figure 6).
Figure 7.
VICTR Associates in Planta with EDS1, PAD4, and eds1L262P.
(A) Myc-tagged EDS1, PAD4, and GUS were transiently coexpressed with HA-VICTR in N. benthamiana leaves. Purified nuclear extracts were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies. Top panel shows the pull-down of PAD4 and EDS1, but not GUS, with VICTR. Bottom two panels show input fractions (n = 3).
(B) eds1L262P does not strongly interact with PAD4 in BiFC assays. Reciprocal BiFC assays of PAD4 with wild-type EDS1 and eds1L262P were performed by Agrobacterium-mediated transient expression in N. benthamiana. Reconstituted YFP is indicated by yellow fluorescence. Bar values are indicated.
(C) VICTR interacts with eds1L262P. Co-IP was performed as described above. Top panel is the co-IP of PAD4, EDS1, and eds1L262P, but not GUS, with VICTR. Bottom two panels show the input fractions. HA-VICTR was detected only after immunoprecipitation with anti-HA antibodies. Migration and sizes (in kilodaltons) of molecular mass standards are at the left of the respective immunoblot panels. Each experiment was performed twice with similar results.
Using a mutated EDS1, eds1L262P, that does not heterodimerize with PAD4 (Rietz et al., 2011), we tested whether VICTR associates with EDS1-PAD4 heterodimers or individually with EDS1. As expected, the eds1L262P mutant showed a strongly diminished interaction with PAD4 in transient N. benthamiana BiFC assays (Figure 7B). We then performed co-IP assays on transiently transformed N. benthamiana cells (Figure 7C). N-terminally HA epitope-tagged VICTR (HA-VICTR) was coexpressed either with cMyc-tagged PAD4, -EDS1, -eds1L262P, or -GUS. Purified nuclei from the infiltrated leaf tissues were isolated and nuclear co-IP assays performed. EDS1 wild-type, eds1L262P, and PAD4, but not GUS, coimmunoprecipitated with VICTR (Figure 7C), supporting that VICTR can form complexes with EDS1 and PAD4 inside nuclei.
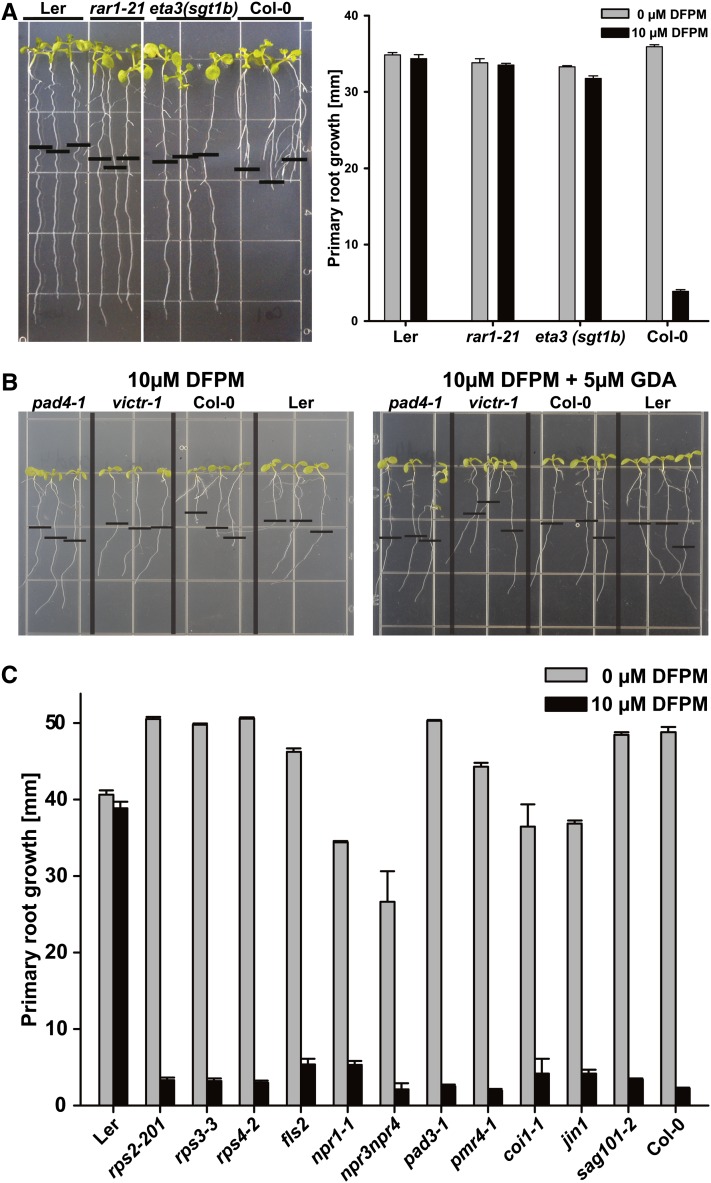
Senescence Associated Gene101 (SAG101) has also been shown to form a protein complex with EDS1 (Feys et al., 2005; Rietz et al., 2011). As eds1-2 and pad4-1 loss-of-function mutants showed a strong DFPM insensitivity, we tested sag101-2 loss-of-function mutants (Feys et al., 2005) for the DFPM-induced root growth arrest response. In contrast with eds1-2, pad4-1, and eds1-2 pad4-1, sag101-2 mutant plants behaved like Col-0 wild-type controls (Figure 8C). Thus, we propose that DFPM may trigger root growth inhibition by activating an SAG101-independent branch of EDS1-PAD4 signaling.
Figure 8.
The DFPM-Induced Signal Transduction Relies on HSP90 and Its Cochaperones but Does Not Require SAG101, SA, and JA Signaling Components.
(A) Chemical signaling by VICTRCol requires functional HSP90 cochaperones RAR1 and SGT1B.
(B) The DFPM signal was blocked by applying additional 5 μM of the HSP90 inhibitor GDA.
(C) Other signaling and response components known to function in pathogen, SA, and JA signal transduction or other NB-LRR genes are not required for the VICTR phenotype. All mutants were tested in Col-0 background. Error bars mean ± sd. Note, some error bars are not visible due to very small sd (n = 3 with 10 plants each).
[See online article for color version of this figure.]
HSP90 Heat Shock Proteins and Their Cochaperones Are Crucial for DFPM Signaling
The HSP90 chaperones functionally stabilize and maintain R proteins in a signaling-competent state, and their activity is dependent on at least two different cochaperones, RAR1 and SGT1B (Kadota et al., 2010). In root assays, the single mutants rar1-21 (Tornero et al., 2002) and sgt1b/eta3 (Gray et al., 2003) exhibited DFPM insensitivity (Figure 8A). Since the Arabidopsis genome contains four cytosolic HSP90 isoforms, with three located in close proximity within a 12-kb region on chromosome 5 (Sangster et al., 2007), the effect of the HSP90 inhibitor geldanamycin (GDA) on the DFPM response was analyzed. GDA inhibits cancer cell proliferation by irreversibly binding to the N terminus of HSP90 proteins (Stebbins et al., 1997). A concentration of 5 µM GDA was sufficient to completely abolish Col-0 specific DFPM root growth inhibition (Figure 8B), consistent with the observed DFPM resistance of rar1-21 and sgt1b/eta3 (Figure 8A). This suggests that HSP90 cooperates with RAR1 and SGT1B to stabilize VICTR as a prerequisite for the DFPM root growth response.
DFPM Response Does Not Require Jasmonic Acid or SA Signaling
The eds5-1 mutant defective in SA biosynthesis (Rogers and Ausubel, 1997) and a SA-depleted 35S:nahG transgenic line (Reuber et al., 1998) were not affected in the DFPM-induced root growth response (Figure 4A). With the exception of the resistance signaling components described above, other known pathogen response loci were also not required for the DFPM-induced root growth arrest. These include the NB-LRR genes RPS2, RPM1, and RPS4, immune signaling and response components NPR1, NPR3-NPR4, PAD3, and PMR4, and a pathogen-associated molecular pattern (PAMP) receptor, FLS2, as well as the jasmonic acid (JA) signaling components COI1 and JAI1/JIN1 (Lorenzo et al., 2004; Zhang et al., 2006; Fonseca et al., 2009; Sheard et al., 2010; Fu et al., 2012) (Figure 8C). Although application of exogenous SA also caused root growth inhibition (see Supplemental Figure 7A online), detailed inspection showed that the SA effect on root growth inhibition is morphologically clearly distinct from the DFPM-induced root growth arrest in several respects. SA blocks general growth of Arabidopsis seedlings (Zhang et al., 2003a), including root elongation, lateral root initiation, and root hair formation (see Supplemental Figure 7B online), in contrast with DFPM (e.g., Figure 4A; see Supplemental Figure 2B online). Also, the SA-induced root growth inhibition is not accession specific because the response to SA was equivalent in Col-0 and Ler.
DISCUSSION
We identified genetic natural variation in the VICTR locus that produces root growth arrest in response to the small molecule DFPM. We report that DFPM perception and signal transduction requires early components of the plant R gene resistance signaling network, including the TIR-NB-LRR protein VICTRCol, EDS1, and PAD4. This response may also involve VICTR stabilization by HSP90 together with RAR1 and SGT1B but operates independently of major defense signaling components in the JA and SA signal transduction pathways. VICTRCol is also required for DFPM-induced inhibition of ABA-induced stomatal closing, further showing that early TIR-NB-LRR signaling rapidly interferes with ABA signal transduction. We further establish in transient protein expression assays that VICTR resides within nuclear protein complexes with EDS1 and PAD4. By contrast, the nucleocytoplasmic TIR-NB-LRR receptors RPS4 and RPS6 were found only in complexes with EDS1 and not PAD4 (Bhattacharjee et al., 2011; Heidrich et al., 2011). Thus, different types of TIR-NB-LRR receptors may operate in distinct resistance signaling complexes containing EDS1 and/or PAD4. Previously, it was shown that the eds1L262P mutant, which does not interact with PAD4 (Figure 7B), has defects in basal immunity to virulent pathogens (Rietz et al., 2011). The retention of a co-IP signal between VICTR and eds1L262P protein in transient expression assays (Figure 7C) suggests that this mutation may not be crucial for TIR-NB-LRR protein–EDS1 complex formation.
A requirement for VICTR interactors EDS1 and PAD4 in the DFPM-induced response and microarray analyses support the notion that a plant effector-triggered immunity signaling pathway (Glazebrook, 2005; Wiermer et al., 2005; Kim et al., 2011) mediates the DFPM response (Figure 4A). Furthermore, the requirement of RAR1 and SGT1B and HSP90 chaperones suggests that VICTR steady state accumulation and possibly also chaperone-mediated conformational changes are important for DFPM-triggered VICTR signaling (Shirasu, 2009) (Figures 8A and 8B). However, the lack of requirement for other analyzed defense signaling elements, including key components of the SA-signaling and JA-signaling pathways (Lorenzo et al., 2004; Zhang et al., 2006; Fu et al., 2012), suggests that a SA/JA-independent disease signaling network participates in the DFPM-induced root growth arrest (Figures 4A and 8C). Since PIN1:GFP and PIN2:GFP expression was decreased after 18 h of DFPM treatment, an involvement of auxin cannot be ruled out and warrants further analysis. Auxin binding–specific fluorescence reporters (Brunoud et al., 2012) with a rapid readout may be required to unequivocally resolve this question.
DFPM may activate TIR-NB-LRR–mediated signal transduction by mimicking a pathogen effector (Belkhadir et al., 2004; Jones and Dangl, 2006; Caplan et al., 2008), analogous to the organophosphate insecticide fenthion that triggers an immune-like response via the kinase Fen and the NB-LRR protein Prf in tomato (Solanum lycopersicum; Pedley and Martin, 2003). Activation of effector-triggered immunity can promote infection by necrotrophic pathogens that feed off dead plant tissue (Glazebrook, 2005). Interestingly, certain necrotrophic fungal pathogens secrete small toxin molecules (such as victorin produced by Cochliobolus victoriae), which are sensed by NB-LRR proteins (Sweat and Wolpert, 2007; Lorang et al., 2012). When primary roots are challenged by soil-borne pathogens, localized VICTR responses may limit damage to the root and function in restriction of primary root growth, thereby protecting roots from further infection. Given the homology of VICTR and its tandem gene VICTL1, biological responses may be mediated redundantly via either of these two proteins. Chemical genetics provides a powerful approach for identification of redundant signaling genes and their linked phenotypes by protein-specific selective activation, as in the case of the discovery of the redundant PYR1 ABA receptor (Park et al., 2009). Whether VICTR and VICTL1 function in concert in biologically induced root growth arrest will require identification of natural effectors that stimulate this root response in Arabidopsis.
DFPM was identified as a small molecule that rapidly inhibits ABA responses (Kim et al., 2011). DFPM interference with ABA signal transduction also required PAD4, EDS1, RAR1, and SGT1B but not SA or JA signal transduction components (Kim et al., 2011). As shown in stomatal movement analyses, DFPM inhibition of ABA-induced stomatal closing is impaired in victr T-DNA insertion mutant stomata (Figure 4B), supporting the previous model that DFPM mediates inhibition of ABA signal transduction via early effector-triggered immune signaling (Kim et al., 2011). A requirement for VICTR in this response is consistent with data sets showing VICTR expression in leaves (Schmid et al., 2005; Winter et al., 2007).
TIR-NB-LRR genes mainly function in resistance against pathogens. Retention of nonfunctional R gene alleles and a statistically significant increase in the occurrence of R gene polymorphisms is hypothesized to be an outcome of fitness tradeoffs in evolving a large array of recognition specificities in a plant population (Tian et al., 2003; Weigel, 2012). An additional consequence of natural variants in NB-LRR genes can be environmentally conditioned hybrid necrosis (Bomblies et al., 2007; Alcázar et al., 2009). TIR-NB-LRR–mediated root growth arrest has not been reported previously. Here, we show that DFPM inhibits root growth via VICTRCol in an EDS1/PAD4/RAR1/SGT1B/HSP90-dependent manner. These data suggest a direct link between R gene–related signaling and root growth arrest. DFPM causes a rapid root growth arrest along with a decreased cell division rate in the primary root meristematic zone in Col-0. However, the quiescent center cells and cortical stem cells appear unaffected after 24 h DFPM treatment in both resistant and sensitive genotypes, a time point at which initial root growth inhibition becomes visible. These findings and the DFPM-induced VICTR promoter activity in wild-type plants suggest a role for VICTR protein levels in determining this interesting root meristem-localized EDS1- and PAD4-dependent response. As described for delocalized or overexpressed R proteins (Zhang et al., 2003b; Yi and Richards, 2007; Kim et al., 2010), ectopic VICTR expression also leads to stunted growth phenotypes.
Identification of the small molecule DFPM as a trigger of an effector-triggered immune-related signaling pathway that causes a strong visual and quantifiable primary root growth arrest can provide a powerful tool for further genetic dissection of R gene–mediated signaling. This is demonstrated here by the identification of the VICTR gene and findings showing association of PAD4 and EDS1 within VICTR protein complexes. Also, genetic variation in this small molecule response in Arabidopsis accessions is consistent with the hypothesis that the repertoire of TIR-NB-LRR genes provides activation mechanisms to diverse cues and here via a root meristem-targeted response.
METHODS
Chemicals and Plant Materials
DFPM (ID 6015316) was isolated from screening of 9600 compounds in ChemBridge’s DIVERSet E library. DFPM (ID 6015316), DFPM-1 (ID 5649754), DFPM-2 (ID 5872683), DFPM-3 (ID 6881095), DFPM-5 (ID 6017927), and DFPM-7 (ID 5143713) were purchased from ChemBridge. Transgenic reporter lines were in the Col-0 background or were crossed into the Col-0 background. Arabidopsis thaliana T-DNA lines were obtained from the ABRC (Ohio State University).
Arabidopsis ecotype set CS22660, victr-1 (Salk_123918), victr-2 (Salk_084068), victr-3 (Salk_072727), victr-4 (Salk_122941), victr-5 (Sail_394_F01), victl1-1 (Salk_097845), eds1-23 (Salk_057149), and Caroline Dean’s 100 Col × Ler RILs (CS1899) (Clarke et al., 1995; Singer et al., 2006) were obtained from the ABRC (Ohio State University). The eds1-23 T-DNA mutant (Salk_057149) has a T-DNA insertion in At3g48090, one of a tandem pair of EDS1 homologs in accession Col-0. Another T-DNA insertion mutant (Salk_071051) in the same gene, denoted eds1-22, substantially rescues the growth phenotype of bon1-1 constitutive resistance plants (Yang and Hua, 2004). pDR5:GFP, pPIN1:PIN1:GFP, and pPIN2:PIN2:GFP were kindly provided by Yunde Zhao (University of California at San Diego). pSCR:GFP and pCyclinB1:GUS were kindly provided by Jeff Long (Salk Institute) and Jeff Harper (Univ. of Nevada), respectively. Mutants eds1-2 (Bartsch et al., 2006) and sag101-2 (Feys et al., 2005), pad4-1 and pmr4-1 (Nishimura et al., 2003), rar1-21 (Tornero et al., 2002), eta3/sgt1b (Gray et al., 2003), fls2 (Gómez-Gómez and Boller, 2000), eds5-1 (Rogers and Ausubel, 1997) 35S:nahG line (Reuber et al., 1998), and npr3-2npr4-2 (Fu et al., 2012) were kindly provided by Jane Glazebrook (University of Minnesota), Jeff Dangl (University of North Carolina), William Gray (University of Minnesota), Shauna Somerville (University of California at Berkeley), Sheng Yang He (University of Michigan), Mary Wildermuth (University of California at Berkeley), and Xin Li (University of British Columbia, Canada), respectively.
Root Growth Assays
Sterilized Arabidopsis seeds were germinated on general growth medium (1% Suc, 0.5× Murashige and Skoog [MS] salts, 0.05% MES, and 0.8% plant agar, pH 5.8) after 2 d of stratification and vertically grown for 10 d before transfer to new plates containing the indicated chemicals. After transfer onto new plates, the end of each root was marked as a starting point, and root growth arrest was monitored after 6 d. Root morphology and GUS staining were examined by differential interference contrast microscopy (Leica DM5000B), and fluorescence reporter lines were analyzed by confocal microscopy (Nikon TE2000U) with propidium iodide staining.
Confocal Microscopy
Col-0, PIN1:GFP, and PIN2:GFP plants were grown vertically in half-strength MS medium containing 1% Suc for 5 d and transferred to 10 μM DFPM half-strength MS medium. At the indicated time, primary roots were stained with propidium iodide and observed using a Zeiss LSM 710 confocal microscope with spectral settings for excitation at 488 nm/emission at 493 to 555 nm for the GFP signal and excitation 514 nm and emission at 596 to 719 nm for propidium iodide. A constant gain value was used for each genotype. For root meristem cell quantification, Col-0, victr-1, and victr-2 plants were prepared as described above. Meristem cells in the division zone were counted using Image J (Schneider et al., 2012). A single file of cortex cells was analyzed in all roots. The division zone was defined as the region made up of isodiametric cells between the quiescent center up to the cell, which was twice the length of the cell immediately before it (González-García et al., 2011). analysis of variance and Tukey test statistical analyses were conducted using the Origin-Pro version 8.6 package. Ten seedlings per genotype and treatment were analyzed.
QTL Analyses and VICTR Cloning
For initial mapping and QTL analyses, the DFPM-induced VICTR phenotype was scored on the 100 Col × Ler RILs. QTL analyses were performed using QTL package (version 1.11-12) that uses R software (version 2.81) based on the QTL package manual (Broman et al., 2003). Parameters used for LOD estimation were step = 1 and error probability = 0.01. Haldane map function was used as a mapping algorithm. Genotyping information of each Col × Ler RILs was retrieved from the microarray-based physical genomic mapping data set (Singer et al., 2006).
For marker-based cloning of the VICTRCol locus, the recessive Ler root development phenotype was screened from a mapping population of ∼2000 F2 plants of Col crosses to Ler. The VICTRCol locus was mapped to an ∼5-centimorgan interval on the lower arm of chromosome 5 based on linkage to PCR-based markers 18.1 Mb (5′-GTCGTAGAAGTAGCAGCTTGTACGAAGT-3′/5′-TCTCAATCCAATGTTCAGGTGACACACT-3′) and 19.1 Mb (5′-GAATCTCTAACGATGCTAACACAAGGA-3′/5′-CTGCAATCACAGTTTAAGTCATGAACT-3′). Within the 100-kb region defined by markers 18.86 and 18.95 Mb, 21 T-DNA insertion lines targeting 14 genes among 18 predicted genes (At5g46490 to At5g46650) were analyzed for the root VICTR phenotype.
Deletion of VICTR in certain ecotypes (see Supplemental Figure 4 online) was examined by PCR amplification of genomic DNA templates using LA Taq and Ex Taq polymerases (Takara Bio). Control primers are 5′-ATGGTTGCGATACCCAATATTAAGA-3′ and 5′-CTAAAGGATCTCTCCTTCCTCAAGATCGT-3′. Internal primers are 5′-CGGGTAGAAATATTGTACGCTCACAGT-3′ and 5′-CCAAATGGTTGAGAGTCAGATGTTC-3′. Note that internal primers anneal to both VICTR and VICTL1. External primers are 5′-GGACGCTTTGTTTCCAGCTTGTGGGGCATTAAATCGAG-3′ and 5′-CACTTCGTAGGGCGTTGGGGCAGACATTGCCTTCTC-3′.
Transgenic Studies
To generate VICTR promoter-driven reporter lines, a 2.5-kb fragment from the start codon of Col-0 and Bay-0 was PCR amplified from genomic DNA using primers (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAGAACCAGTACACAATGGCAGGTAAG-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAAGAAGCCATAGAGAGAGTTAGAAGGAGGA-3′) and cloned into pBGGUS and pHGY (vector plasmids were kindly provided by Taku Demura from RIKEN) (Kubo et al., 2005). The VICTRCol gene was PCR amplified using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCCCTGTGGTATCTGGTAATCAAAATTCTGT-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGCTACAAAGCAAGTCAGTCGTTCGTTG-3′. The VICTRCol coding region for the 35S promoter driven transgenic line was PCR amplified using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCTCCTTCTAACTCTCTCTATGGCTTC-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCATATGAAGTGGTGTAGCTGCAAAAG-3′. For recapitulation of the Col-0 VICTR phenotype in Ler and C24, the VICTRCol gene including the promoter and coding region was inserted into pHGY. For generation of 35S promoter–driven VICTRCol transgenic lines, the VICTRCol coding region, including introns, was cloned into pH35GS. Transformed lines with empty vector were used as controls for phenotypic analyses.
For subcellular localization analysis in onion (Allium cepa) epidermal cells (see Supplemental Figure 4D online), full-length VICTRCol cDNA was inserted into pH35GY and pH35YG to construct plasmids encoding VICTR-YFP and YFP-VICTR, respectively. Transient expression by biolistic bombardment (using PDS-1000/He; Bio-Rad) was performed in onion epidermal cells.
Localization and Immunodetection of VICTR in Nicotiana benthamiana Cells
VICTR cDNA in the entry vector (pENTR-TOPO-VICTR) was transferred via Clonase LR recombination system (Invitrogen) into the Gateway-compatible destination vector pMDC43 (Curtis and Grossniklaus, 2003), resulting in a GFP-VICTR expression vector. The RFP vector used in colocalization analysis was described earlier (Bhattacharjee et al., 2011). N. benthamiana leaves were infiltrated with the indicated Agrobacterium tumefaciens strains at an OD600 of 0.3 for each strain. Two days after infiltration, tissue sections from the infiltrated patches were analyzed by a two-photon equipped Zeiss confocal microscope. Three 7-mm-diameter leaf punches were excised from the infiltrated tissues and ground in 100 μL of 8 M urea and subjected to immunoblotting with anti-GFP antibodies (Sigma-Aldrich).
BiFC Assays
The pENTR-TOPO-VICTR clone was recombined into the Gateway-compatible BiFC vectors pMDC-nVenus or pMDC-cCFP (Bhattacharjee et al., 2011) using the Clonase LR recombination system. BiFC vectors for EDS1, PAD4, and GUS expression were used as in previous studies (Bhattacharjee et al., 2011). Agroinfiltration and confocal microscopy analysis were performed as indicated earlier. For fusion protein detection, three 7-mm-diameter leaf discs were excised from the infiltrated patches, ground in 8 M urea and analyzed by immunoblotting using anti-GFP antibodies (Santa Cruz Biotechnology).
Coimmunoprecipitation Analyses
HA-tagged VICTR was generated using pENTR-TOPO-VICTR and the destination vector HA-pBA (Kim et al., 2010). Myc-tagged EDS1, PAD4, and GUS have been used before (Bhattacharjee et al., 2011). The eds1L262P mutant described by Rietz et al. (2011) was generated by overlap PCR using the primers 5′-TTCCCTGAGCTAAGTCCTTATA-3′ and 5′-GCTCAGGGAAACTAGAAAGGGT-3′ and replacing the wild-type fragment in the entry clone of EDS1. Myc-eds1L262P was generated by Clonase recombining the entry clone into the Myc-pBA binary vector. Agrobacterium-infiltrated N. benthamiana leaves were processed for nuclear extracts as described by Palma et al. (2007). The resulting nuclear lysates were centrifuged at 14,000g for 10 min to remove nuclear debris. Immunoprecipitation was performed as published before (Bhattacharjee et al., 2011). The immunoprecipitates and the input fractions were analyzed with anti-HA (Roche) or anti-Myc (Santa Cruz Biotechnology) antibodies.
Quantitative Real-Time RT-PCR
Col-0, C24, Bay-0, and one line of Bay-0 expressing VICTRCol-0 were grown in half-strength MS media for 14 d and were transferred to half-strength MS containing 10 μM DFPM. After 24 h of DFPM treatment, total RNA was extracted, and real-time RT-PCR was performed to quantify the transcript levels of VICTR. Primers were designed with QuantPrime (Arvidsson et al., 2008): VICTR-qRT2-F, 5′-AGAGACCGGTTCATCAGCAGAG-3′;VICTR-qRT2-R, 5′-CCATATTGCCTTCTTCGGCTTGAG-3′. Amplified samples were normalized against PDF2 levels: PDF2_F, 5′-TAACGTGGCCAAAATGATGC-3′; PDF2_R, 5′-GTTCTCCACAACCGCTTGGT-3′.
Stomatal aperture measurements were performed as described before (Kim et al., 2011; Hubbard et al., 2012). Student’s t tests were performed for n = 3 independent experiments with 42 to 50 stomata analyzed for each condition.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: VICTR, At5g46520; VICTL1, At5g46510; EDS1, At3g48090; and PAD4, AT3G52430.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Functional Characterization of Modified DFPM Structures and 35S:VICTRCol Ectopic Expression Caused Stunted Growth.
Supplemental Figure 2. Gene Expression and Morphological Changes of DFPM-Treated Seedlings.
Supplemental Figure 3. DFPM Reduces the Number of Meristem Cells in the Wild Type, but Not in victr Mutant Alleles.
Supplemental Figure 4. Characterization of eds1-23 T-DNA Insertion Line and victl1-1, Subcellular Localization of VICTR, and VICTR Deletion in Other Accessions.
Supplemental Figure 5. VICTR Gene Sequence Comparison of Col-0 with Bay-0 and Kin-0.
Supplemental Figure 6. VICTR Does Not Associate with GUS.
Supplemental Figure 7. Root Growth Inhibition by Salicylic Acid Differs from the DFPM-Induced Primary Root Growth Arrest Response.
Supplemental Table 1. List of Arabidopsis Accessions That Did Not Produce the Root Developmental Arrest in Response to DFPM Treatment.
Supplemental Table 2. DFPM-Induced Root Growth Arrest VICTR Phenotype in 100 Col × Ler Recombinant Inbred Lines.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 4B.
Supplemental Data Set 2. Text File of the Alignment in Supplemental Figure 5.
Acknowledgments
We thank Chris Somerville (University of California at Berkeley) for providing access to the chemical library of 9600 compounds, Jeff Dangl for providing rar1-21 seeds, and Amber Ries [University of California at San Diego (UCSD)] for independent blinded stomatal movement analyses. We thank Rosangela Sozzani (University of Pavia), Yunde Zhao, Sangho Jeong, Aurelien Boisson-Dernier, Noriyuki Nishimura and Carolyn Rasmussen at UCSD for comments on the article and helpful discussions. This research was supported by the National Institutes of Health (R01GM060396; J.I.S.), the National Science Foundation (MCB0918220, J.I.S.; IOS1121114, W.G.), and the Max-Planck Society and Deutsche Forschungsgemeinschaft 'SFB 670' (J.E.P) grants. Root growth analyses were in part funded by a grant from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (DE-FG02-03ER15449; J.I.S.) and in part by the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012-0002781 and 2012-041653) to T.-H.K. H.-H.K. was funded by the Human Frontier Science Program and the Alexander von Humboldt Foundation.
AUTHOR CONTRIBUTIONS
T.-H.K. and J.I.S. designed the experiments. T.-H.K. performed most of the experiments. S.B., H.-H.K., W.G., and J.I.S. designed BiFC and co-IP experiments. S.B. and H.-H.K prepared constructs and S.B. conducted BiFC and co-IP experiments. H.-H.K. worked on GDA/DFPM interference and distributed material. A.L. conducted stomatal movement analyses. T.-H.K., F.H., H.-H.K., W.G., J.E.P., and J.I.S. analyzed the data. C.E. analyzed meristematic cell division, J.P. contributed quantitative PCR, pPIN∷PIN:GFP results, and repeated pDR5:GFP data. S.B., H.-H.K., W.G., and J.E.P. contributed to the eds1L262P experiment. T.H. assisted with the VICTR cloning. T.-H.K., H.-H.K., and J.I.S. wrote the article with input from other coauthors.
Glossary
- ABA
abscisic acid
- DFPM
[5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione
- NB
nucleotide binding
- LRR
Leucine-rich repeat
- TIR
Toll-Interleukin1 Receptor
- Col-0
Columbia-0
- Ler
Landsberg erecta
- Kin-0
Kindalville-0
- Bay-0
Bayreuth-0
- GFP
green fluorescent protein
- RIL
recombinant inbred line
- QTL
quantitative trait locus
- GUS
β-glucuronidase
- YFP
yellow fluorescent protein
- SA
salicylic acid
- RFP
red fluorescent protein
- BiFC
bimolecular fluorescence complementation
- co-IP
coimmunoprecipitation
- HA
hemagglutinin
- JA
jasmonic acid
- GDA
geldanamycin
- MS
Murashige and Skoog
References
- Alcázar R., García A.V., Parker J.E., Reymond M. (2009). Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. USA 106: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J.I., Yuan S., Dale J.M., Tanner V.N., Theologis A. (2004). Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riano-Pachon D.M., Mueller-Roeber B. (2008). QuantPrime--A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Subramaniam R., Dangl J.L. (2004). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7: 391–399 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Halane M.K., Kim S.H., Gassmann W. (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., Dangl J.L., Weigel D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhan M.H., Holub E.B., Beynon J.L., Rozwadowski K., Rimmer S.R. (2004). The Arabidopsis TIR-NB-LRR gene RAC1 confers resistance to Albugo candida (white rust) and is dependent on EDS1 but not PAD4. Mol. Plant Microbe Interact. 17: 711–719 [DOI] [PubMed] [Google Scholar]
- Broman K.W., Wu H., Sen S., Churchill G.A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Caplan J., Padmanabhan M., Dinesh-Kumar S.P. (2008). Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clarke J.H., Mithen R., Brown J.K.M., Dean C. (1995). QTL analysis of flowering time in Arabidopsis thaliana. Mol. Gen. Genet. 248: 278–286 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682 [DOI] [PubMed] [Google Scholar]
- DeYoung B.J., Innes R.W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Fernández-Marcos M., Sanz L., Lewis D.R., Muday G.K., Lorenzo O. (2011). Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. USA 108: 18506–18511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Wiermer M., Bhat R.A., Moisan L.J., Medina-Escobar N., Neu C., Cabral A., Parker J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S.H., Tada Y., Zheng N., Dong X. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Zook M., Mert F., Kagan I., Rogers E.E., Crute I.R., Holub E.B., Hammerschmidt R., Ausubel F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- González-García M.-P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Gray W.M., Muskett P.R., Chuang H.W., Parker J.E. (2003). Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-L., Fitz J., Schneeberger K., Ossowski S., Cao J., Weigel D. (2011). Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol. 157: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L., Parker J.E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Kerppola T.K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K.E., Siegel R.S., Valerio G., Brandt B., Schroeder J.I. (2012). Abscisic acid and CO2 signalling via calcium sensitivity priming in guard cells, new CDPK mutant phenotypes and a method for improved resolution of stomatal stimulus-response analyses. Ann. Bot. (Lond.) 109: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Guerois R. (2010). NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 35: 199–207 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Gao F., Bhattacharjee S., Adiasor J.A., Nam J.C., Gassmann W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kwon S.I., Saha D., Anyanwu N.C., Gassmann W. (2009). Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 150: 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-H., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J., Kidarsa T., Bradford C.S., Gilbert B., Curtis M., Tzeng S.-C., Maier C.S., Wolpert T.J. (2012). Tricking the guard: Exploiting plant defense for disease susceptibility. Science 338: 659–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno M.A., Van Norman J.M., Moreno A., Zhang J., Ahnert S.E., Benfey P.N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M.T., Stein M., Hou B.H., Vogel J.P., Edwards H., Somerville S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Palma K., Zhao Q., Cheng Y.T., Bi D., Monaghan J., Cheng W., Zhang Y., Li X. (2007). Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21: 1484–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Raghava G.P.S., Barton G.J. (2006). Quantification of the variation in percentage identity for protein sequence alignments. BMC Bioinformatics 7: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber T.L., Plotnikova J.M., Dewdney J., Rogers E.E., Wood W., Ausubel F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16: 473–485 [DOI] [PubMed] [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., Medina-Escobar N., Vlot A.C., Feys B.J., Niefind K., Parker J.E. (2011). Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Rogers E.E., Ausubel F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T.A., Bahrami A., Wilczek A., Watanabe E., Schellenberg K., McLellan C., Kelley A., Kong S.W., Queitsch C., Lindquist S. (2007). Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE 2: e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S.L. (2000). Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287: 1964–1969 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.H., Schulze-Lefert P. (2007). Rumble in the nuclear jungle: Compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Singer T., Fan Y., Chang H.S., Zhu T., Hazen S.P., Briggs S.P. (2006). A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Russo A.A., Schneider C., Rosen N., Hartl F.U., Pavletich N.P. (1997). Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell 89: 239–250 [DOI] [PubMed] [Google Scholar]
- Sweat T.A., Wolpert T.J. (2007). Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 19: 673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Thierry J.C., Poch O. (2003). RASCAL: Rapid scanning and correction of multiple sequence alignments. Bioinformatics 19: 1155–1161 [DOI] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Tornero P., Merritt P., Sadanandom A., Shirasu K., Innes R.W., Dangl J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012). Natural variation in Arabidopsis: From molecular genetics to ecological genomics. Plant Physiol. 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J., Parker J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Yang S., Hua J. (2004). A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. (2007). A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-C., Gassmann W. (2007). Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 145: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y.T., Qu N., Zhao Q., Bi D., Li X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48: 647–656 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X. (2003b). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M.J., Lassner M., Li X. (2003a). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski P.A., Koczyk G., Galganski L., Sadowski J. (2009). Genome sequence comparison of Col and Ler lines reveals the dynamic nature of Arabidopsis chromosomes. Nucleic Acids Res. 37: 3189–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J., Hicks G.R., Raikhel N.V. (2004). Sorting inhibitors (Sortins): Chemical compounds to study vacuolar sorting in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 9497–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]