Abstract
Animals establish their body plans in embryogenesis, but only a few animals can recapitulate this signaling milieu for regeneration after injury. In planarians, a pluripotent stem cell population and perpetual signaling of polarity axes collaborate to direct a steady replacement of cells during homeostasis and to power robust regeneration after even severe injuries. Several studies have documented the roles of conserved signaling pathways in maintaining and resetting axial polarity in planarians, but it is unclear how planarians reestablish polarity signaling centers after injury and whether these centers serve to influence identity decisions of stem cell progeny during their differentiation. Here we find that a planarian Follistatin homolog directs regeneration of anterior identity by opposing an Activin/ActR-1/Smad2/3 signaling pathway. Follistatin and Notum, a Wnt inhibitor, are mutually required to reestablish an anterior signaling center that expresses both cues. Furthermore, we show that the direction of cells down particular differentiation paths requires regeneration of this anterior signaling center. Just as its amphibian counterpart in the organizer signals body plan and cell fate during embryogenesis, planarian Follistatin promotes reestablishment of anterior polarity during regeneration and influences specification of cell types in the head and beyond.
Keywords: nervous system regeneration, Schmidtea mediterranea
Well-known signaling cascades continuously coordinate axes of polarity during homeostasis and regeneration in planarians (1). Wnt signaling through β-catenin induces posterior identity (2–5), guiding the animal’s body plan during homeostasis and, after injury, supporting regeneration programs and directing reestablishment of axial polarity (6, 7). Hedgehog signaling also promotes posteriorization, partly through regulation of Wnt gene expression (8, 9), whereas the α/β-hydrolase Notum, an inhibitor of Wnt signaling, plays a critical role in anterior polarity reestablishment (10). The differentiated cells that secrete polarity cues must be respecified after injury, a process that can occur in the absence of planarian stem cells (5, 6, 11), pluripotent cells called neoblasts (12). Although the mechanisms underlying the restoration of polarity after amputation are unknown, they likely involve interpretation of existing gradients of polarity, as evidenced by the observation that very thin fragments with insufficient gradient depth fail to reestablish polarity (13). Nonetheless, robust regeneration of polarity gradients must involve redundant signaling; for example, concomitant knockdown of wnt1 with notum negates its headless phenotype, leading to faithful resetting of anterior polarity through some other, unknown mechanism (10).
Results and Discussion
Characterization of Planarian Follistatin.
To better understand reestablishment of anterior polarity, we have characterized a Follistatin homolog in the planarian species Schmidtea mediterranea. Smed-follistatin mRNA is evident in a ventrally enriched punctate distribution throughout the body of the animal, with a strong focus of expression in a small cluster of several cells at the anterior midline tip, beneath the epithelium (Fig. 1 A and B). Consistent with a previously published study (7), follistatin expression is up-regulated after injury, with more puncta evident as early as 6 h after a cut in the side of the animal and continuing to at least 2 d after amputation (Fig. 1A and Fig. S1A). Double fluorescence in situ hybridization (FISH) experiments showed that the cluster of follistatin+ cells in the head of the planarian also expresses notum (Fig. 1B and Fig. S1B), whereas follistatin+ cells outside this region are notum−. After amputation, follistatin+/notum+ cells appeared near the wound within 1 d and an anterior focus of double-positive cells is clear within 2 d (Fig. S1 C and D). follistatin+ cells outside the anterior center often resided in or near the central nervous system (Fig. S1E). Double FISH indicated that these cells often abutted neurons [prohormone convertase-2 (PC2)+ or choline acetyltransferase (ChAT)+ cells], but that they were not themselves neural in nature (Fig. 1C and Fig. S1 E and F). However, follistatin+/ChAT+ cells were infrequently observed in regenerating organisms (Fig. S1G), suggesting a possible transient overlap in the two differentiation trajectories.
Fig. 1.
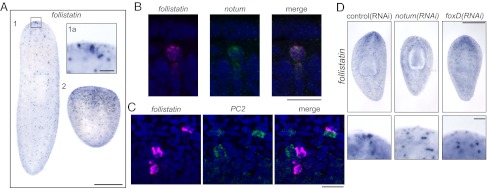
Planarian follistatin is expressed in the head. (A) Whole-mount in situ hybridization with a follistatin probe (1) reveals strong expression in a few anterior cells (boxed area, 1a) and weaker expression in numerous cells throughout the planarian body. Puncta of follistatin expression are markedly increased in a tail piece regenerating a new head 2 d after amputation (2). (B) follistatin and notum transcripts, viewed by FISH, are present in the same few cells in the anterior of the planarian. The follistatin transcripts are visualized in magenta, whereas notum transcripts are in green. (C) follistatin-expressing cells (magenta) and PC2-expressing cells (green) are often near each other, but coexpression of follistatin and PC2 was not observed by FISH. (D) follistatin expression in the tip of the head is absent or disorganized in 17 of 20 notum(RNAi) and 6 of 8 foxD(RNAi) animals after 5 d of regeneration. Expression of follistatin in the remainder of the animal is not affected. [Scale bars, 500 µm (A and D), 20 µm (B and C), and 50 µm (A, Inset).] The anterior of each animal is oriented toward the top.
We next investigated whether follistatin expression in the head depends on Notum. After notum(RNAi), follistatin expression in the tip of the head was absent or disorganized (Fig. 1D). In contrast, expression of follistatin outside the anterior focus was unaffected (Fig. 1D). The group of follistatin+/notum+ cells in the planarian head also evoked the expression pattern of foxD, a forkhead transcription factor of unknown function that was identified in the planarian Dugesia japonica (14) (Fig. S1B). The forkhead transcription factor FoxL2 had previously been shown to regulate follistatin expression in the context of the murine ovary and pituitary (15, 16), so we evaluated whether the anterior group of follistatin+ cells depended on Smed-foxD. Indeed, we found that foxD(RNAi) animals had absent or disorganized anterior follistatin foci (Fig. 1D) that were diminished even without amputation (Fig. S1A). Neither foxD(RNAi) nor notum(RNAi) treatment eliminated early follistatin up-regulation after a superficial cut (Fig. S1A), suggesting that foxD and notum function more specifically in reestablishment and/or maintenance of follistatin expression in the anterior-most focus and that other regulatory mechanisms underly additional follistatin expression. foxD(RNAi), like notum(RNAi) (10), led to aberrant eyespot number and altered nervous system morphology after amputation (Fig. S1 H and I), indicating that both gene products play roles in proper anterior regeneration.
Follistatin Is Essential for Planarian Head Regeneration.
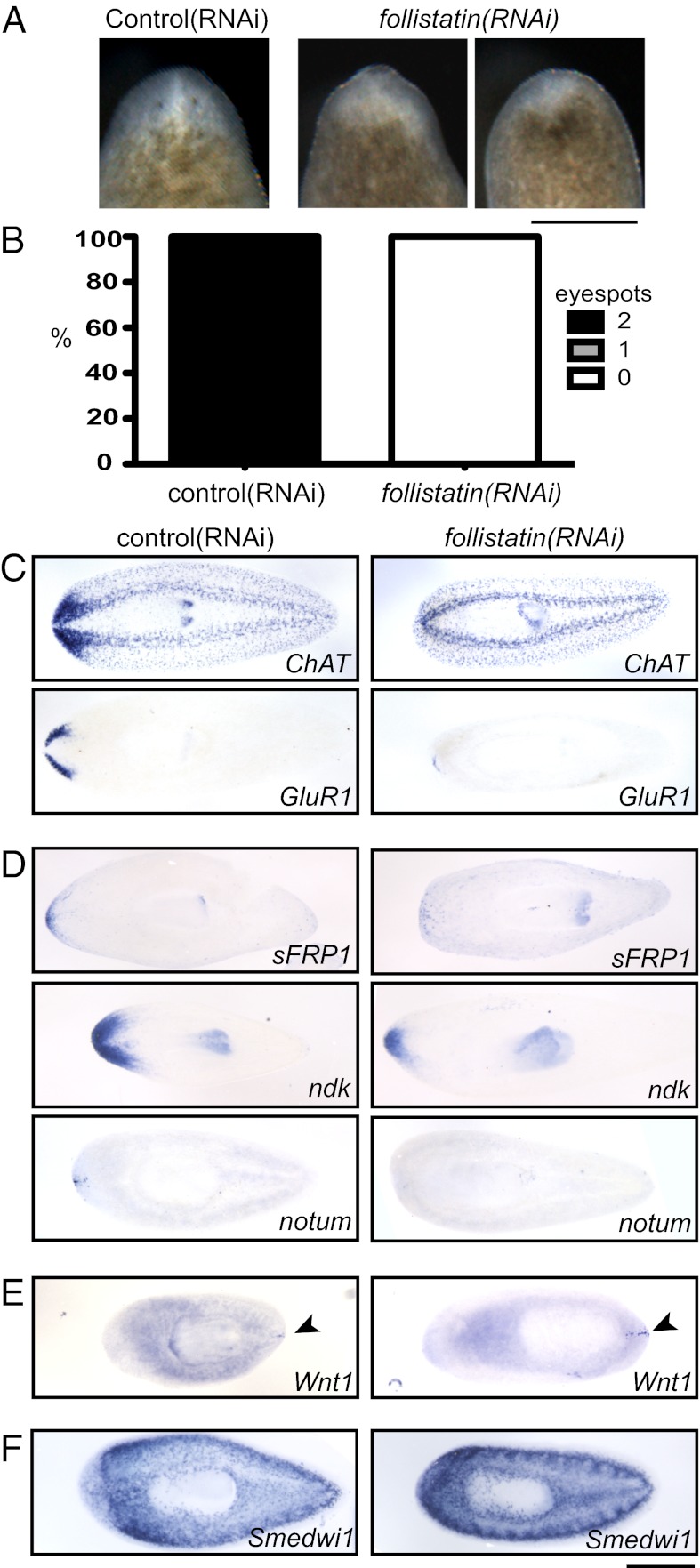
We next tested whether Follistatin itself is required for proper head regeneration. follistatin(RNAi) animals were amputated and, after 5 d of regeneration, these animals were significantly impaired in their ability to regenerate heads compared with controls. Outwardly, follistatin(RNAi) animals had small blastemas that were devoid of regenerated eyespots (Fig. 2 A and B and Fig. S2 A and B). In situ hybridization for ChAT and glutamate receptor (GluR1), which are expressed in the entire nervous system and branches of the cephalic ganglia (brain), respectively (17, 18), indicated impairment of cephalic ganglia regeneration after follistatin(RNAi) (Fig. 2C and Fig. S2 C and F). These defects appeared to be contingent upon amputation, as long-term treatment with follistatin(RNAi) caused animals to display slightly regressed tips of the head without an overall deterioration of the central nervous system (Fig. S2 D and E).
Fig. 2.
follistatin plays a critical role in anterior regeneration. (A) After 5 d of head regeneration, follistatin(RNAi) animals display small blastemas compared with control animals and are missing eyespots. (B) Nearly all control animals regenerated eyespots within 5 d of amputation, but eyespots were missing from all follistatin(RNAi) animals (n ≥ 50 each). (C) Cephalic ganglia are absent or dramatically reduced in size in follistatin(RNAi) animals 5 d after amputation of the head. Both control(RNAi) and follistatin(RNAi) animals were subjected to in situ hybridization with ChAT and GluR1 probes to mark the entire central nervous system and the brain branches, respectively. (D) Anterior marker expression was reduced after 5 d of head regeneration in follistatin(RNAi) animals. sFRP-1, ndk, and notum probes each mark different anterior cell populations. (E) A posterior marker, wnt1, was expressed in an expanded posterior region (arrowheads) but was not expressed inappropriately in the anterior of follistatin(RNAi) animals. (F) In situ hybridization with a probe for a neoblast marker, Smedwi-1, indicates that neoblasts were present after follistatin(RNAi). (Scale bars, 500 µm.) Anterior is up (A) or to the left (C–F).
A defect in neural regeneration could be due to failure of either neurogenesis or respecification of anterior polarity. To determine whether anterior polarity was reestablished after amputation, follistatin(RNAi) animals were subjected to in situ hybridization to detect transcripts with roles in anterior polarity (2, 3, 10, 19). secreted frizzled-related protein 1 (sFRP-1) and nou darake (ndk) mRNAs showed reduced expression in follistatin(RNAi) planarians (Fig. 2D). We also observed that notum expression was disrupted in follistatin(RNAi) animals (Fig. 2D), indicating a reciprocal requirement between Notum and Follistatin. The interdependence of follistatin and notum signals at the tip of the head could assist in focusing the anterior signaling center, ensuring accurate and robust organization of the body after dramatic injury.
One explanation for the follistatin(RNAi) phenotypes was that RNAi resulted in posteriorization of the planarians, but this seemed not to be the case, as the zone of wnt1 expression was broader but still confined to the true posterior end of treated animals (Fig. 2E and Fig. S2F). Furthermore, we ruled out the possibility that defects in regeneration were due to neoblast depletion or lack of mitotic cells in follistatin(RNAi) planarians (Fig. 2F and Fig. S2G). Finally, ventral nerve cord regeneration after tail amputation occurred (Fig. S2 C and F), indicating that some nerve tissue regeneration could occur after follistatin(RNAi). Taken together, our data indicate that Follistatin plays a positive role in establishing anterior polarity during planarian regeneration.
Follistatin Antagonizes Activin Signaling.
Follistatin homologs in vertebrates and Drosophila bind to and inhibit members of the TGF-β family, in particular Activin and bone morphogenetic protein (BMP) family members (20–24). No planarian Activin homolog has been described, but BMP signaling drives dorsal polarity and is important for maintenance of the midline (11, 25, 26). To determine whether planarian Follistatin functions by inhibiting one of these pathways, we performed RNAi of activin and BMP4 along with follistatin(RNAi). activin(RNAi) animals regenerated anterior structures and concomitant RNAi targeting both activin and follistatin rescued the phenotype of follistatin(RNAi), both in terms of cephalic ganglia regeneration (assayed by strong ChAT expression) and anterior polarity specification (reflected by sFRP-1 expression) (Fig. 3A). Quantitatively, 90% of activin(RNAi); follistatin(RNAi) animals regenerated two eyespots within 5 d of amputation, compared with zero planarians treated with follistatin(RNAi) alone (Fig. 3B and Fig. S3A). In contrast, BMP4(RNAi) did not rescue follistatin(RNAi) in terms of nervous system regeneration, anterior polarity specification, or eyespot formation (Fig. 3 A and B).
Fig. 3.
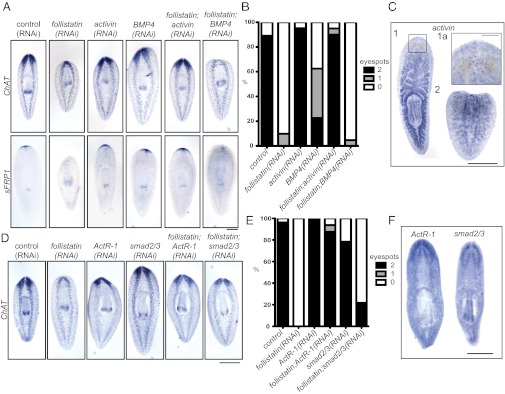
Planarian follistatin inhibits activin. (A) activin(RNAi) combined with follistatin(RNAi) rescues nervous system regeneration and anterior polarity, whereas a combination of BMP4(RNAi) and follistatin(RNAi) does not lead to rescue of these phenotypes. In situ hybridization with ChAT and sFRP-1 probes illustrates nervous system and anterior polarity, respectively, in 5-d regenerates. (B) Concomitant activin(RNAi) and follistatin(RNAi) rescues the follistatin(RNAi) phenotype, as evidenced by eyespot regeneration (n ≥ 40 each). (C) Whole-mount in situ hybridization with an activin probe indicates expression in the gut and pharynx (1) and in ventral spots along the animal and in the head (boxed area, 1a). The punctate expression pattern is dominant in 2-d regenerating tail pieces (2). (D) Simultaneous knockdown of follistatin with ActR-1 or smad2/3 by RNAi rescues the follistatin(RNAi) phenotype. dsRNA-fed animals 5 d after amputation were subjected to in situ hybridization with the ChAT probe to illustrate regeneration of the nervous system. (E) Eyespot quantitation after RNAi and 5 d of head regeneration. Concomitant treatment with ActR-1(RNAi) rescues the follistatin(RNAi) phenotype, and treatment with smad2/3(RNAi) partially rescues the follistatin(RNAi) phenotype (n ≥ 30 each). (F) ActR-1 and smad2/3, detected by whole-mount in situ hybridization, are broadly expressed but neurally enriched. [Scale bars, 500 µm (A, C, D, and F) and 100 µm (C, Inset).] Anterior is up.
Knockdown of planarian activin alone did not result in a dramatic phenotype, but activin(RNAi) animals had a diminished capacity to regenerate posterior tissues, sometimes regenerating with notched tails or regenerating with a smaller wnt1-expressing domain (Fig. S3 B and D). activin is expressed in the planarian gut and pharynx during homeostasis, as well as in cells distributed along the ventral side of the animal (Fig. 3C). These ventral puncta become the dominant expression pattern during regeneration (Fig. 3C). Although the patterns of activin and follistatin expression in regenerating worms appear superficially similar (Figs. 1A and 3C), the two genes are not expressed in the same cells (Fig. S3C).
Activin signals are transduced by receptor serine/threonine kinases, which phosphorylate and activate Smad2/3 transcription factors (for a review, see ref. 27). We cloned eight planarian TGF-β receptors and one Smad2/3 homolog. We found that knockdown of one type I Activin/TGF-β receptor (ActR-1) also rescued the follistatin(RNAi) phenotype. ActR-1(RNAi); follistatin(RNAi) animals showed near-control regeneration of the cephalic ganglia (Fig. 3D), and 87.5% of these animals regenerated two eyespots (Fig. 3E and Fig. S3E). RNAi against planarian smad2/3 also partially rescued the follistatin(RNAi) phenotype (Fig. 3 D and E and Fig. S3E), although with less efficiency. Knockdown of ActR-1 alone led to a behavioral phenotype, with worms adopting a flattened posture (Fig. S3F) and movement defects, consistent with a previous report (28). Additionally, ActR-1(RNAi) animals amputated in front of and behind the pharynx occasionally regenerated with supernumerary pharynges (Fig. S3D). Knockdown of smad2/3 alone resulted in a mild head regeneration phenotype, with cyclopia or absence of eyes in some regenerating animals (Fig. 3E and Fig. S3G). Consistent with these phenotypes, ActR-1 and smad2/3 expression was broad, with an enrichment of expression in the eyes and cephalic ganglia (Fig. 3F). Whereas double-RNAi experiments indicate that ActR-1 and Smad2/3 function downstream of Activin, detection of secondary phenotypes in ActR-1(RNAi) and smad2/3(RNAi) animals that were not present in activin(RNAi) animals suggests that the receptor and downstream transcription factor also play roles in mediating the effects of additional TGF-β family members.
Knockdown of notum or follistatin results in the absence of head regeneration after amputation, but simultaneous knockdown of wnt1 or activin, respectively, rescues anterior polarity (10). Taken together, these data suggest that either pair of signals is dispensable as long as other redundant polarity cues remain. However, the interdependence of follistatin and notum expression in the tip of the head and the influence of Follistatin and Activin on wnt1 expression in the tail imply cross-talk between the two pathways. The benefit of parallel signals of anterior polarity could be to reinforce and refine polarity reestablishment after injury, thus supporting the robustness of planarian regeneration through verification of cellular position. Both Follistatin and Notum function as inhibitors, blocking Activin and Wnt signaling, respectively. It remains unclear whether cells of the planarian body (neoblast progeny in particular) receive and integrate signals from one or both pathways, although the ActR-1 transcript is not enriched in cycling neoblasts themselves (Fig. S3H). Our determination that ActR-1 is expressed broadly suggests that a wide variety of cell types could be responding to the Follistatin–Activin balance, and the large number of Frizzled-related receptors of Wnt signals also suggests a wide audience of cells able to respond to the level of Wnt signaling (2). The impact of Hedgehog on Wnt signaling as well (8, 9) further illustrates the complex mechanisms underlying assignment of place in the anteroposterior axis.
Follistatin Directs Fate Decisions in Planarians.
Although it is known that axial gradients organize the body plan during regeneration and homeostasis in the planarian, it is not clear how these gradients are received and interpreted in other cells. Of critical importance, it is not clear whether neoblast progeny respond to Wnt, TGF-β, or other polarity signals (directly or indirectly) to determine migration direction or cell identity. By contrast, a wealth of literature documents the effects of similar ligands (and their inhibitors) during development or for differentiation and maintenance of pluripotent stem cells (for a review, see ref. 29). For example, inhibition of Activin and/or BMP signaling (by Noggins, Follistatin, Chordin, or chemical inhibitors) drives neuronal fate of pluripotent stem cells in vitro and also during development of a range of vertebrate and invertebrate organisms (22, 30–39). The expression of follistatin in cells near the nervous system and the absence of cephalic ganglia in regenerating follistatin(RNAi) animals led us to investigate whether Follistatin could be inducing specific fates (neural or otherwise) in planarians as well.
Because molecular markers along the path to neural differentiation have not yet been described in planarians, we first used markers that are available to label subtypes of differentiating cells. A previous report documented a number of genes (called category 3 genes) that are down-regulated 7 d after ablation of neoblasts by irradiation (40). Although gene expression in differentiated cells remains unchanged at this time point, neoblast progeny would be depleted. Thus, down-regulated genes of this category were interpreted to label cells during the process of differentiation. Furthermore, because expression of pairs of category 3 genes often do not overlap (40), each category 3 gene might represent neoblast progeny within a certain fate trajectory or during a certain window of differentiation (Fig. S4).
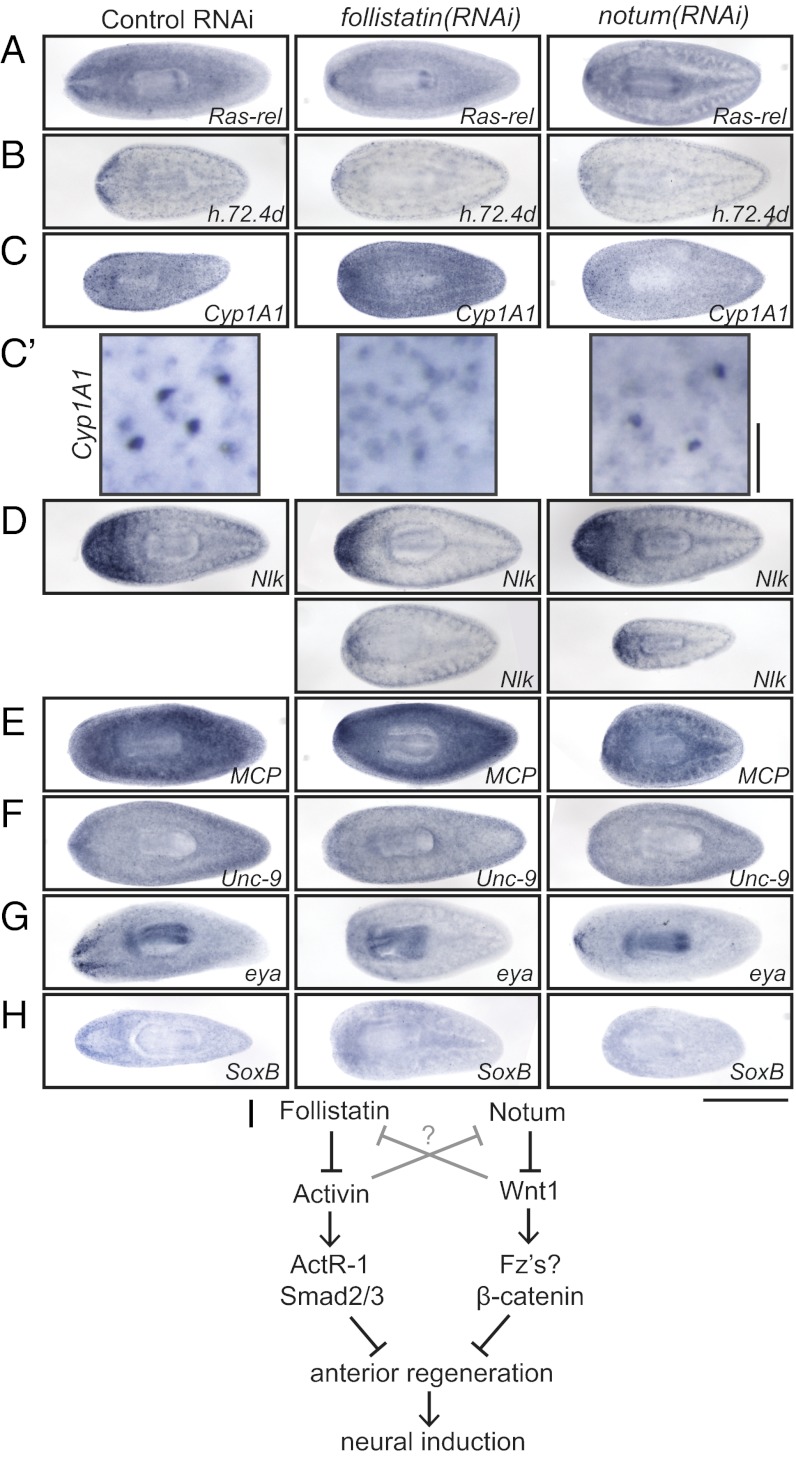
We used genes in this category to evaluate whether specific populations of differentiating cells were affected after knockdown of follistatin or notum. We found that Ras-related and h.72.4d, category 3 genes with expression near the nervous system, were indeed expressed in fewer cells in follistatin(RNAi) and notum(RNAi) animals (Fig. 4 A and B). We next examined the expression of Cyp1A1, a category 3 gene that marks two populations of cells throughout the planarian body. Surprisingly, one of the two populations of cells (the darker-staining population present ventrally) disappeared in follistatin(RNAi) animals but not in notum(RNAi) animals (Fig. 4 C and C′). This population was absent not only anteriorly but in posterior regions as well, indicating that the effect of Follistatin on this population of cells is Notum-independent and is likely not acting through Follistatin’s role in anterior polarity alone. Nemo-like kinase, a gene expressed in neoblasts and the brain (41), also showed a mildly to moderately reduced domain of expression in both follistatin(RNAi) and notum(RNAi) animals (Fig. 4D). Some subtypes of differentiating cells were insensitive to follistatin(RNAi) treatment, including those expressing mitochondrial carrier protein (MCP) and unc-9 (Fig. 4 E and F), indicating that Follistatin is not responsible for instruction of cell differentiation in general.
Fig. 4.
Follistatin influences fate decisions in planarians. In situ hybridization of control(RNAi), follistatin(RNAi), or notum(RNAi) animals 5 d after head amputation, using Ras-related (A), h.72.4d (B), Cyp1A1 (C; higher magnification in C′), nemo-like kinase (nlk; D), MCP (E), Unc-9 (F), eya (G), or SoxB (H) probes. (I) Model illustrating the role of Follistatin and Notum signals in providing a positive feedback loop to stabilize signaling of anterior polarity in planarians. [Scale bars, 500 µm (A–H) and 50 µm (C′).] Anterior is to the left.
Although the influence of Follistatin on category 3 gene-expressing cells suggests a role for Follistatin in differentiation, we were specifically interested in whether Follistatin might drive cells toward neuronal fates. Toward this end, we examined the expression of eyes absent (eya), a transcription factor and phosphatase expressed in differentiating photoreceptors (among other cell types) (42, 43), and found that eya expression was diminished in follistatin(RNAi) and notum(RNAi) animals (Fig. 4G). Sox transcription factors often function in neural induction in other organisms (44), with Xenopus SoxD and Drosophila SoxN being inhibited by BMP/Dpp signaling and Xenopus Sox-2 being induced by Chordin (45–47). We identified a planarian Sox transcription factor expressed in the nervous system, smed-SoxB (48), that is also dramatically down-regulated in follistatin(RNAi) and notum(RNAi) animals (Fig. 4H). Together, these results demonstrate that planarian Follistatin does influence differentiation, possibly directing some neoblast progeny toward anterior fates.
Conclusion
Organization of the body plan and determination of cell fates during embryonic development often rely on signaling centers such as the amphibian organizer, which secretes BMP/Activin inhibitors—including Follistatin—and Wnt inhibitors (49). In this study, we suggest that regenerating organisms use and reuse similar mechanisms, with follistatin and notum expression within a population of cells with organizer-like activity that promotes anterior identity and morphogenesis of the cephalic ganglia and eyespots (Fig. 4I). Here we have characterized the function of planarian Follistatin, a signaling molecule with a critical role in head regeneration. We have further investigated the mechanism of Follistatin activity, using double-RNAi rescue experiments to reveal that Follistatin inhibits an Activin/ActR-1/Smad2/3 signaling pathway that itself represses anterior regeneration (Fig. 4I). Together, our results indicate that redundancy in signals that promote anterior polarity sustains the body plan in planarians, with signals reinforcing one another to ensure accurate organization of the body, especially after severe injury. Furthermore, the intimate association between follistatin+ cells and neurons raises the intriguing possibility that Follistatin could be influencing neuronal fate and function directly. In the future, it will be important to dissect direct and indirect functions of Follistatin and to separate, if possible, the roles of follistatin+ cells in the anterior versus the remainder of the planarian body.
Methods
Planarian Experiments.
A clonal line of asexual planarians (ClW4) was maintained as previously described (50), except that animals were kept in Instant Ocean salts (Spectrum Brands) at 0.5 g/L in ultrapure water and 50 µg/mL gentamicin (Gemini Bio-Products). Each riboprobe or dsRNA was synthesized as previously described from 400- to 500-bp fragments of genes cloned into the pJC53.2 vector (51, 52). For single-RNAi experiments, animals were fed 3 µg dsRNA in 35 µL of a liver:salts puree (3:1). For double-RNAi rescue experiments, animals were fed a total of 4 µg dsRNA (either 4 µg control, 2 µg control + 2 µg experimental, or 2 µg each experimental). Volumes were also kept constant in these experiments. For all experiments, unless otherwise noted, animals were fed dsRNA on days 0, 6, and 12, before being cut prepharyngeally on day 17. On day 22 (day 5 postamputation), regenerating animals were evaluated for phenotypes (counting eyespots or imaging) or were killed and fixed (for in situ hybridizations).
In situ hybridizations were performed as per ref. 53 with some modifications, either by hand or using an InsituPro VS (Intavis). For immunofluorescence experiments, planarians were killed in 2% (vol/vol) HCl, fixed for 1 h at 4 °C in 4% (vol/vol) formaldehyde in PBS, and then bleached in 6% (vol/vol) H2O2 in PBS overnight. Animals were blocked in 6 mg/mL BSA (Jackson ImmunoResearch) and 0.45% fish gelatin (Sigma-Aldrich) in PBS + 0.3% Triton X-100. Animals were incubated with rabbit anti–phospho-histone H3 (S10; Cell Signaling) primary antibodies at a 1:5,000 dilution overnight at 4 °C. The secondary antibody (Molecular Probes) was used at a 1:2,500 dilution.
RT-quantitative PCR and irradiation experiments were performed as previously described (51, 54), except for an increase to 100-Gy dosage for irradiation.
Image Acquisition and Processing.
Live animals and chemically developed in situ hybridization experiments were imaged with a Leica M205A stereomicroscope running LAS software 3.6.0 (Leica). Immunofluorescence and fluorescence in situ hybridization experiments were imaged using a Zeiss LSM 710 confocal microscope with either a 20× (Plan-Apochromat 206/0.8) or a 63× objective (Plan-Apochromat 636/1.4) using Zen software (Carl Zeiss).
The complete mRNA sequence for S. mediterranea follistatin, as well as incomplete mRNA sequences for activin, ActR-1, and smad2/3, have been deposited in GenBank under accession numbers KC161222–5.
Supplementary Material
Acknowledgments
We are grateful to members of the P.A.N. laboratory for advice and technical help during the course of this project. We also thank Bret Pearson for suggesting the use of Instant Ocean salts and Ryan King for suggested improvements to the in situ protocol. R.H.R.-G. is a fellow of The Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by National Institutes of Health Grant R01 HD043403 (to P.A.N.). P.A.N. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC161222, KC161223, KC161224, and KC161225).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214053110/-/DCSupplemental.
References
- 1.Forsthoefel DJ, Newmark PA. Emerging patterns in planarian regeneration. Curr Opin Genet Dev. 2009;19(4):412–420. doi: 10.1016/j.gde.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319(5861):323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319(5861):327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 4.Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135(7):1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- 5.Gurley KA, et al. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 2010;347(1):24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci USA. 2009;106(40):17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26(9):988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326(5958):1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yazawa S, Umesono Y, Hayashi T, Tarui H, Agata K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci USA. 2009;106(52):22329–22334. doi: 10.1073/pnas.0907464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen CP, Reddien PW. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 2011;332(6031):852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134(22):4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- 12.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332(6031):811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan T. Experimental studies of the regeneration of Planaria maculata. Arch Entwicklungsmech Org. 1898;7(2-3):364–397. [Google Scholar]
- 14.Koinuma S, Umesono Y, Watanabe K, Agata K. The expression of planarian brain factor homologs, DjFoxG and DjFoxD. Gene Expr Patterns. 2003;3(1):21–27. doi: 10.1016/s1567-133x(02)00097-2. [DOI] [PubMed] [Google Scholar]
- 15.Blount AL, et al. FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J Biol Chem. 2009;284(12):7631–7645. doi: 10.1074/jbc.M806676200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashimada K, et al. FOXL2 and BMP2 act cooperatively to regulate follistatin gene expression during ovarian development. Endocrinology. 2011;152(1):272–280. doi: 10.1210/en.2010-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura K, Kitamura Y, Taniguchi T, Agata K. Analysis of motor function modulated by cholinergic neurons in planarian Dugesia japonica. Neuroscience. 2010;168(1):18–30. doi: 10.1016/j.neuroscience.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Cebrià F, et al. The expression of neural-specific genes reveals the structural and molecular complexity of the planarian central nervous system. Mech Dev. 2002;116(1-2):199–204. doi: 10.1016/s0925-4773(02)00134-x. [DOI] [PubMed] [Google Scholar]
- 19.Cebrià F, et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419(6907):620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, et al. Activin-binding protein from rat ovary is follistatin. Science. 1990;247(4944):836–838. doi: 10.1126/science.2106159. [DOI] [PubMed] [Google Scholar]
- 21.Fainsod A, et al. The dorsalizing and neural inducing gene follistatin is an antagonist of BMP-4. Mech Dev. 1997;63(1):39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto M, et al. Follistatin is a developmentally regulated cytokine in neural differentiation. J Biol Chem. 1992;267(11):7203–7206. [PubMed] [Google Scholar]
- 23.Bickel D, Shah R, Gesualdi SC, Haerry TE. Drosophila Follistatin exhibits unique structural modifications and interacts with several TGF-beta family members. Mech Dev. 2008;125(1-2):117–129. doi: 10.1016/j.mod.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iemura S, et al. Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc Natl Acad Sci USA. 1998;95(16):9337–9342. doi: 10.1073/pnas.95.16.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina MD, Saló E, Cebrià F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol. 2007;311(1):79–94. doi: 10.1016/j.ydbio.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Gaviño MA, Reddien PW. A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr Biol. 2011;21(4):294–299. doi: 10.1016/j.cub.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Y, Schneyer AL. The biology of activin: Recent advances in structure, regulation and function. J Endocrinol. 2009;202(1):1–12. doi: 10.1677/JOE-08-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8(5):635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132(4):661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, et al. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28(10):1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JR, et al. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol. 2008;313(1):107–117. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Bradham CA, et al. Chordin is required for neural but not axial development in sea urchin embryos. Dev Biol. 2009;328(2):221–233. doi: 10.1016/j.ydbio.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77(2):283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 34.Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77(2):273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 35.Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann’s organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8(3):401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 36.François V, Bier E. Xenopus chordin and Drosophila short gastrulation genes encode homologous proteins functioning in dorsal-ventral axis formation. Cell. 1995;80(1):19–20. doi: 10.1016/0092-8674(95)90446-8. [DOI] [PubMed] [Google Scholar]
- 37.Holley SA, et al. A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin. Nature. 1995;376(6537):249–253. doi: 10.1038/376249a0. [DOI] [PubMed] [Google Scholar]
- 38.Sasai Y, et al. Xenopus chordin: A novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79(5):779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rentzsch F, Guder C, Vocke D, Hobmayer B, Holstein TW. An ancient chordin-like gene in organizer formation of Hydra. Proc Natl Acad Sci USA. 2007;104(9):3249–3254. doi: 10.1073/pnas.0604501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3(3):327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10(3):299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapan SW, Reddien PW. dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet. 2011;7(8):e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mannini L, et al. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev Biol. 2004;269(2):346–359. doi: 10.1016/j.ydbio.2004.01.042. [DOI] [PubMed] [Google Scholar]
- 44.Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15(1):7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Mizuseki K, Kishi M, Shiota K, Nakanishi S, Sasai Y. SoxD: An essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron. 1998;21(1):77–85. doi: 10.1016/s0896-6273(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 46.Crémazy F, Berta P, Girard F. Sox neuro, a new Drosophila Sox gene expressed in the developing central nervous system. Mech Dev. 2000;93(1-2):215–219. doi: 10.1016/s0925-4773(00)00268-9. [DOI] [PubMed] [Google Scholar]
- 47.Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125(4):579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- 48.Lapan SW, Reddien PW. Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep. 2012;2(2):294–307. doi: 10.1016/j.celrep.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinstein DC, Hemmati-Brivanlou A. Neural induction. Annu Rev Cell Dev Biol. 1999;15:411–433. doi: 10.1146/annurev.cellbio.15.1.411. [DOI] [PubMed] [Google Scholar]
- 50.Cebrià F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132(16):3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- 51.Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA. PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development. 2012;139(6):1083–1094. doi: 10.1242/dev.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins JJ, III, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8(10):e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearson BJ, et al. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238(2):443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller CM, Newmark PA. An insulin-like peptide regulates size and adult stem cells in planarians. Int J Dev Biol. 2012;56(1-3):75–82. doi: 10.1387/ijdb.113443cm. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.