Abstract
Gram-negative bacteria have an outer membrane containing LPS. LPS is constituted of an oligosaccharide portion and a lipid-A moiety that embeds this molecule within the outer membrane. LPS is a pathogen-associated molecular pattern, and several pathogens modify their lipid-A as a stealth strategy to avoid recognition by the innate immune system and gain resistance to host factors that disrupt the bacterial cell envelope. An essential feature of Salmonella enterica Typhimurium pathogenesis is its ability to replicate within vacuoles in professional macrophages. S. Typhimurium modifies its lipid-A by hydroxylation by the Fe2+/α-ketoglutarate-dependent dioxygenase enzyme (LpxO). Here, we show that a periplasmic protein of the bacterial oligonucleotide/oligosaccharide-binding fold family, herein named virulence and stress-related periplasmic protein (VisP), on binding to the sugar moiety of peptidoglycan interacts with LpxO. This interaction inhibits LpxO function, leading to decreased LpxO-dependent lipid-A modifications and increasing resistance to stressors within the vacuole environment during intramacrophage replication promoting systemic disease. Consequently, ΔvisP is avirulent in systemic murine infections, where VisP acts through LpxO. Several Gram-negative pathogens harbor both VisP and LpxO, suggesting that this VisP-LpxO mechanism of lipid-A modifications has broader implications in bacterial pathogenesis. Bacterial species devoid of LpxO (e.g., Escherichia coli) have no lipid-A phenotypes associated with the lack of VisP; however, VisP also controls LpxO-independent phenotypes. VisP and LpxO act independently in the S. Typhimurium murine colitis model, with both mutants being attenuated for diverging reasons; ΔvisP is less resistant to cationic antimicrobial peptides, whereas ΔlpxO is deficient for epithelial cell invasion. VisP converges bacterial cell wall homeostasis, stress responses, and pathogenicity.
Keywords: QseC, chemical signaling
Bacterial–host relationships lead to different outcomes, ranging from beneficial to pathogenic interactions. Bacterial cells search for energy sources and niches for colonization within the host using either mutually beneficial or aggressive strategies that lead to pathogenesis. The host keeps these relationships balanced through the immune system, which is constantly surveying these interactions with microbes. Salmonella is a human pathogen responsible for food poisoning and typhoid fever (1). The pathogenesis of S. Typhimurium is a complex process. An essential feature of S. Typhimurium pathogenesis is its ability to replicate within vacuoles in macrophages (2).
During its evolution, S. Typhimurium acquired many pathogenicity islands (3). The main islands involved in S. Typhimurium infection are Salmonella pathogenicity island 1 (SPI-1) and SPI-2, both of which encode a type three secretion system (T3SS), a syringe-like apparatus used by the bacteria to inject effectors into the host cell. Both of these T3SSs are essential for S. Typhimurium virulence (4–8). The SPI-1 T3SS is required for invasion of the intestinal epithelium (4), whereas the SPI-2 T3SS is essential for S. Typhimurium replication and survival within macrophages and systemic murine infection (8–10). S. Typhimurium is a Gram-negative bacterium, and in addition to the expression of specialized apparatus such as T3SS, it also uses additional strategies to promote host infection (11). Gram-negative bacteria have LPS in their outer membrane. Although LPS is a pathogen-associated molecular pattern recognized by toll-like receptor 4 (TLR4) (12), several pathogens modify its lipid-A domain to either avoid recognition by the innate immune system and/or gain resistance to host factors that compromise the integrity of the bacterial cell envelope (11). S. Typhimurium expresses machinery to modify its lipid-A by addition or removal of acyl chains, addition of phosphoethanolamine or amino-arabinose, and hydroxylation by the Fe2+/α-ketoglutarate-dependent dioxygenase enzyme (LpxO) (11).
Bacterial pathogens rely on tightly regulated and complex signaling systems to express virulence traits. Virulence gene expression is an expensive process and should only occur within a suitable host. Hence, pathogens evolved to sense diverse environmental cues to promote host colonization (13). Such cues include the hormones epinephrine and norepinephrine that play a central role in stress responses in mammals (14). Stress responses have a profound effect on an organism’s physiology and survival, and many pathogens, including S. Typhimurium, use the Quorum-sensing Escherichia C (QseC) adrenergic sensor to activate virulence (15, 16). S. Typhimurium relies on QseC to cause systemic disease and activates its virulence repertoire when replicating in the spleen and liver (16).
Here, we show that a gene within the QseC regulon in S. Typhimurium encodes a periplasmic protein, renamed virulence and stress-related periplasmic protein (VisP), which binds to peptidoglycan and interacts with the LpxO–lipid-A–modifying enzyme inhibiting its function. This inhibition of LpxO-mediated lipid-A modifications is essential for resistance to intravacuolar stressors to promote intramacrophage replication and systemic disease. However, both VisP and LpxO have independent functions in colitis.
Results
VisP Is a Member of the QseC Regulon.
QseC plays an important role in S. Typhimurium pathogenesis by regulating transcription of SPI-1 and sifA (encodes an effector for the SPI-2 T3SS) genes (16). To gain a global view of the QseC regulon in S. Typhimurium, we performed transcriptomic studies comparing WT S. Typhimurium with a ΔqseC. These studies, as well as a previous report (17), identified the gene ygiW, encoded upstream of the qseBC operon, as a member of the QseC regulon (Fig. S1). YgiW (protein ygiW precursor E. coli) as cryptic gene function encodes a periplasmic protein of the bacterial oligonucleotide/oligosaccharide-binding fold (BOF) family (Figs. S1–S3) (18). BOF proteins (cl01196) have five antiparallel β-strands forming a closed or partly opened barrel, and six conserved pairs of aspartic acid or glutamic/aspartic acid residues are located within the last three β-strands preceding the C terminus (Fig. S2). BOF proteins are predicted to bind oligosaccharides (18), and the crystal structure of the Escherichia coli YgiW has been solved (Protein Data Bank ID code 1NNX). YgiW is prevalent in several bacterial species (Table 1 and Fig. S2), and it is involved in antimicrobial resistance in S. Typhimurium (19). The ΔygiW(ΔvisP) was more sensitive than WT to several stressors, such as cadmium chloride, acidic pH, and hydrogen peroxide (Fig. 1 A–D). Additionally, the ΔvisP grows poorly under low magnesium (MgCl2) conditions (20 μM), but its growth is similar to WT under high MgCl2 concentrations (200 μM) (Fig. 1E). S. Typhimurium is a facultative intracellular pathogen, and its adaptation to acidic pH, low magnesium (20 μM), and reactive oxygen species vacuolar conditions is essential for intramacrophage survival (2). The ΔvisP presented a four orders of magnitude decrease in intramacrophage survival compared with WT (Fig. 1F), congruent with its increased sensibility to vacuolar stressors. Because YgiW plays a role in stress resistance and virulence in S. Typhimurium, we renamed this protein VisP.
Table 1.
Distribution of VisP and LpxO among different bacterial species
| Bacterial species | VisP | LpxO |
| Erwinia spp | + | + |
| Pectobacteria spp | + | − |
| Pantoea spp | + | + |
| Enterobacter spp | + | + |
| Serratia spp | + | + |
| Brucella spp | − | + |
| Ralstonia spp | − | + |
| Salmonella spp | + | + |
| E. coli | + | − |
| Yersinia spp | + | + |
| Pseudomonas spp | + | + |
| Klebsiella pneumoniae | + | + |
| Burkholderia spp | − | + |
| Bordetella spp | − | + |
| Acinetobacter spp | − | + |
| Xylella spp | − | + |
| Haemophilus influenzae | + | − |
| Pasteurella multocida | + | − |
| Shigella spp | + | − |
| Citrobacter spp | + | − |
| Hafnia alvei | + | − |
Fig. 1.

VisP promotes resistance to stressors, virulence, and cell envelope remodeling in bacteria. (A) S. Typhimurium WT, ΔvisP, and complemented strains survival of CdCl2 (4 mg mL−1). (B) Survival of HCl (pH 5.0) (C) Survival of HCl (pH 2.5). (D) Survival of H2O2 (34 nmol L−1). (E) Bacterial growth under MgCl2 low (20 μM) and high (200 μM) concentrations. (F) Intramacrophage replication of S. Typhimurium WT, ΔvisP, ΔlpxO, ΔlpxO/visP, and complemented strains (+) in J774 macrophage-like cells. (G) LpxO-mediated modifications on the lipid-A chemical structure. (H) TLC of 32P-labeled lipid-A from E. coli K12 W3110 and WD101 (controls), S. Typhimurium WT, ΔvisP, ΔlpxO, ΔlpxO/visP, and complemented strains. Modifications assessed are aminoarabinose (l-Ara4N), 1-diphosphate (1-PP), phosphoethalonamine (pEtn), double lipid-A modifications (double), and LpxO-mediated dioxigenase (OH). W3110 was the E. coli strain used as a control to visualize lipid-A (1,4 bisphosphorylated) and 1-PP species; WD101 was the E. coli strain used as phosphoethalonamine and lipid-A double controls. *P < 0.01, **P < 0.001.
VisP and Lipid-A Modifications.
LPS modification is a common strategy used by many Gram-negative pathogens to survive stress responses (20). Of note, the LpxO enzyme is absent in E. coli, which explains the observations that an enterohemorragic E. coli (EHEC) ΔvisP is indistinguishable from WT concerning lipid-A modifications (Fig. S4), stress resistance (Fig. S4), and virulence [ability to express virulence genes and form attaching and effacing lesions on epithelial cells, a signature of EHEC gastrointestinal (GI) infection in mammals] (Fig. S4) phenotypes. S. Typhimurium expresses machinery to modify its lipid-A by addition or removal of several chemical groups, including hydroxylation by the LpxO enzyme (11). The ΔvisP presented significant differences in lipid-A modifications (Fig. 1H). Detailed analysis by MS (Fig. S5) identified that the differences in the ΔvisP were LpxO-mediated (Fig. 1 G and H and Fig. S5) (21), specifically an S-2-hydroxymyristate moiety dioxygenase modification, known to be an Fe2+/α-ketoglutarate–dependent dioxygenase homolog (21). These data indicate that VisP inhibits LpxO. To better elucidate the VisP/LpxO relationship, we generated ΔlpxO and ΔlpxO/visP strains. S-2-hydroxymyristate moiety dioxygenase modification to lipid-A was observed in the ΔvisP by the characteristically lower migration in separation by TLC (Fig. 1H) as well as a mass increase of ∼16 m/z units to the lipid-A analyzed by MS (Fig. S5). These effects were absent both in ΔlpxO and ΔlpxO/visP and restored on complementation (Fig. 1H and Fig. S4). Intramacrophage survival was also significantly decreased two orders of magnitude in ΔlpxO and ΔlpxO/visP, although not as drastically as in ΔvisP (Fig. 1F). VisP is a periplasmic protein, and LpxO is an inner membrane protein. Far Western blots using VisP-His and LpxO-Flag tagged proteins show that these proteins interact with each other but not with the QseB protein that was used as a negative control (Fig. 2 A–C). The interaction between VisP and LpxO was also independently confirmed through coimmunoprecipitation (Fig. S6B). Hence, these results suggest that VisP acts on LpxO by directly interacting with LpxO to modulate its function (Fig. 2 A–C and Fig. S6B).
Fig. 2.
VisP–LpxO interactions. (A) Control far Western blot of immobilized LpxO-Flag, VisPWT-His, VisPE110A-His, and QseB (untagged) probed with ΔvisPΔlpxO whole-cell lysate and then probed with anti-His antiserum. (B) Control far Western blot of immobilized LpxO-Flag, VisPWT-His, VisPE110A-His, and QseB (untagged) probed with ΔvisPΔlpxO whole-cell lysate and then probed with anti-Flag antiserum. (C) Far Western blot of immobilized LpxO-Flag, VisPWT-His, VisPE110A-His, and QseB (untagged) probed with ΔvisPΔlpxO expressing LpxO-Flag whole-cell lysate and then probed with anti-Flag antiserum. (D) Superposition of VisP modeled S. Typhimurium structure to the E. coli VisP structure (1NNX). E. coli structure is represented in red, S. Typhimurium is blue, three modified residues/structures are shown in yellow, and six site-directed point mutants are in green: VisPD88A, VisPD90A, VisPE110A, VisPD112A, VisPE119A, and VisPD121A. (E) Intramacrophage replication of ΔvisP complemented with WT VisP or the VisP point mutants in J774 macrophage-like cells. *P < 0.01.
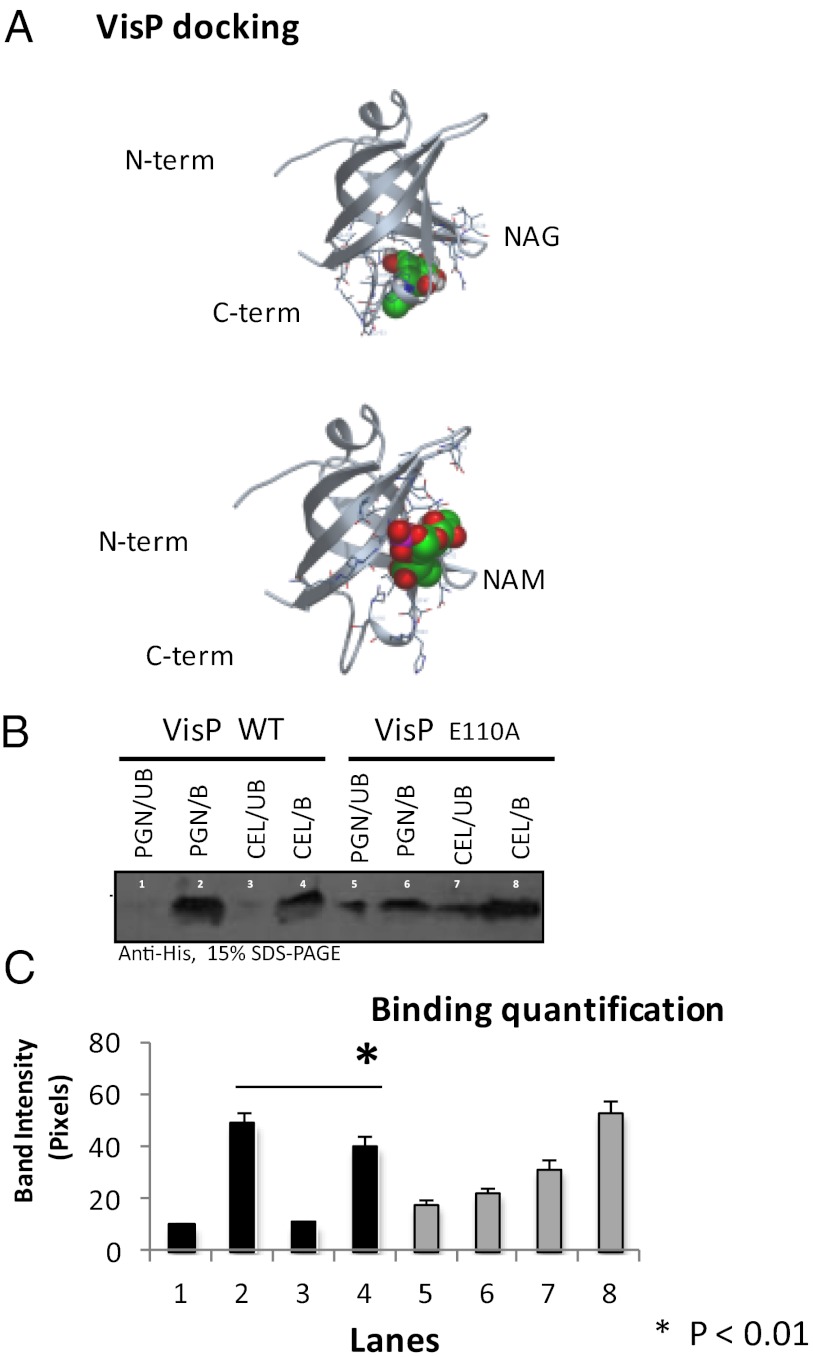
The crystal structure of the E. coli VisP is solved (Protein Data Bank ID code 1NNX), and the modeled S. Typhimurium VisP structure suggests that this protein is conserved in these two organisms and that the six amino acid residues (D88, D90, E110, D112, E119, and D121) of the BOF binding pocket are conserved between them (Fig. 2D). Because these six residues in the binding pocket are involved in ligand binding, which can modulate the function of these proteins (18), we performed site direct mutagenesis of these six residues and showed that E110 is critical for the role of VisP in intramacrophage replication (Fig. 2E). Of note, expression and stability of all six VisP point mutants are indistinguishable from the WT protein (Fig. S6C). The VisPE110A mutant is unable to rescue intracellular replication in ΔvisP, and the VisPD90A rescues this phenotype, albeit to a lesser extent than the other four mutants, where complementation restores intramacrophage replication to WT levels (Fig. 2E). BOF proteins bind to oligosaccharides, and a major source of oligosaccharides in the periplasmic space is peptidoglycan. Peptidoglycan is composed of repeating units of N-acetyl-glucosamine (NAG) and N-acetyl-muramic acid (NAM). In silico analysis of the modeled VisP docking to NAG and NAM (Fig. 3A and Dataset S1) (www.dockingserver.com) shows that binding of these sugars, specially for NAM, is near to the predicted binding pocket. The binding of VisP for carbohydrates was also assayed using insoluble peptidoglycan (PGN) and cellulose (Fig. 3B). VisP binding was evaluated by pull-down assays to these insoluble fractions. The VisPWT bound better to PGN than VisPE110A, whereas VisPE110A bound better to cellulose (Fig. 3B). These data show that VisP binds to peptidoglycan in the periplasmic space. Importantly, this binding seems to alter VisP function, given that the VisPE110A mutant, which has its ability to bind to PGN decreased, no longer interacts with LpxO (Fig. 2 A–C), and is defective for intramacrophage replication (Fig. 2D). Availability of peptidoglycan sugars in the periplasmic space enhances during cell wall remodeling, which occurs during environmental stress conditions, as well as during expression of T3SSs, which use peptidoglycan hydrolases during their assembly (22). Of note, T3SSs are used by S. Typhimurium to survive within macrophages (23).
Fig. 3.
VisP PG binding. (A) Docking modeling of S. Typhimurium VisP molecule against NAG and NAM. (B) Pull-down binding assays of purified VisPWT and VisPE110A with insoluble PGN or cellulose (CEL). B, bound fraction; UB, unbound fraction. (C) Quantification of pull-down binding assays (performed in triplicates). *P < 0.01.
VisP in S. Typhimurium Murine Systemic Disease.
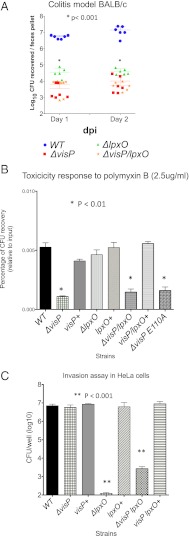
An in vivo systemic infection model (12) in BALB/c mice, (through an i.p. infection route) was used to assess virulence of VisP and LpxO individually and coupled. The ΔvisP mutant was avirulent in the systemic model, given that all animals survived infection by ΔvisP, whereas infection with WT led to 100% mortality by day 2 postinfection (Fig. 4A). Additionally, ΔvisP infected animals presented a decrease of four orders of magnitude in bacterial loads in spleens and livers compared with WT infected animals (Fig. 4B). The ΔlpxO and double ΔlpxO/visP were also attenuated, albeit to a lesser extent in these animal infections, compared with ΔvisP (Fig. 4). In the systemic model, VisP is dispensable for virulence if LpxO is absent, which is consistent with a negative regulatory role of VisP on LpxO function. These in vivo results are also congruent with the ability of these strains to replicate within macrophages, where ΔvisP replication is strikingly decreased, whereas ΔlpxO and the double mutant, although also decreased for intracellular survival compared with WT, are not as attenuated as ΔvisP (Fig. 1F). The ΔlpxO and the double mutant intermediary attenuation, compared with ΔvisP, suggests that absence of LpxO–lipid-A modifications hampers pathogenesis, but the complete deregulation of this phenotype (increased modification of lipid-A) in the absence of VisP has even more profound effects in pathogenesis. It has been proposed that LpxO–lipid-A modifications enhance S. Typhimurium membrane permeability, although a ΔlpxO presented permeability similar to WT (24), and it was also as resistant to detergent treatments as WT (Fig. S6A), suggesting that loss of LpxO does not have a profound effect in membrane permeability. Our data suggest that, although 2-hydroxymyristilation of lipid-A during intramacrophage replication leads to optimal resistance to intramacrophage stressors, enhancement of this modification in the absence of VisP inhibition of LpxO activity is deleterious to survival within these cells. These data suggest that VisP/LpxO-dependent remodeling of the bacterial cell wall within macrophages is an important feature of S. Typhimurium systemic infection (Fig. 4).
Fig. 4.
VisP and LpxO in S. Typhimurium systemic murine infection. (A) Survival curves of mice infected i.p. with S. Typhimurium [days postinfection (dpi)]. (B) Organ loads of mice infected i.p. with S. Typhimurium 20 h postinfection (hpi). *P < 0.001, **P < 0.01.
LpxO-Independent VisP Phenotypes.
Because S. Typhimurium also causes gastroenteritis, we assessed whether VisP and/or LpxO played any role in the S. Typhimurium colitis model (oral inoculation of BALB/c mice pretreated with streptomycin) (25). In contrast to the systemic murine infections, in the colitis model, all three mutants (ΔvisP, ΔlpxO, and ΔlpxO/visP) were attenuated three to four orders of magnitude compared with WT (Fig. 5A). These data indicate that, although both VisP and LpxO have important roles in S. Typhimurium infection of the GI tract, they may play independent roles in this host compartment. During GI infection, S. Typhimurium encounters cationic antimicrobial peptides, which are an important host defense in the GI tract (2). The visP mutant is more susceptible to cationic peptides (e.g., polymyxin B) (Fig. 5B) (19), whereas ΔlpxO presented similar susceptibility to WT (Fig. 5B). The double ΔlpxO/visP mutant also presented reduced resistance to polymyxin B, further supporting the hypothesis that VisP also performs LpxO-independent functions. The ΔvisP-increased susceptibility to polymyxin B could be rescued on complementation with WT VisP but not with the VisPE110A mutant, suggesting that this mutation in the VisP ligand binding pocket also affects LpxO-independent phenotypes (Fig. 5B). Although the increased susceptibility to cationic antimicrobial peptides could substantiate the attenuation of ΔvisP in the colitis model, it cannot explain the attenuation of ΔlpxO. However, another important facet of S. Typhimurium GI infection relies in the ability of this pathogen to invade intestinal epithelial cells (2). The ΔvisP mutant invades epithelial cells at similar levels to WT, whereas both ΔlpxO and ΔlpxO/visP have decreased ability (four to five orders of magnitude) to invade epithelial cells compared with WT (Fig. 5C). These data suggest that VisP is important for antimicrobial peptide resistance, whereas LpxO is required for efficient epithelial cell invasion by S. Typhimurium in the GI compartment.
Fig. 5.
VisP and LpxO in S. Typhimurium murine colitis. (A) CFUs of S. Typhimurium in feces days 1 and 2 postinfection (dpi). *P < 0.001. (B) Resistance to polymyxin B. *P < 0.01. (C) Invasion of HeLa cells. **P < 0.001.
Discussion
BOF proteins are present in a number of Gram-negative pathogenic bacterial strains and have unknown function. The association of genes encoding BOF proteins with prophage inserts and virulence plasmids suggests that these genes were inherited horizontally through mobile genetic elements, providing a mechanism for rapid evolution and adaptation to different niches. The BOF family lacks highly conserved amino acid residues involved in nucleotide binding, suggesting that these proteins can bind oligosaccharides but not nucleotides (18). All BOF proteins also harbor sec-dependent secretion signal sequences and are predicted to be periplasmic proteins (18). An important source of free saccharides within the periplasmic space, especially within stress environments or expression of T3SSs that induces remodeling of the bacterial cell wall, is peptidoglycan. Here, we have described that a previously uncharacterized BOF protein YgiW, herein renamed VisP, binds to the sugar moiety of peptidoglycan (Fig. 3). All BOF proteins have very conserved amino acid residues within their ligand binding pockets (18), and disruption of one of these residues (E110), although not altering protein expression/stability, alters its binding to peptidoglycan and prevents its function (Figs. 2, 3, and 5).
VisP binds to the inner membrane protein LpxO (Fig. 2), whose function is to promote an S-2-hydroxymyristate moiety dioxygenase modification on lipid-A (21). The GI pathogen S. Typhimurium harbors LpxO, and in addition to LpxO lipid-A modifications, it is also known to use a variety of other modifications to promote adaptations to different environments (11). Although LpxO-mediated lipid-A modifications have been exquisitely defined biochemically (21, 26), the biological function of this modification has remained a mystery. Here, we show that controlling the LpxO–lipid-A modifications is key for S. Typhimurium pathogenesis. VisP and LpxO mediate resistance to stressors and virulence in S. Typhimurium (Figs. 1, 2, 3, 4, and 5). VisP binds to PG. Decreased peptidoglycan (PG) binding alters the ability of VisP to interact with LpxO and LpxO function (Figs. 2 and 3), suggesting that VisP PG binding plays a role in decreasing LpxO-dependent lipid-A modifications to increase resistance to stressors within the vacuole environment during S. Typhimurium intramacrophage replication promoting systemic disease (Figs. 1, 2, 3, and 4).
However, VisP also has LpxO-independent functions (Fig. 5), suggesting that it could either have self-sufficient activities or engage with other protein partners. The distribution of VisP and/or LpxO varies among different Gram-negative species, with them either occurring together or by themselves (Table 1). Hence, these two proteins could perform synergistic or individual functions in niche/host adaptations, suggesting that they may have broader implications in bacterial pathogenesis. VisP and LpxO relationships converge bacterial cell wall homeostasis, stress responses, and pathogenicity. Importantly, expression of visP itself is regulated by host stress molecules (epinephrine and norepinephrine) sensed by the QseC bacterial adrenergic receptor (Fig. S1), further linking bacterial/host stress in modulation of pathogenesis. This study highlights the coevolution and the fundamental relationship between mammals and microbes, and it highlights that these signaling systems fully integrate bacterial and mammalian cell behavior.
Materials and Methods
Strains and Plasmids.
All strains and plasmids used in this study are listed on Table S1. Recombinant DNA and molecular biology techniques were performed as previously described (27). All oligonucleotides used are listed on Table S2.
Construction of the Isogenic Mutants.
Construction of isogenic nonpolar S. Typhimurium SL1344 visP and lpxO single mutants, visP/lpxO double KO, and EHEC ΔvisP (CP171) was achieved using λ-red mutagenesis (28). The visP mutant (CGM300) was complemented with the visP gene cloned into pBADMycHisA (Invitrogen) (20 copies per cell) vector, generating strain CGM301. The lpxO mutant (CGM302) was complemented with the lpxO gene cloned into pBAD33 vector (29) (15 copies per cell), generating strain CGM303. The visP/lpxO mutant (CGM304) was double complemented with pBADMycHisA and pBAD33c constructions, generating strain CGM305.
Microarray Analysis.
The microarray study compared the qseC mutant with WT in LB broth. The Salmonella arrays were performed as previously described (30). The Gene Expression Omnibus database accession number for the microarray results is GSE38353.
Quantitative Real-Time RT-PCR.
Quantitative RT-PCRs were performed as previously described (31).
Macrophage Infection.
Intramacrophage replication of S. Typhimurium in J774 murine macrophage-like cells was performed as previously reported (32–34).
HeLa Invasion and Adhesion Assays.
Invasion of HeLa cells was performed as previously reported (9, 24, 33–35).
Docking Assays.
Docking modeling was performed according to www.dockingserver.com instructions (36–39).
Structure Prediction of Salmonella VisP.
A structure for the BOF-containing fragment of Salmonella VisP was predicted using MODELER v9.10 (40, 41).
Protein Purification.
Performed according to PeriPreps kit manufacturer's instructions (EPICENTRE Biotechnologies). The His-tagged and Flag-tagged protein fractions were isolated as previously described (27), whereas the ANTI-FLAG M2 affinity gel (Sigma) was employed according to manufacturer's directions.
Far Western.
Same equimolar amounts of purified His-tagged protein were separated on an SDS gel and transferred to membranes. Replicate purified fractions were then probed with whole-cell lysates of the ΔvisP/lpxO double mutant or the double mutant overexpressing either Flag-tagged LpxO or His-tagged VisP. As another (negative) control, a replicate membrane was left unprobed by the whole-cell lysate. All membranes were then probed with either anti-His or anti-Flag primary antibodies and then incubated with a secondary antibody. ECL reagent (GE) was added, and membranes were exposed to film to detect interacting proteins.
PGN Binding Assays.
These binding assays were performed as previously described (42).
Stress Response Assays.
The resistance assays were performed in the presence of hydrogen peroxide and cadmium chloride (survival test; adapted from previous studies) (43). The acid resistance assay was performed as previously reported (44).
Polymyxin B Sensitivity Assay.
The sensitivity assays were performed as previously reported (32–34).
Site Direct Mutagenesis.
VisP site-directed mutagenesis was performed using the QuikChange II Site Directed Mutagenesis kit (Stratagene) using the manufacturer’s recommendations.
Isolation of Labeled Lipid-A.
Lipid-A isolations were performed as previously reported (45).
MS.
Lipid-A was isolated for analysis by MA as previously described (46). Lipid samples were analyzed using an MALDI-TOF/TOF (ABI 4700 Proteomics Analyzer) mass spectrometer in the negative ion linear mode as previously described (47).
Fluorescent Actin Staining Assay.
Fluorescent actin staining was performed as previously described (48).
Colitis Model and Systemic Infections with S. Typhimurium.
Mice (BALB/c, 7- to 9-wk old, female) were infected orally for the colitis model as previously described (49). The systemic infections were also performed with mice (BALB/c, 7- to 9-wk old, female) infected using an i.p. route as previously described (32).
Supplementary Material
Acknowledgments
We thank Lora Hooper for help with protocols and reagents and critique of this manuscript. This work was supported by National Institutes of Health Grants AI064184 (to M.S.T.), AI76322 (to M.S.T), UO1-AI053067 (to V.S.), and AI053067 (to V.S.) and the Burroughs Wellcome Fund (V.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38353).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215416110/-/DCSupplemental.
References
- 1.Boyle EC, Bishop JL, Grassl GA, Finlay BB. Salmonella: From pathogenesis to therapeutics. J Bacteriol. 2007;189(5):1489–1495. doi: 10.1128/JB.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6(1):53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 3.Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. Salmonella pathogenicity islands: Big virulence in small packages. Microbes Infect. 2000;2(2):145–156. doi: 10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 4.Galán JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86(16):6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groisman EA, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12(10):3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galán JE. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20(2):263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 7.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93(6):2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93(15):7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30(1):175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 10.Hensel M, et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30(1):163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 11.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 13.Hughes DT, Sperandio V. Inter-kingdom signalling: Communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6(2):111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: The language of hormones. Proc Natl Acad Sci USA. 2003;100(15):8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasko DA, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321(5892):1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78(3):914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merighi M, et al. Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol. 2009;9:42. doi: 10.1186/1471-2180-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginalski K, Kinch L, Rychlewski L, Grishin NV. BOF: A novel family of bacterial OB-fold proteins. FEBS Lett. 2004;567(2–3):297–301. doi: 10.1016/j.febslet.2004.04.086. [DOI] [PubMed] [Google Scholar]
- 19.Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J Bacteriol. 2009;191(23):7243–7252. doi: 10.1128/JB.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12(4):205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons HS, Lin S, Cotter RJ, Raetz CR. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem. 2000;275(42):32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- 22.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 23.Hensel M, et al. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269(5222):400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 24.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189(20):7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hapfelmeier S, et al. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72(2):795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons HS, Reynolds CM, Guan Z, Raetz CR. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella lipid A. Biochemistry. 2008;47(9):2814–2825. doi: 10.1021/bi702457c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 28.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson AR, et al. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell Host Microbe. 2011;10(1):33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walters M, Sperandio V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun. 2006;74(10):5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol. 2003;48(2):385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 33.Fierer J, et al. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect Immun. 1993;61(12):5231–5236. doi: 10.1128/iai.61.12.5231-5236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun. 1999;67(11):5690–5698. doi: 10.1128/iai.67.11.5690-5698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finlay BB, Ruschkowski S, Dedhar S. Cytoskeletal rearrangements accompanying salmonella entry into epithelial cells. J Cell Sci. 1991;99(Pt 2):283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 36.Hazai E, Kovács S, Demkó L, Bikádi Z. DockingServer: Molecular docking calculations online. Acta Pharm Hung. 2009;79(1):17–21. [PubMed] [Google Scholar]
- 37.Halgren TA. Merck molecular force field. I. Basis, form, scope, parametrization, and performance of MMFF94. J Comput Chem. 1998;17(5–6):490–519. [Google Scholar]
- 38.Morris GM, et al. Automated docking using a Lamarckian genetic algorithm and and empirical binding free energy function. J Comput Chem. 1998;19(14):1639–1662. [Google Scholar]
- 39.Solis FJ, et al. Minimization by random search techniques. Math Oper Res. 1981;6(1):19–30. [Google Scholar]
- 40.Eswar N, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;5(Oct):5.6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martí-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 42.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang XS, García-Contreras R, Wood TK. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol. 2007;189(8):3051–3062. doi: 10.1128/JB.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol. 2003;48(3):699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274(26):18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 46.Tran AX, et al. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol. 2006;188(12):4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hankins JV, et al. Elucidation of a novel Vibrio cholerae lipid A secondary hydroxy-acyltransferase and its role in innate immune recognition. Mol Microbiol. 2011;81(5):1313–1329. doi: 10.1111/j.1365-2958.2011.07765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knutton S, Baldwin T, Williams PH, McNeish AS. New diagnostic test for enteropathogenic Escherichia coli. Lancet. 1988;1(8598):1337. doi: 10.1016/s0140-6736(88)92152-6. [DOI] [PubMed] [Google Scholar]
- 49.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71(5):2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.