Abstract
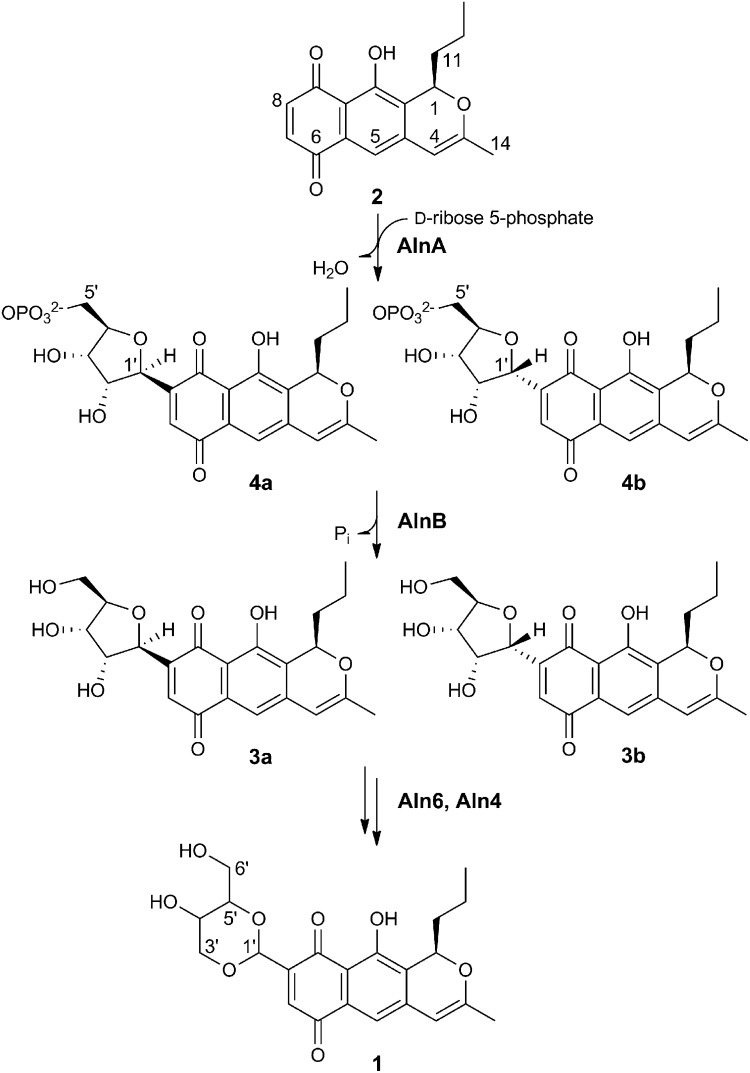
Alnumycin A is an exceptional aromatic polyketide that contains a carbohydrate-like 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane moiety attached to the aglycone via a carbon−carbon bond. Recently, we have identified the D-ribose-5-phosphate origin of the dioxane unit and demonstrated that AlnA and AlnB are responsible for the overall C-ribosylation reaction. Here, we provide direct evidence that AlnA is a natural C-glycosynthase, which catalyzes the attachment of D-ribose-5-phosphate to prealnumycin by formation of the C8−C1′ bond as demonstrated by the structure of the intermediate alnumycin P. This compound is subsequently dephosphorylated by AlnB, an enzyme of the haloacid dehalogenase superfamily. Structure determination of the native trimeric AlnA to 2.1-Å resolution revealed a highly globular fold encompassing an α/β/α sandwich. The crystal structure of the complex with D-ribose-5-phosphate indicated that the phosphosugar is bound in the open-chain configuration. Identification of residues E29, K86, and K159 near the C-1 carbonyl of the ligand led us to propose that the carbon−carbon bond formation proceeds through a Michael-type addition. Determination of the crystal structure of the monomeric AlnB in the open conformation to 1.25-Å resolution showed that the protein consists of core and cap domains. Modeling of alnumycin P inside the cap domain positioned the phosphate group next to a Mg2+ ion present at the junction of the domains. Mutagenesis data were consistent with the canonical reaction mechanism for this enzyme family revealing the importance of residues D15 and D17 for catalysis. The characterization of the prealnumycin C-ribosylation illustrates an alternative means for attachment of carbohydrates to natural products.
Keywords: enzyme mechanism, natural product biosynthesis, protein structure
Many important natural products are decorated with carbohydrate moieties, and these include all of the major metabolite classes such as the polyketides, nonribosomal peptides, terpenoids, and flavonoids (1–3). The carbohydrate units are often essential for the biological activity and solubility of the compounds (1). For instance, many compounds in clinical use, such as the antibiotic erythromycin (4) and the anthracycline anticancer agent doxorubicin (5), owe their activities to deoxysugar units. These observations have generated considerable interest in the scientific community toward research on the biosynthesis, engineering, and attachment of deoxysugars onto natural products. Moreover, studies have revealed that the carbohydrate moieties are modified as 4-keto-6-deoxy-D-hexose nucleoside diphosphates before attachment by glycosyl transferases (GTs) (6, 7).
The 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane unit of the aromatic polyketide alnumycin A (1; Fig. 1), which is produced among others by Streptomyces sp. CM020, presents an unconventional solution for formation of carbohydrate-like structures in natural products. Intriguingly, the unit is connected via a C8–C1′ bond to the prealnumycin (2; Fig. 1) chromophore (8, 9). This mode of attachment is in contrast to the majority of GTs that most often attach carbohydrates through an O-glycosidic bond but that are then susceptible to hydrolysis in the acidic environment of the stomach or by the action of glycosidases in the small intestine (10). The more chemically stable C-glycosylated metabolites (11) are, by comparison, rare and only found in selected natural products (12, 13), such as the polyketide antibiotic urdamycin (14).
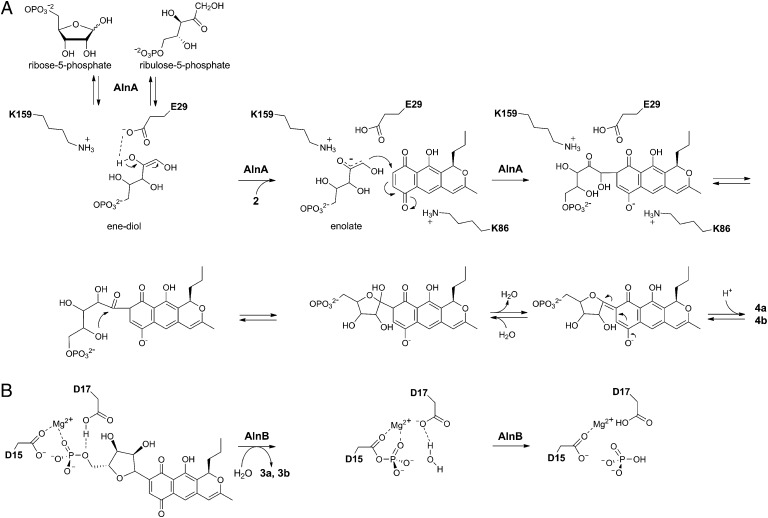
Fig. 1.
Model pathway for the biosynthesis of the carbohydrate-like 4′-hydroxy-5′-hydroxymethyl-2′,7′-dioxane. The structures of alnumycin A (1), prealnumycin (2), alnumycin C1 (3a), C2 (3b), P1 (4a), and P2 (4b) are shown.
Recent studies have established that the biosynthesis of 1 proceeds through C-ribosylation of 2 by the concerted action of two enzymes: AlnA and AlnB (15, 16). When the two enzymes were incubated with 2 and D-ribose-5-phosphate, alnumycin C (3; Fig. 1) could be isolated from the reaction mixture (16). The formation of the C8−C1′ bond was annotated to AlnA, which is homologous to pseudouridine C-glycosidases such as YeiN (17) and TM1464 (17, 18) from Escherichia coli and Thermotoga maritima, respectively, whereas AlnB, which is a predicted member of the haloacid dehalogenase (HAD) superfamily (19, 20), was thought to be responsible for dephosphorylation of the attached D-ribose-5-phosphate unit.
In this paper, we established the functions of the two enzymes through enzymatic synthesis of a phosphorylated intermediate and stepwise characterization of the reactions catalyzed by AlnA and AlnB. Furthermore, to understand the molecular basis for the C-ribosylation reaction, we determined the structures of the two enzymes by protein crystallography. Finally, structure/function studies of the enzymes enabled us to propose a mechanistic model for the unusual attachment of carbohydrates to natural products.
Results and Discussion
Identification of the Functions of AlnA and AlnB.
When AlnA was incubated together with 2 (0.5 mM) and a 150-fold excess of D-ribose-5-phosphate (75 mM), a previously undetected compound was observed by HPLC (Fig. S1). The product was confirmed as a true intermediate of the pathway, because it was readily converted in an independent reaction further to 3 by AlnB (Fig. S1). Despite the poor chromatographic behavior and instability of the compound, multiple enzymatic reactions provided sufficient material for structure elucidation by high resolution electrospray ionization mass spectrometry (HR-ESI-MS) and both 1H and 31P NMR (SI Text). The data ascertained that the product was in fact an epimeric pair of two unique compounds differing in their stereochemistry at C-1′, denoted as alnumycin P1 (4a, major isomer 84%; Fig. 1) and P2 (4b, 16%; Fig. 1), which contained D-ribose-5-phosphate attached to the expected C-8 position of 2. The experiments therefore confirmed unequivocally the functions of the two enzymes: AlnA is a natural C-glycosynthase (21) responsible for formation of the C8−C1′ bond, whereas AlnB is a phosphatase in accordance to the function predicted by the sequence analysis (20). Kinetic characterization of AlnA (Fig. S2) revealed that the rate of the reaction was modest in regard to utilization of the cosubstrates D-ribose-5-phosphate (apparent Km of 74 µM and kcat of 0.58 s−1) and prealnumycin (apparent Km of 39 µM and kcat of 0.37 s−1).
Structure of the Prealnumycin C-Glycosynthase AlnA.
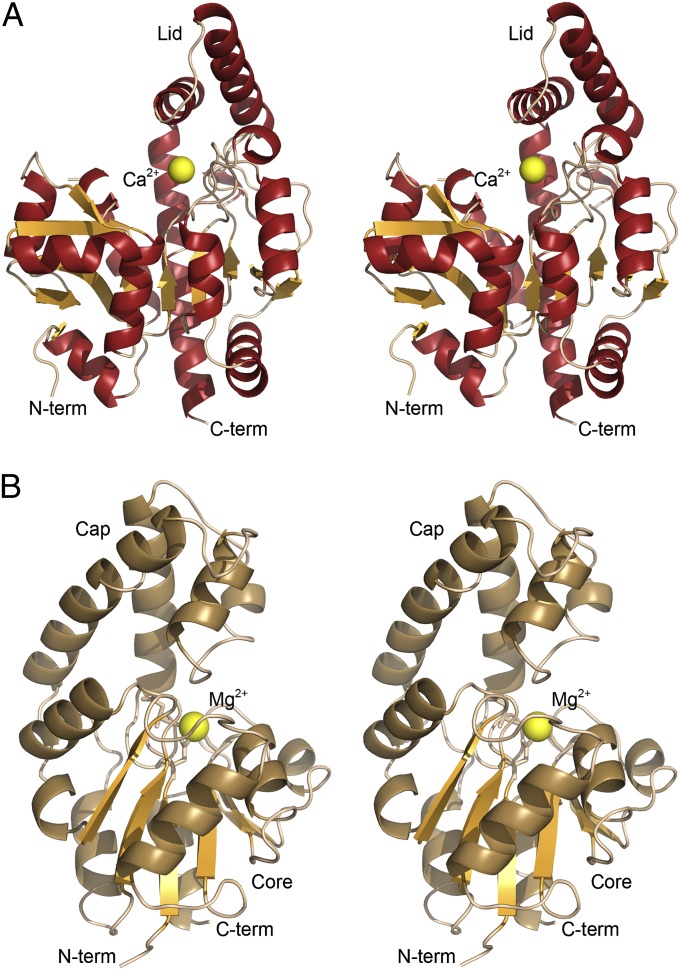
The structure of the 32.9-kDa AlnA was determined by molecular replacement using the pseudouridine glycosidase TM1464 (18) as a search model and refined to 2.1-Å resolution. The quality of the electron density map and refinement statistics (Table S1) were in good agreement with the obtained resolution, and the electron density map could be traced for most of the polypeptide chain. One surface loop region comprising residues 37 and 38 was less well defined, and only the Cα backbone could be traced with confidence through a helix-turn-helix motif containing residues 251−261. AlnA is a trimer both in solution and in the crystal structure based on gel filtration experiments and calculated crystal contacts, respectively (Fig. S3).
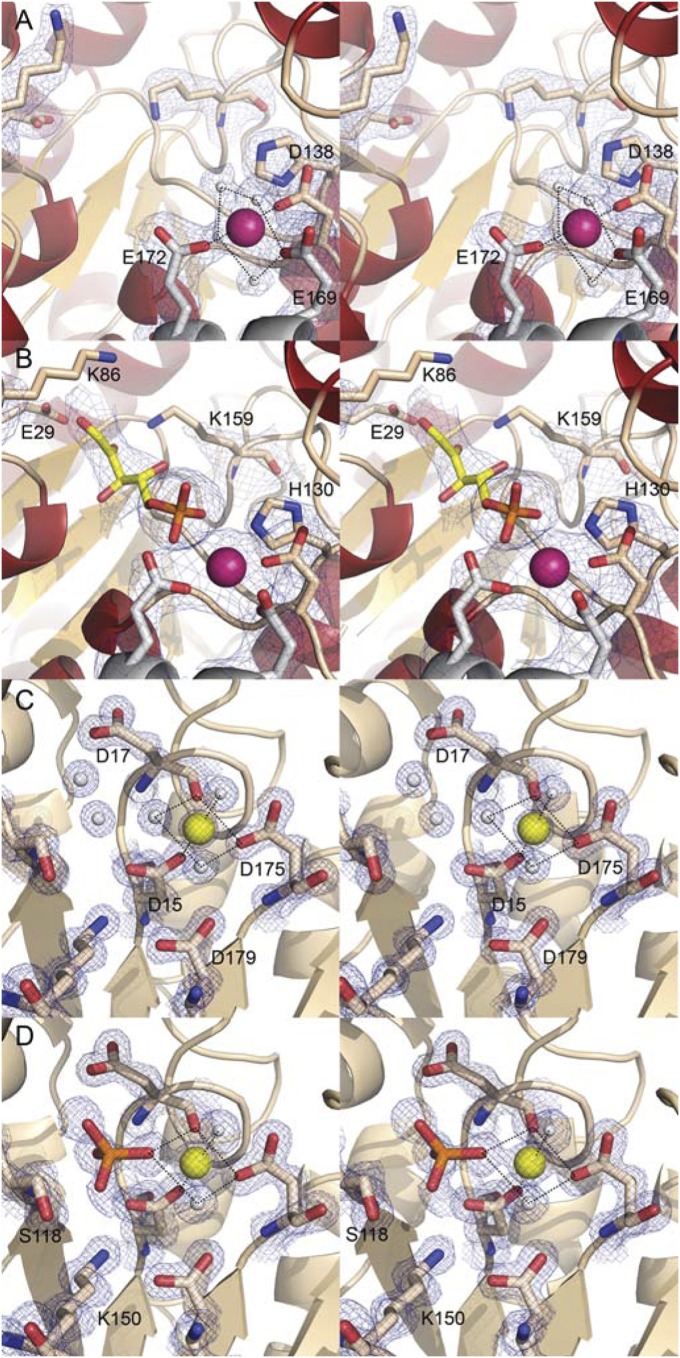
Overall, AlnA folds into a α/β/α domain, which is dominated by a twisted, nine-stranded mixed β-sheet (Fig. 2A). One of these β-strands is very elongated and mediates contacts with two other β-strands that lie on top of the central β-sheet. Four and three α-helices, respectively, pack against each side of this β-sheet. A structural similarity search performed using the DALI server (22) indicated that AlnA is significantly similar in structure and fold only to the pseudouridine glycosidase TM1464 (18) with a root mean square deviation (RMSD) of 2.0 Å (Z-score, 37.3). A discernible feature in the structure is a long helix-turn-helix motif, which forms a lid for a deep crevice situated perpendicular to the central β-sheet (Fig. 2A). These helices lie in a tilted angle, which leaves the putative active site open to bulk solvent. The crevice spans nearly the whole width of the polypeptide, measuring 24 Å from a metal ion situated at the front of the active site to the C-terminal helix that forms the back end. Based on the electron density and the pentagonal bipyramidal configuration, the metal ion is likely to be calcium, taken up from the crystallization conditions. The coordination to the Ca2+ is formed through four H2O molecules, D138 from the same subunit and two residues, E169 and E172, from the neighboring polypeptide chain (Fig. 3A). Difference electron density in the putative active site of AlnA was observed on soaking of crystals with 100 mM D-ribose-5-phosphate (Table S1). The electron density could be best fitted with a model of D-ribulose-5-phosphate in a linear configuration, which is an alternative cosubstrate for AlnA (16). However, at the medium resolution of the electron density map, the differences between models of linear D-ribulose-5-phosphate and cyclic D-ribose-5-phosphate are minor (Fig. S4). Therefore, to obtain supporting evidence for our interpretation, we next performed binding experiments by NMR, which demonstrated that D-ribulose-5-phosphate, which only exists in a linear configuration, does indeed bind to AlnA (Fig. S5). Finally, the structure of the related pseudouridine glycosidase TM1464 contained an unidentified linear phosphosugar “larger than glycerol-3-phosphate with density extending beyond the carbon 1 atom” (18), which is thus in line with our structural analysis. The crystal structure revealed that the phosphate group of the ligand formed contacts with the calcium ion and residues H130 and S140, whereas the C-1 hydroxyl and C-2 carbonyl groups of D-ribulose-5-phosphate are within hydrogen bonding distance to E29 and K159, respectively (Fig. 3B). Residues E29, K86, and K159 form a putative triad, which could be involved in the catalytic reaction (see below).
Fig. 2.
Schematic views of AlnA and AlnB. Stereoview images of the overall structures of (A) AlnA and (B) AlnB.
Fig. 3.
Molecular architectures of the active sites of AlnA and AlnB. Stereo views of (A) native AlnA at 2.1-Å resolution (σ = 1.2), (B) AlnA in complex with D-ribose-5-phosphate (yellow) at 3.15-Å resolution (σ = 1.2), (C) native AlnB in the open conformation at 1.25-Å resolution (σ = 2.0), and (D) AlnB in complex with phosphate (orange) at 1.5-Å resolution (σ = 1.5). The ligands were omitted from calculation of the electron density maps (2Fo-Fc) to reduce model bias. The metal ions Ca2+ (purple) and Mg2+ (yellow) and water molecules (white) are shown as spheres.
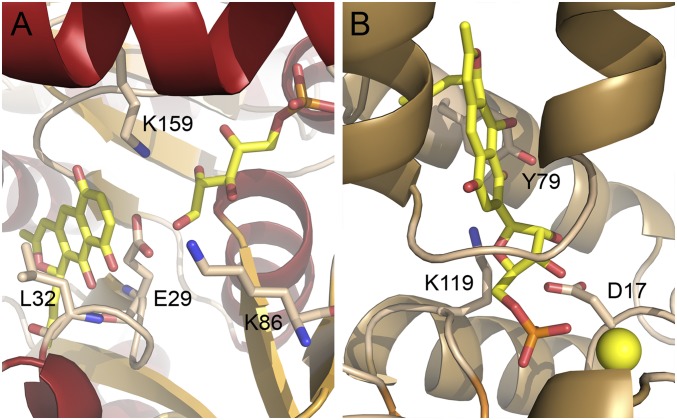
The back end of the active site is lined with several hydrophobic residues, e.g., L28, L32, I45, and L285, which is consistent with the predominantly hydrophobic nature of the polyaromatic cosubstrate 2. The ligand was computationally docked into the AlnA structure (Fig. 4A), because experimental procedures were hindered either by degradation of the ligand in cocrystallization and loss of crystal diffraction after soaking experiments. The calculations positioned the propyl side chain of the polyketide in a hydrophobic pocket at the bottom of the binding site and the backbone amide nitrogen of L32 was within hydrogen bonding distance to the C-9 quinone oxygen and the C-10 hydroxyl group. The reactive C-8 carbon of 2 could thus be docked near the C-1 group of D-ribose-5-phosphate.
Fig. 4.
Docking of (A) prealnumycin (2) into the active site of AlnA containing bound D-ribose-5-phosphate and (B) alnumycin P1 (4a) inside the four-helix bundle of AlnB.
Structure of the Alnumycin P Phosphatase AlnB.
The structure of the 24.1-kDa AlnB was determined by molecular replacement using the core domain of a putative phosphatase of unknown function from Haemophilus somnus (PDB ID code 2HSZ) as a search model. The asymmetric unit of the orthorhombic crystal form contained a single AlnB molecule, and gel filtration results suggested that AlnB was also a monomer in solution. The excellent quality of the electron density map (Fig. 3C) was in good agreement with the 1.25-Å high-resolution diffraction data (Table S1), and the electron density was continuous for the whole protein apart from 12 N-terminal residues, including the His-tag, and the last two C-terminal residues.
The AlnB monomer is comprised of two distinct domains (Fig. 2B): a helical cap domain consisting of a four-helix bundle and an α/β core domain. The core domain is composed of a centrally located six-stranded parallel β-sheet, which is surrounded by three and four α-helices on either side of the β-sheet. In general, the core domain forms a modified Rossmann fold and is similar to other members of the HAD family, where the major differences arise from variation in the size and shape of the cap domain (20). A DALI search revealed that AlnB was most similar to a predicted phosphatase from Clostridium acetobutylicum (PDB ID code 2MC1) with an RMSD of 2.4 Å (Z-score, 27.3). The electron density observed for a metal ion, which is likely to be Mg2+ taken up from the crystallization media, at the junction of the cap and core domains was in line specifically with findings from related HAD phosphatases (23).
The canonical active site in HAD enzymes is formed between the core and cap domain interphase, where four loop regions from the core domain protrude providing a platform for binding of the Mg2+ ion and the phosphate group of the ligand (19). In AlnB, the octahedral coordination geometry to the metal cofactor is generated via three water molecules, the carboxylate groups of D15 and D175, and the carbonyl oxygen of D17 (Fig. 3C). The complex structure of AlnB with free phosphate at 1.5-Å resolution revealed the expulsion of three water molecules on ligand binding and hydrogen bonds of the ligand phosphate group to S118 and K150 in addition to coordination to the Mg2+ ion (Fig. 3D).
Although the core domain provides catalytically important residues, the cap domain is mainly responsible for substrate recognition and is more varied in HAD enzymes. In AlnB, the putative substrate binding pocket within the four-helix cap domain is surprisingly small, considering the structure of the substrate 4. However, the cavity is lined mostly with hydrophobic residues such as I27, I47, L48, V51, and L55, which are suitable for binding of the polyaromatic substrate. The only nonhydrophobic exception that may be important for substrate recognition is K119, which protrudes inside the four-helix bundle from the core domain. The ligand could be computationally docked inside this four-helix bundle (Fig. 4B), which positioned the phosphate group of 4a next to the Mg2+ ion in the same position as in the ternary complex with free phosphate.
In Vitro Mutagenesis and Implications for the Catalytic Mechanisms.
AlnA catalyzes an unusual reaction: the attachment of D-ribose-5-phosphate to the polyketide aglycone via a C8−C1′ bond. The protein bears no similarity in sequence or fold to glycosyl transferases, and moreover, it does not use nucleoside diphosphates for catalysis, which indicates that the reaction mechanism is likely to differ from C-glycosyl transferases (7). To date, no mechanistic studies have been conducted for this enzyme family, and therefore several residues in the active site were probed by mutagenesis (Table 1) to gain insight into the C-glycosynthase reaction.
Table 1.
Activity of AlnA and AlnB variants determined with the Malachite green assay
| Variant | Released PO4 (nmol) | Activity (%) |
| AlnA | ||
| Native | 9.2 ± 0.4 | 100 |
| E29A | 0.39 ± 0.51 | 4.2 |
| E29Q | 0.05 ± 0.49 | 0.5 |
| K86A | 0.48 ± 0.51 | 5.2 |
| H130A | 1.1 ± 0.5 | 12 |
| D138A | 1.1 ± 0.3 | 12 |
| K159A | 0.35 ± 0.52 | 3.8 |
| K159R | 0.39 ± 0.51 | 4.2 |
| AlnB | ||
| Native | 1.2 ± 0.1 | 100 |
| D15A | 0.04 ± 0.02 | 3.3 |
| D15N | 0 | 0 |
| D17A | 0.02 ± 0.01 | 1.7 |
| Y79A | 0.66 ± 0.06 | 55 |
| K119A | 0.05 ± 0.03 | 4.2 |
| K119R | 0.02 ± 0.01 | 1.7 |
The coupled AlnA–AlnB reactions were performed in triplicate, with each individual sample assayed in duplicate, and the value for the blank reaction was subtracted. SD for each assay result is shown in SI Text.
Recent studies have shown that AlnA may use both D-ribose-5-phosphate and D-ribulose-5-phosphate as cosubstrates (16). We propose that the reaction catalyzed by AlnA might proceed through a Michael-type 1,4-addition and that the reactive phosphosugar species would be the linear ene-diol intermediate, which is deprotonated in the initial step at the C-2′ hydroxyl group by E29 and stabilized by the nearby K159 (Fig. 5A). In effect, incubation of AlnA in the presence of D-ribose-5-phosphate alone led to conversion of the substrate to D-ribulose-5-phosphate (Fig. S6), which is consistent with our mechanistic model as such isomerization has been proposed to occur via an ene-diol species (24). The slow rate of the conversion suggests that the riboisomerase activity of AlnA is not a true catalytic activity, but rather a secondary moonlighting effect caused by the release of the ene-diol species in the absence of the cosubstrate 2. The next step would be a nucleophilic attack of the enolate onto the aromatic carbon at C-8. The developing negative charge at the quinone oxygen in the transition state could be stabilized by K86. Residues E29, K86, and K159 appear to be critical for the reaction as demonstrated by the mutagenesis data (Table 1) and suggested by the conservation of equivalent residues in other members of this enzyme family (Fig. S7).
Fig. 5.
Schematic representations of the reactions catalyzed by (A) AlnA and (B) AlnB.
After formation of the C8−C1′ bond, an intermediate suitable for formation of a cyclic hemiketal may be obtained via tautomerization. Subsequent dehydration of the hemiketal and restoration of the polyaromatic chromophore would then lead to the endproducts of the reaction (Fig. 5A). However, the discovery of the two C-1′ epimeric products 4a and 4b of the AlnA reaction suggests that the final cyclization might also occur nonenzymatically. The narrow shape of the active site yields support for this, because it is unlikely that the attached D-ribose-5-phosphate could fold onto itself in the confined space. The benefit of this proposal is that it may also be used to explain the formation of the pathway shunt product alnumycin D (Fig. S8), which contains ribose attached in pyranose form. The considerable enzymatic activity of the H130A (coordination to D-ribose-5-phosphate) and D138A (coordination to the Ca2+ ion) implies that the metal ion is mainly used for binding of the phosphosugar cosubstrate and not in the actual catalytic reaction. This observation is supported by the tolerance of the pseudouridine synthase YeiN toward various divalent metal ions (17).
In contrast to the complex reaction catalyzed by AlnA, the formation of 3 from 4 by AlnB is more straightforward to decipher. The loss of activity through mutagenesis of the conserved aspartic acid residues D15 and D17 imply that the reaction involves canonical nucleophilic chemistry combined with acid/base catalysis (19, 25). In the initial step, the phosphoryl group is likely to be transferred from 4 to D15 of AlnB, producing a labile phospho-aspartyl-enzyme intermediate (Fig. 5B). The dual role of D17 would be to initially act as a general acid to facilitate the release of the product 3 by protonation. In the second step, the same aspartic acid is thought to act as a general base activating a nucleophilic water molecule, which is the ultimate acceptor of the phosphotransfer reaction (19). The decreased activities of K119A and K119R support our docking experiments and suggest that the four-helix bundle is used for substrate recognition. The moderate decrease in activity on the Y79A replacement, however, indicates that this residue is not critical in substrate binding. In general, the overall structure (Fig. 2A), the mutagenesis data (Table 1), and the conservation of characteristic sequence motifs (Fig. S9) provide compelling evidence that AlnB is likely to function in a manner similar to the classical phosphatases of the HAD enzyme family (19, 25).
Concluding Remarks
The vast majority of natural product glycosylations performed by GTs proceed through an O-glycosidic linkage (1). In selected cases, the enzymes have been reported to be capable of forming a hydrolytically resistant C-glycosidic bond (14), but this has been rare by comparison. Here we clarified the structural basis for an alternative means for C-ribosylation found on the biosynthetic pathway of alnumycin A that is mechanistically distinct from GTs. We have shown that the two-step process involves a Michael-type 1,4-addition of D-ribose-5-phosphate to the polyketide aglycone catalyzed by AlnA, which is followed by dephosphorylation of the intermediate alnumycin P by AlnB. The study suggests further that the source of the stereochemical diversity of alnumycins may originate from nonenzymatic cyclization of the AlnA reaction product. The identification of the amino acid residues important for catalysis and substrate recognition for both enzymes paves the way for structure-based protein engineering attempts for expansion of the substrate specificity of the enzymes and the generation of novel C-ribosylated metabolites.
Materials and Methods
Crystallography.
The crystallization of the proteins is described in SI Text. The X-ray data obtained at the European Synchrotron Radiation Facility (Grenoble, France) was processed with the program MOSFLM (26), and the data were scaled using the program SCALA from the CCP4 package (27). Five percent of the reflections, with the same set selected for the native and complex structures, were excluded for calculation of Rfree. Statistics for the data collection are given in Table S1. The structures of AlnA and AlnB were solved by molecular replacement using the program MOLREP (28) in the CCP4 package. For native AlnA, a homologous protein from T. maritima (PDB access code 1VKM) was used as a search model, whereas for native AlnB, the BALBES (29) server was used to construct several molecular replacement templates, from which the core domain of a phosphatase of unknown function from Haemophilus somnus (PDB ID code 2HSZ) yielded the correct solution. For the complex structures, the corresponding native structure was used as a search model. The native data for AlnA and AlnB were submitted for automated model building by ARP/wARP (30), and all structures were finalized by several rounds of manual building in the program COOT (31) and refinement using REFMAC (32). Structure validation was done with the MolProbity web server (33). The figures depicting protein structures were prepared using PyMol (The PyMOL Molecular Graphics System, Version 1.3; Schrödinger).
Docking Calculations.
The PRODRG-server (34) was used to prepare models of the substrates. The ligands were docked in place using the program GOLD (35) integrated into the SOMA2 (36) modeling environment at the CSC–IT Center for Science Ltd (Espoo, Finland). Water molecules were removed from the active sites of the proteins, and several alternative side-chain conformations were manually tested to improve the computational calculations. For docking of 2 into the active site of AlnA in complex with D-ribose-5-phosphate, the conformation of the side-chain of L32 was altered to enable positioning of the propyl side chain of 2 into a hydrophobic pocket at the back of the active site. For docking of 4a into the active site of AlnB, the side-chains of I27, V51, and L59 were altered to allow sufficient space for the ligand inside the cap domain.
In Vitro Mutagenesis and Enzyme Activity Assays.
The AlnA and AlnB mutants were generated by PCR mutagenesis using the four-primer method as described in SI Text. The oligonucleotides used are listed in Table S2. The constructs were cloned into the modified pBADHisB (Invitrogen) vector (37) and verified by DNA sequencing. All variants were produced and purified to at least 95% purity as described previously for AlnB (16), except the D15A variant of AlnB, which required an additional gel filtration step in the storage buffer with a HiLoad 26/60 Superdex 200 column as part of an ÄKTA FPLC system (GE Healthcare), resulting in ∼85% purity.
The activity measurements were conducted in coupled assays that were set up in 12.5 mM Hepes (pH 7.2), 37.5 mM NaCl, 1.25 mM MgCl2, and 27% (vol/vol) glycerol as described above, with the exception that the substrate concentrations were 1 mM D-ribose-5-phosphate and 0.4 mM prealnumycin (2) in 4% (vol/vol) DMSO. The 60 μM AlnA variants and 30 μM AlnA were incubated with 3.75 μM AlnB for 4 h, and the 300 nM AlnB and variants with 40 μM AlnA for 90 min at 288 K, followed by extraction with one to two volumes of CHCl3. The reactions were monitored as endpoint assays through formation of free phosphate in the aqueous phase as detected by use of the Malachite green dye and formation of alnumycin C (3) in the organic phase by HPLC. Because both methods yielded comparable results, final measurements were conducted in triplicate and were based on detection of inorganic phosphate (Table 1). Determination of the initial velocities of the reactions for kinetic characterization of AlnA was accomplished through time-course analysis of formation of inorganic phosphate using endpoint measurements. The kinetic measurements were conducted at 288 K in solutions composed of 50 mM Trizma-HCl (pH 7.2), 50 mM NaCl, 10% (vol/vol) glycerol, 4% (vol/vol) DMSO, 40 µM AlnA, and 1 µM AlnB. The titrations were conducted in 7.5–1,000 µM D-ribose-5-phosphate in 100 µM prealnumycin and 10–500 µM prealnumycin in 1 mM D-ribose-5-phosphate. The oxygen concentration was reduced as described above.
Analysis of Alnumycin P1 and P2.
The AlnA reactions were conducted in solutions composed of 10 mM Trizma-HCl (pH 6.8), 25 mM NaCl, 25 mM KCl, ≥10% (vol/vol) glycerol, 75 mM D-ribose-5-phosphate, 0.5 mM prealnumycin (2) in 5% (vol/vol) DMSO, and 23 μM AlnA and incubated for 3 h at 288 K. The oxygen concentration was reduced using the glucose oxidase (100 nM)–catalase (1.5 μM) system in 60 mM D-glucose. The products 4a and 4b were recovered using Sep-Pak PLUS C18 cartridges (Millipore, Waters) and purified by a SCL-10Avp HPLC system equipped with a SPD-M10Avp diode array detector (Shimadzu) and a Discovery HSC18 column (5 μm, 5 cm × 4.6 mm; Supelco), eluted with 20 mM aqueous ammonium acetate (pH 3.6) and a 70–100% (vol/vol) methanol gradient. The same HPLC conditions were applied for acquiring high-resolution HPLC-ESI-MS data using a MicrOTOF-Q mass spectrometer (Bruker) with 4-kV capillary voltage, 573 K dry heater temperature, and a nebulizer pressure of 1.6 bar coupled to a 1200 Series HPLC system equipped with a diode array detector (Agilent Technologies). NMR spectra were acquired using a Bruker Avance NMR spectrometer operating at 500 and 200 MHz for 1H and 31P nuclei, respectively. A full description on the production, isolation, and structure determination is presented in SI Text and in Table S3.
Protein Data Bank Accession Codes.
The crystallographic data have been deposited in the RCSB Protein Data Bank with the accession codes 4EX8, 4EX9, 4EX6, and 4EX7 for native AlnA, AlnA ligand complex, native AlnB, and AlnB in complex with phosphate, respectively.
Supplementary Material
Acknowledgments
We acknowledge access to synchrotron radiation at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) and CSC–IT Center for Science Ltd. for computing resources. We thank Dr. Olli Martiskainen for technical assistance with the high-resolution mass spectrometry measurements. This study was supported by the National Graduate School in Informational and Structural Biology, the Academy of Finland (Grants 121688, 136060, and 127844), and the Swedish Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4EX6–4EX9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207407110/-/DCSupplemental.
References
- 1.Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14(2):99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 2.Pfander H, Stoll H. Terpenoid glycosides. Nat Prod Rep. 1991;8(1):69–95. doi: 10.1039/np9910800069. [DOI] [PubMed] [Google Scholar]
- 3.Veitch NC, Grayer RJ. Flavonoids and their glycosides, including anthocyanins. Nat Prod Rep. 2011;28(10):1626–1695. doi: 10.1039/c1np00044f. [DOI] [PubMed] [Google Scholar]
- 4.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413(6858):814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 5.Cipollone A, et al. Novel anthracycline oligosaccharides: Influence of chemical modifications of the carbohydrate moiety on biological activity. Bioorg Med Chem. 2002;10(5):1459–1470. doi: 10.1016/s0968-0896(01)00411-4. [DOI] [PubMed] [Google Scholar]
- 6.Luzhetskyy A, Méndez C, Salas JA, Bechthold A. Glycosyltransferases, important tools for drug design. Curr Top Med Chem. 2008;8(8):680–709. doi: 10.2174/156802608784221514. [DOI] [PubMed] [Google Scholar]
- 7.Thibodeaux CJ, Melançon CE, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446(7139):1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 8.Bieber B, Nüske J, Ritzau M, Gräfe U. Alnumycin a new naphthoquinone antibiotic produced by an endophytic Streptomyces sp. J Antibiot (Tokyo) 1998;51(3):381–382. doi: 10.7164/antibiotics.51.381. [DOI] [PubMed] [Google Scholar]
- 9.Naruse N, Goto M, Watanabe Y, Terasawa T, Dobashi K. K1115 A, a new anthraquinone that inhibits the binding of activator protein-1 (AP-1) to its recognition sites. II. Taxonomy, fermentation, isolation, physico-chemical properties and structure determination. J Antibiot (Tokyo) 1998;51(6):545–552. doi: 10.7164/antibiotics.51.545. [DOI] [PubMed] [Google Scholar]
- 10.Kren V, Martínková L. Glycosides in medicine: “The role of glycosidic residue in biological activity”. Curr Med Chem. 2001;8(11):1303–1328. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]
- 11.Weatherman RV, Mortell KH, Chervenak M, Kiessling LL, Toone EJ. Specificity of C-glycoside complexation by mannose/glucose specific lectins. Biochemistry. 1996;35(11):3619–3624. doi: 10.1021/bi951916z. [DOI] [PubMed] [Google Scholar]
- 12.Bililign T, Griffith BR, Thorson JS. Structure, activity, synthesis and biosynthesis of aryl-C-glycosides. Nat Prod Rep. 2005;22(6):742–760. doi: 10.1039/b407364a. [DOI] [PubMed] [Google Scholar]
- 13.Hultin PG. Bioactive C-glycosides from bacterial secondary metabolism. Curr Top Med Chem. 2005;5(14):1299–1331. doi: 10.2174/156802605774643015. [DOI] [PubMed] [Google Scholar]
- 14.Mittler M, Bechthold A, Schulz GE. Structure and action of the C-C bond-forming glycosyltransferase UrdGT2 involved in the biosynthesis of the antibiotic urdamycin. J Mol Biol. 2007;372(1):67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Oja T, et al. Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem Biol. 2008;15(10):1046–1057. doi: 10.1016/j.chembiol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Oja T, et al. Biosynthetic pathway toward carbohydrate-like moieties of alnumycins contains unusual steps for C-C bond formation and cleavage. Proc Natl Acad Sci USA. 2012;109(16):6024–6029. doi: 10.1073/pnas.1201530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preumont A, Snoussi K, Stroobant V, Collet JF, Van Schaftingen E. Molecular identification of pseudouridine-metabolizing enzymes. J Biol Chem. 2008;283(37):25238–25246. doi: 10.1074/jbc.M804122200. [DOI] [PubMed] [Google Scholar]
- 18.Levin I, et al. Crystal structure of an indigoidine synthase A (IndA)-like protein (TM1464) from Thermotoga maritima at 1.90 A resolution reveals a new fold. Proteins. 2005;59(4):864–868. doi: 10.1002/prot.20420. [DOI] [PubMed] [Google Scholar]
- 19.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: ERvolution of a catalytic scaffold. Trends Biochem Sci. 2004;29(9):495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361(5):1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 21.Hancock SM, Vaughan MD, Withers SG. Engineering of glycosidases and glycosyltransferases. Curr Opin Chem Biol. 2006;10(5):509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20(11):478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 23.Cho H, et al. BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase. Proc Natl Acad Sci USA. 2001;98(15):8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R-G, et al. The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction. J Mol Biol. 2003;332(5):1083–1094. doi: 10.1016/j.jmb.2003.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang WR, et al. Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol. 2002;319(2):421–431. doi: 10.1016/S0022-2836(02)00324-8. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AGW, Powell HR. In: Evolving Methods for Macromolecular Crystallography. Read RJ, Sussman JL, editors. Heidelberg: Springer; 2007. pp. 41–51. [Google Scholar]
- 27.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 29.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 35.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52(4):609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 36.Lehtovuori PT, Nyrönen TH. SOMA—Workflow for small molecule property calculations on a multiplatform computing grid. J Chem Inf Model. 2006;46(2):620–625. doi: 10.1021/ci050388n. [DOI] [PubMed] [Google Scholar]
- 37.Kallio P, Sultana A, Niemi J, Mäntsälä P, Schneider G. Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: Implications for catalytic mechanism and product stereoselectivity. J Mol Biol. 2006;357(1):210–220. doi: 10.1016/j.jmb.2005.12.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.