Abstract
The polyketide natural product Leptomycin B inhibits nuclear export mediated by the karyopherin protein chromosomal region maintenance 1 (CRM1). Here, we present 1.8- to 2.0-Å-resolution crystal structures of CRM1 bound to Leptomycin B and related inhibitors Anguinomycin A and Ratjadone A. Structural and complementary chemical analyses reveal an unexpected mechanism of inhibition involving covalent conjugation and CRM1-mediated hydrolysis of the natural products’ lactone rings. Furthermore, mutagenesis reveals the mechanism of hydrolysis by CRM1. The nuclear export signal (NES)-binding groove of CRM1 is able to drive a chemical reaction in addition to binding protein cargos for transport through the nuclear pore complex.
Keywords: exportin-1, Xpo-1, lactone hydrolysis, Michael addition, KPT inhibitor
The polyketide natural product Leptomycin B (LMB) has intrigued chemists and biologists with its highly complex structure, anticancer properties, and biological activity as an efficient and selective inhibitor of nuclear export mediated by the chromosomal region maintenance 1 protein (CRM1) (1–7). Its frequent use as a cell biological tool has led to the discovery of hundreds of broadly functioning nuclear export cargos (2–4), which bind a hydrophobic groove of CRM1 through their nuclear export signals (NESs) (8–11). LMB is a 540-Da polyketide containing an α,β-unsaturated δ-lactone, two conjugated dienes, a β-hydroxy-ketone moiety, and a terminal carboxylate (Fig. 1A). LMB binds covalently to Cys-528 in the human CRM1 (HsCRM1) NES-binding groove through a Michael reaction at its α,β-unsaturated δ-lactone moiety (Fig. 1B) (1). Although LMB is predicted to occupy at least part of the groove (9, 11), it is unclear how it interacts with CRM1, if it is an NES mimic, or if it changes the CRM1 groove conformation.
Fig. 1.
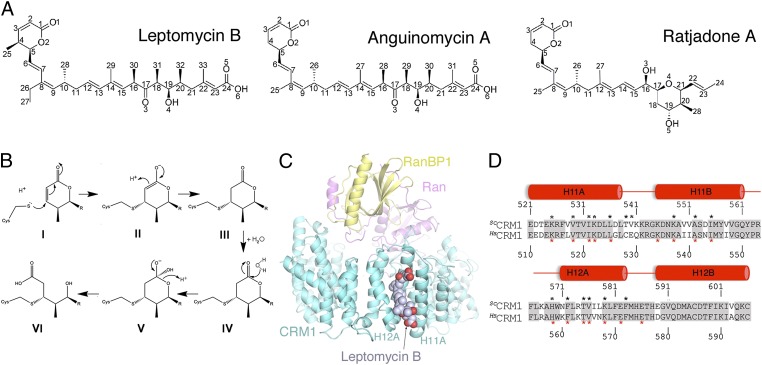
Chemical structures of the α,β-unsaturated lactone polyketide inhibitors and crystal structure of the LMB-bound ScCRM1*-HsRan-ScRanBP1 complex. (A) Chemical structures of inhibitors LMB, AGA, and RJA. (B) Michael addition and hydrolysis reactions of the LMB lactone. The polyketide chain of LMB is represented as R. (Upper) The deprotonated reactive cysteine of CRM1 attacks (Left) the β-alkene of the lactone (I), generating a saturated lactone that is conjugated to (Right) the cysteine (III). (Lower) A water molecule attacks the carbonyl carbon of the conjugated saturated lactone to form (Center) a tetrahedral oxyanion intermediate (V) followed by (Left) the breaking of ester bond and formation of the hydroxy acid product (VI). (C) Overall structure of LMB (space-filling representation) bound to the ternary complex of ScCRM1* (aquamarine), HsRan (magenta), and ScRanBP1 (yellow). The proteins are shown in cartoon representation. (D) Sequence alignment of the NES-binding grooves (HEAT repeats H11 and H12) of ScCRM1 and HsCRM1 (81% sequence identity). Identical residues are shaded gray. Residues that contact LMB are marked with black asterisks, and residues that contact the PKINES (Protein Data Bank ID code 3NBY) are marked with red asterisks.
Results and Discussion
Overall Structures of Inhibitor-Bound CRM1 Complexes.
We present the 1.8- to 2.0-Å-resolution crystal structures of LMB and related inhibitors Ratjadone A (RJA) (12, 13) and Anguinomycin A (AGA) (14) bound to the ternary complex of Saccharomyces cerevisiae CRM1 (ScCRM1), RanBP1, and human Ran•GppNHp (Table S1). CRM1-LMB complexes did not crystallize, whereas the CRM1-Ran-RanBP1 complex bound LMB and formed crystals in 1–2 d. The ternary protein complex was, therefore, used solely to obtain high-resolution crystals of LMB-bound CRM1. The overall structure of the LMB-bound complex is shown in Fig. 1C. We mutated Thr-539 of LMB-insensitive ScCRM1 (equivalent to Cys-528 of HsCRM1) to cysteine for covalent modification by inhibitors, and the mutant is named ScCRM1*. HsCRM1 and ScCRM1 grooves differ in only a few residues (Fig. 1D). Structure of the yeast groove with swapped human residues is virtually unchanged, thus validating the ScCRM1* complex as a mimic of the human complex (SI Results and Discussion and Figs. S1 and S2 A and B).
CRM1 is composed of 21 tandem HEAT repeats (HEAT named after the proteins Huntingtin, Elongation factor 3, protein phosphatase 2A, and TOR1 kinase), each designated H1–H21 and containing a pair of antiparallel helices A and B. The NES-binding groove of CRM1 is located between HEAT repeats H11 and H12 (9–11). The three inhibitor-bound structures are virtually identical (Cα rmsds of 0.2–0.3 Å), with inhibitors occupying ∼70% of the NES-binding groove (Fig. 1C). LMB, AGA, and RJA each bury 738, 704, and 663 Å2 of the groove, respectively. Overall structures of the inhibitor-bound ScCRM1*-Ran-RanBP1 complexes are very similar to the previously reported ScCRM1-Ran-RanBP1 structure (all residues Cα rmsds of 0.7–0.8 Å) (15).
Conformational Plasticity of the CRM1 Groove.
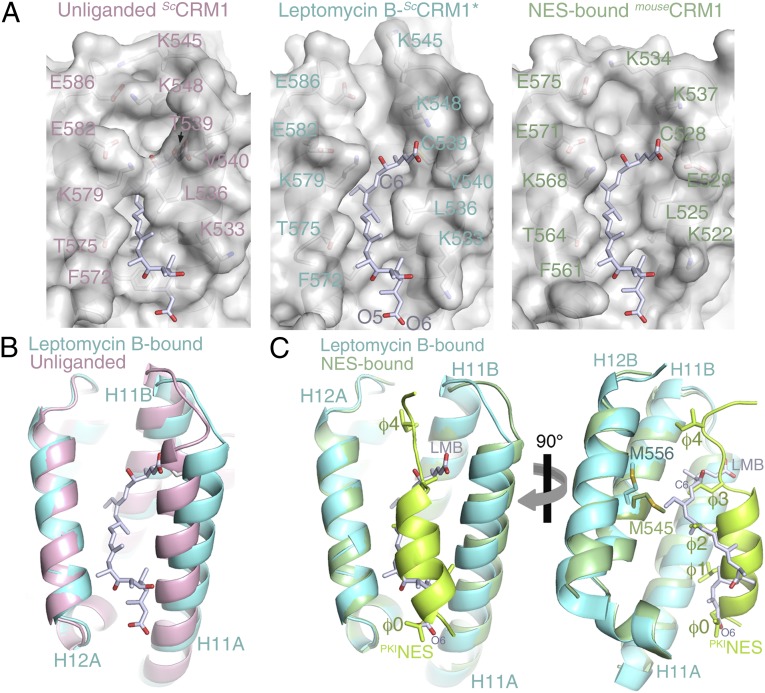
In the absence of inhibitors, the ScCRM1* groove is closed as observed previously in the ScCRM1-Ran-RanBP1 complex (Fig. 2A, Fig. S2 C and D, and Table S2) (Cα rmsd is 0.3 Å for groove residues 521–605) (15). However, the CRM1 grooves open to bind the lactone polyketide inhibitors (Fig. 2A and Figs. S2 A and B and S3) (Cα rmsds of 0.8–1.0 Å for superpositions of groove residues 521–605). Interestingly, covalent conjugation is not strictly required for LMB binding or opening of the CRM1 groove, because the groove is also open in a complex of LMB with CRM1 that lacks the reactive cysteine (Fig. S4 and Table S2). Each of the three inhibitor-bound CRM1 grooves adopts conformation that is intermediate between the closed groove of inhibitor-free ScCRM1-Ran-RanBP1 (15) and the slightly wider grooves of NES-bound CRM1 (9–11) (Fig. 2 A–C) (Cα rmsds of 0.8–1.1 Å for 85 groove residues). Conformational plasticity explains why computational modeling of LMB into a rigid NES-bound groove produced a model that is quite different from our crystal structures (14) (SI Results and Discussion).
Fig. 2.
Comparison of the unliganded and LMB- and NES-bound CRM1 grooves. (A) Surface representations of (Left) the unliganded NES-binding groove of ScCRM1-Ran-RanBP1 (Protein Data Bank ID code 3M1I) (15), (Center) the LMB-bound ScCRM1* groove, and (Right) the PKINES-bound HsCRM1 groove (Protein Data Bank ID code 3NBY) (10). The unliganded groove contains no LMB, which is superimposed and shown only as a reference. The PKINES peptide has been removed from the PKINES-bound HsCRM1 groove, and a superimposed LMB is shown as a reference. (B) Superposition of the unliganded (pink) and LMB-bound (aquamarine) grooves. (C) Superposition of the LMB- (aquamarine) and PKINES-bound (green) grooves.
Conformational differences between empty and inhibitor- and NES-bound grooves result from both helix reorientation (Fig. 2 B and C) and rearrangements of a few sidechains, including Arg-543, Lys-545, Lys-548, Phe-572, Glu-582, and Phe-583 (Fig. S2 A and C). When LMB is bound, groove residues Met-556 and Met-594 sidechains (HsCRM1 Met-545 and Met-583) also each rotate ∼90° away from the groove surface to deepen the groove, thus allowing the inhibitor to penetrate much deeper into the groove than the NES peptide (Fig. 2C) (9–11). LMB occupies the same space as four of five hydrophobic PKIαNES residues (ϕ0, ϕ1, ϕ2, ϕ3, and ϕ4) (Fig. 2C) (10). Extensive inhibitor–NES overlap and inhibitor occupation of most of the groove suggest that LMB will displace most NES peptides, thus explaining its broad spectrum of nuclear export block.
Covalent Conjugation and CRM1-Mediated Hydrolysis.
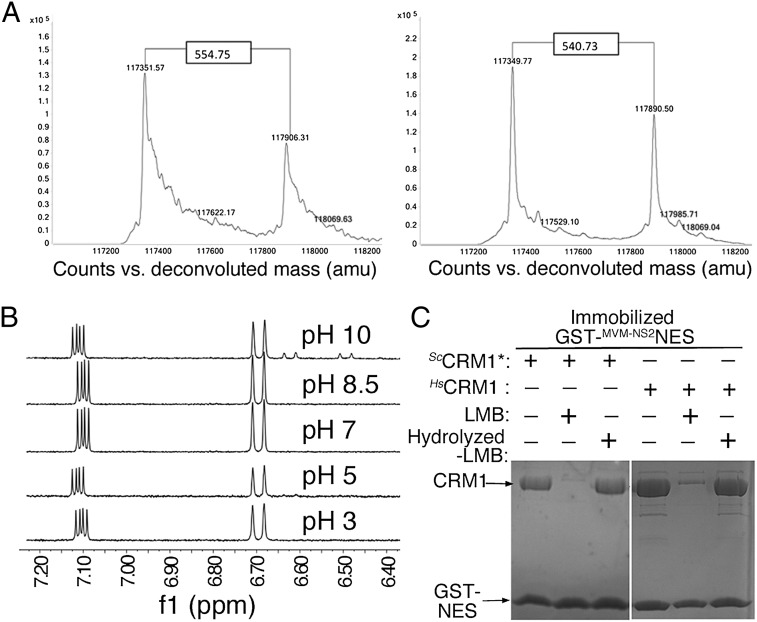
LMB, RJA, and AGA are all covalently conjugated to Cys-539 of ScCRM1* (Fig. 3 and Figs. S2 A and B and S3). Most strikingly, electron densities clearly show that, in each case, the lactone ring has been hydrolyzed to a hydroxy acid, although hydrolysis of α,β-unsaturated lactone compounds is disfavored at neutral pH (16) (Figs. 1B and 3 and Figs. S2A and S3 A and C) (MS data in Fig. 4A). In fact, 1H-NMR analysis of LMB at pH values 3, 5, 7, and 8.5 showed no detectable lactone hydrolysis; LMB hydrolysis only begins to be observable at pH 10.0 (Fig. 4B). Therefore, without CRM1, less than 1% (limit of detection of NMR analysis) of LMB is hydrolyzed in our crystallization buffer of pH 6.6, suggesting that CRM1 stabilizes hydrolyzed lactone (Fig. 4B). Furthermore, comparison of LMB and a chemically hydrolyzed LMB showed that the latter does not inhibit CRM1 (Fig. 4C), suggesting that hydrolysis likely follows Michael addition. Consistent with this argument, Michael addition to LMB is more facile, because the β-carbon of its activated alkene is more electrophilic than the counterpart in the hydrolyzed LMB carboxylate.
Fig. 3.
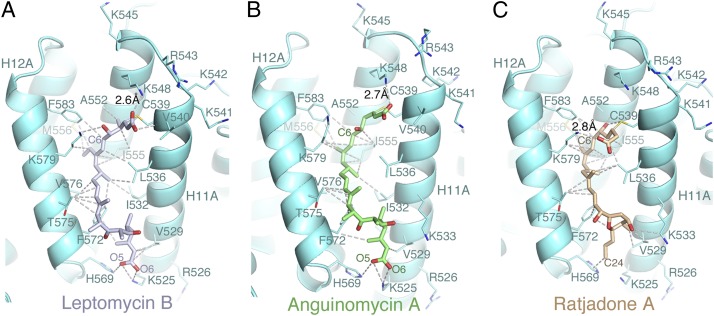
Interactions of the inhibitors with the CRM1 grooves. Cartoon representation of the ScCRM1 grooves (aquamarine) bound with (A) LMB (light blue), (B) AGA (green), and (C) RJA (brown). Select inhibitor–CRM1 interactions (<4 Å) are shown with dashed lines.
Fig. 4.
LMB hydrolysis and CRM1 inhibition. (A) Intact mass analysis of excess ScCRM1* treated with LMB. Both samples were prepared and analyzed using the exact same protocol. The deconvoluted mass spectra show two strong protein peaks with mass shifts of 554.75 (Left) and 540.73 (Right), respectively. Although the unmodified form of ScCRM1* was detected with high mass accuracy and precision (average theoretical mass of 117,350.39 ± 0.4 Da), modified CRM1 showed a significant variation that can only be explained by molecular instability of the CRM1-bound LMB during the MS experiment. (B) 1H-NMR spectra of LMB. Lactone hydrolysis is observed only at pH 10 with ∼10% conversion to hydrolysis product after 10 min (new 1H signals at 6.50 and 6.65 ppm). (C) Pull-down inhibition assays using immobilized GST-MVM-NS2NES, ScCRM1*, HsCRM1, and LMB or hydrolyzed LMB (Coomassie-stained). LMB-modified CRM1 proteins do not bind GST-NES. Chemically hydrolyzed LMB does not inhibit ScCRM1* or HsCRM1.
LMB, AGA, and RJA bind the CRM1 groove through extensive electrostatic and hydrophobic interactions. The carboxyl groups of the hydrolyzed lactones or hydroxy acid moieties of LMB and AGA form salt bridges with Lys-548, polar interactions with the amide of Val-540, and long-range electrostatic interactions with Arg-543 (Fig. 3 A and B). The hydroxyl groups of the hydrolyzed lactones of LMB and AGA are near the Lys-579 sidechain, whereas that group in RJA adopts an alternative conformation, rotating ∼150° to hydrogen bond with Ala-552 and folding its carboxylate to the groove opening for a salt bridge with Lys-579 (Fig. 3C). The polyketide chain of each inhibitor binds within the groove in a similar fashion, making numerous hydrophobic contacts to protein sidechains that also contact NESs (Fig. 3) (9–11). With the exception of the β-hydroxy-ketone groups of LMB and AGA, almost every carbon atom of each inhibitor contacts CRM1. The terminal carboxylates of LMB and AGA make several electrostatic interactions with Lys-525 and His-569 at the bottom of the groove to provide a second electrostatic anchor at the opposite end of the inhibitors (Fig. 3 A and B).
Mutagenesis Identifies CRM1 Residues Critical for Lactone Hydrolysis.
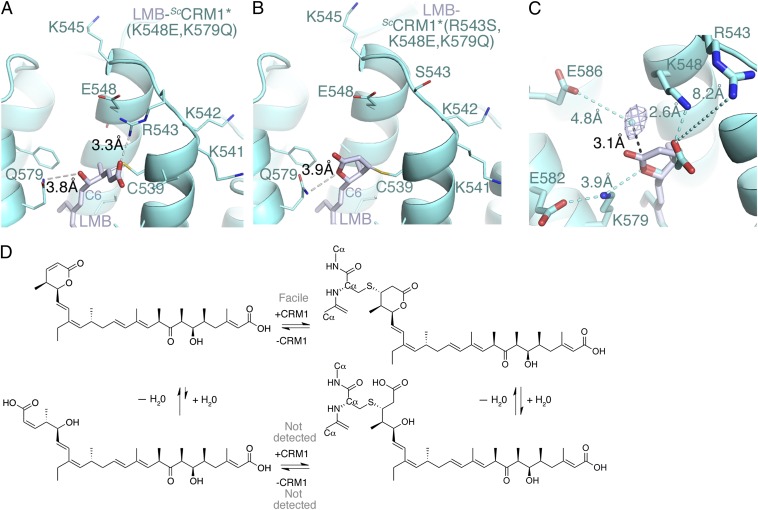
The hydrolyzed lactone of LMB seems to be stabilized by Lys-548 and Lys-579. Four additional basic residues (Lys-541, Lys-542, Arg-543, and Lys-545) nearby could potentially reach into the groove near the hydrolysis site (Fig. 3A). Electrostatic surface potential of the CRM1 groove is shown in Figs. S2B and S3 B and D. We mutated these basic residues and solved structures of five different LMB-ScCRM1 mutants to look for effects in conjugation and lactone hydrolysis (Fig. 5 A and B, Figs. S5, S6, S7, and S8, and Tables S3 and S4). We changed K548, K579, or both residues together [ScCRM1*(K548A), ScCRM1*(K579A), and ScCRM1*(K548E,K579Q)]; in all cases, LMB or RJA was conjugated to Cys-539, and the lactone ring was hydrolyzed (Fig. 5A and Figs. S5, S6, and S8 A and B). When Lys-548 is mutated, the Arg-543 sidechain moves into the groove, substituting for the missing lysine (Fig. 5A). Thus, it seems that only one of Arg-543, Lys-548, and Lys-579 or even simply, a general positive charge near the lactone may be sufficient to drive ester hydrolysis (Fig. 3A and Figs. S2B and S3 B and D). We tested this hypothesis by mutating Arg-543, Lys-548, and Lys-579 in mutant ScCRM1*(R543S,K548E,K579Q) and removing all positively charged residues near the reaction site in mutant ScCRM1*(K541Q,K542Q,R543S,K545Q,K548Q,K579Q). Structures of both mutants showed closed lactone rings conjugated to Cys-539, suggesting that we had trapped a covalently linked saturated lactone intermediate (Fig. 5B and Figs. S7 and S8C). Electron densities of the closed-ring lactone intermediate are shown in Fig. S7 A and C. The presence of any one of Arg-543, Lys-548, or Lys-579 seems sufficient to drive LMB hydrolysis on conjugation of the α,β-unsaturated lactone ring to CRM1. These three basic residues are also important for NES binding (Fig. S9). Structures of the CRM1 mutants conjugated to closed saturated lactone intermediates (Fig. 5B and Fig. S8C) also suggest that conjugation alone is insufficient for lactone hydrolysis. Consistent with this argument, we observed that only a small amount (<10%) of DTT-conjugated LMB alone is hydrolyzed after 26 h (Fig. S10 and Table S5).
Fig. 5.
Structures of LMB-bound ScCRM1 mutants and the mechanism of CRM1 inhibition. (A and B) Reaction sites in ScCRM1 double mutant ScCRM1(K548E,K579Q) and triple mutant ScCRM1(R543S,K548E,K579Q), respectively. CRM1 in aquamarine, and LMB is light blue. (C) Superposition of LMB bound to ScCRM1* and mutant ScCRM1*(R543S,K548Q,K579Q). Omit map density (1σ cutoff) is shown for the only water molecule in the reaction site. (D) A model showing the equilibria of conjugation and hydrolysis for CRM1 inhibition by LMB.
Superposition of the LMB-bound grooves of ScCRM1* and ScCRM1*(R543S,K548E,K579Q) informed on potential nucleophile and stabilization of the tetrahedral intermediate of hydrolysis (Fig. 5C). A bound water molecule is located 3.1 Å from the carbonyl carbon of the superimposed saturated lactone intermediate. Its almost perpendicular position to the plane of the ester is optimal for nucleophilic attack on the carbonyl carbon (Figs. 1B and 5 C and D). Arg-543, Lys-548, and Lys-579 are poised to form an oxyanion hole that could stabilize the resulting anionic tetrahedral intermediate and lower the energy barrier for hydrolysis. Lys-548 and Lys-579 sidechains could approach as close as 2.5 Å to the carbonyl carbon of the conjugated lactone, whereas Arg-543 may get within 4 Å of the group. The three basic residues may also contribute to hydrolysis through stabilization of the final anionic product. Chemically hydrolyzed LMB slowly reverses back to ring-closed LMB in the absence of CRM1 (Fig. S11). Although not commonly observed, a similar hydrolysis reaction was recently shown to occur with a cyclic imide, which is formed by Michael addition of a cysteine to a succinimide ring, in a positively charged binding site of an engineered antibody (17).
Structural Comparison with KPT-185 and KPT-251 Inhibitors.
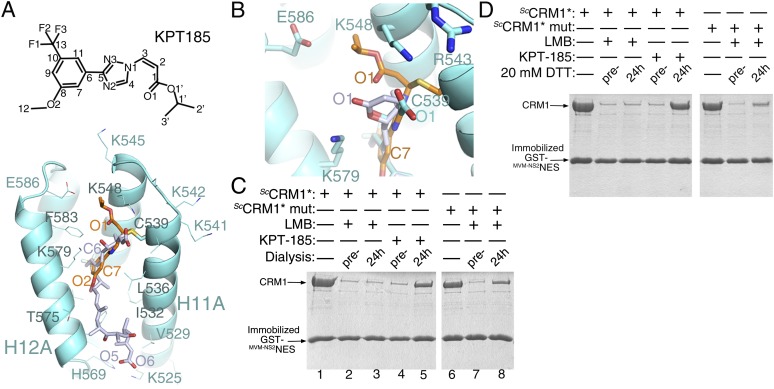
KPT-185 and KPT-251 (Karyopharm Therapeutics, Natick, MA) are members of a new class of drug-like small-molecule compounds that were designed to bind the NES groove of CRM1. We recently reported crystal structures of ScCRM1*-Ran-RanBP1 complexes bound to the prototype compounds KPT-185 and KPT-251, respectively (molecular masses of 353.3 and 375.2 Da, respectively) (Fig. 6A) (18, 19). The KPT inhibitors share a trifluoromethyl phenyl triazole scaffold. Both compounds also contain Michael acceptors, an isopropyl acrylate in KPT-185, and an alkyl oxadiazole in KPT-251 for covalent conjugation to the reactive cysteine of CRM1. Because the two KPT compounds bind CRM1 very similarly, we compare only CRM1-bound LMB with KPT-185, as the latter also contains a reactive enone moiety.
Fig. 6.
The LMB- vs. KPT-185–bound CRM1 grooves and the stability of inhibitor conjugation to CRM1. (A) Chemical structure of KPT-185 and crystal structure of the KPT-185-bound ScCRM1* groove. (B) Superposition of KPT-185– and LMB-bound ScCRM1* with LMB bound to mutant ScCRM1*(R543S,K548Q,K579Q). (C and D) ScCRM1* or mutant ScCRM1*(K541Q,K542Q,R543S,K545Q,K548Q,K579Q) were incubated with LMB or KPT-185 to achieve full CRM1 inhibition before dialysis of the samples (C) or treatment with 20 mM DTT (D) to remove excess unbound inhibitor. The extent of CRM1 inhibition was determined using pull-down inhibition assays with immobilized GST-NES, and the proteins were separated by SDS/PAGE and visualized with Coomassie staining.
Like LMB, KPT-185 binds in the NES groove, and the reactive alkene of its enone forms a covalent bond with Cys-539 of ScCRM1*. However, although LMB fills most of the groove, the smaller KPT-185 occupies only ∼40% of the groove. Much of the space filled by the polyketide chain of LMB or by the PKIαNES helix remains unoccupied in the KPT-185–bound groove (Fig. 6A) (18). Instead, the isopropyl acrylate portion of KPT-185 sits in the narrow channel above Cys-539 that is not occupied by LMB. Unlike LMB, which binds CRM1 through extensive electrostatic and hydrophobic interactions, KPT-185 binds almost solely through hydrophobic interactions.
KPT-185 is not hydrolyzed when bound to CRM1 (Fig. 6 A and B) (18). Its isopropyl acrylate binds deeper in the CRM1 groove than either the intact or opened lactone of LMB. The closed-ring LMB lactone is positioned near the opening of the NES groove with an ordered water molecule nearby as the potential nucleophile (Figs. 5C and 6B). In contrast, the carbonyl carbon of the KPT-185 enone is close to the floor of the groove surrounded by hydrophobic sidechains (Fig. 6 A and B). In addition to the lack of potential nucleophiles in this hydrophobic environment, the intact KPT-185 isopropyl acrylate (potential hydrolysis substrate) likely makes many more contacts with CRM1 than the cleaved hydrolysis product. This situation contrasts with LMB, where the hydrolysis product forms many more interactions with CRM1 than the lactone substrate. Thus, in the case of KPT-185, substrate stabilization may further decrease the likelihood of enone hydrolysis.
Lactone Hydrolysis Decreases Reversibility of Covalent Conjugation.
Lactone hydrolysis of LMB results in electrostatic anchoring of the inhibitor at both termini of the CRM1 groove, and the anionic hydrolysis product complements a highly basic pocket in the groove, possibly contributing significant additional binding energy beyond covalent conjugation at the single cysteine site (Fig. 3A and Fig. S2B). Furthermore, by analogy with other Michael addition reactions, reversibility of the conjugate addition of cysteine should be kinetically controlled, with deprotonation of the inhibitor α-proton as the rate-determining step (Fig. 5D). The α-proton of the hydrolyzed (carboxylate) inhibitor should be appreciably less acidic than the α-proton of the lactone (20, 21). Thus, ring opening should enable lactone-based inhibitors to attach more persistently to CRM1 than analogous inhibitors without this capability.
We compared the stability of LMB conjugation with ScCRM1* vs. mutant ScCRM1*(K541Q,K542Q,R543S,K545Q,K548Q,K579Q), which does not hydrolyze LMB, to test the effect of lactone hydrolysis on the reversibility of covalent conjugation (Fig. 6 C and D and Fig. S12). LMB-conjugated proteins were either dialyzed or treated with 20 mM DTT to remove unbound inhibitors, and the extent of LMB conjugation was determined by a CRM1 inhibition assay using immobilized NES. LMB persistently bound and fully inhibited ScCRM1*, even after dialysis and DTT treatment, indicating no detectable deconjugation. Interestingly, the hydrolysis incompetent CRM1 mutant showed decreased inhibition after removal of unbound inhibitor (Fig. 6 C and D), suggesting that ∼20–30% of previously bound LMB is no longer conjugated to CRM1 (Fig. S12). Similarly, KPT-185, which contains a reactive enone but is not hydrolyzed by CRM1, also significantly decreased inhibition after dialysis or DTT treatment (Fig. 6 C and D); ∼40–60% of previously bound KPT-185 seems to be no longer conjugated to CRM1 (Fig. S12), suggesting that the inhibitor binds CRM1 in a slowly reversible fashion. These results support the notion that lactone hydrolysis decreases reversibility of the Michael addition to enable persistent binding of LMB to CRM1. A similar protein-driven hydrolysis of a succinimide ring that is conjugated to an engineered antibody also resulted in increased stability of the protein conjugate (17).
Timescale of CRM1-Mediated Hydrolysis of LMB.
The main evidence for hydrolysis of LMB by CRM1 derives from LMB-ScCRM1*-Ran-RanBP1 crystals that were typically frozen in liquid nitrogen within 24 h of LMB addition to CRM1. Because we have been unable to analyze the kinetics of lactone hydrolysis of the LMB-CRM1 conjugate in solution, our resolution in quantifying the rate of this process is limited by the timescale of crystallization. Thus, we conservatively place the timescale of this reaction in the range of hours, although it could be significantly faster. Lactone or enone hydrolysis is not crucial for LMB or KPT-185 to inhibit NES recognition by CRM1, but CRM1-mediated lactone hydrolysis seems to be significant for long-lived inhibition by LMB and other α,β-unsaturated lactone polyketide inhibitors (Fig. 6 C and D). Such persistent inhibition may contribute to the long-lived clinical toxicity previously observed for LMB, even several days after removal of the drug (5).
Conclusion
In summary, LMB is targeted to the NES-binding groove of CRM1 through covalent conjugation to a reactive cysteine residue (Fig. 5D). Subsequent lactone hydrolysis by CRM1 optimizes LMB–CRM1 interactions and irreversibility of conjugation and thus, inhibitor potency. A karyopherin protein, which normally binds transport cargos and other protein ligands, has been shown here to drive a chemical reaction. An intriguing question to address in the future is whether CRM1 has analogous catalytic activities with endogenous biological substrates other than the α,β-unsaturated lactone polyketide inhibitors.
Materials and Methods
Detailed materials and methods are described in SI Materials and Methods. Briefly, (i) ScCRM1 proteins, Ran and RanBP1, were purified separately and mixed, and the complex was purified by gel filtration and finally incubated with excess inhibitors. (ii) Crystals grew in 1–2 d after conditions similar to the conditions used in ref. 15. (iii) Structures were solved by molecular replacement using ScCRM1-ScRan-ScRanBP1 (Protein Data Bank ID code 3M1I) (15) as search model. (iv) 1H-NMR spectra of LMB in D2O crystallization buffer at pH values 3.0, 5.0, 7.0, 8.5, and 10 were measured at 600 MHz. (v) LC-MS analysis of LMB + DTT and LMB (no DTT) in buffer was performed using a Phenomenex C18 Luna HPLC column, and the molecules were detected at 254 nm and with MS [M + H]+. (vi) ScCRM1* and LMB-ScCRM1* were analyzed by Q-TOF MS. (vii) LMB was chemically hydrolyzed with LiOH, purified by RP-HPLC, and analyzed by LC-MS. (viii) CRM1-binding/inhibition assays were performed using immobilized GST-MVM-NS2NES, and the proteins were visualized by SDS/PAGE and Coomassie staining. To assess the reversibility of inhibitor conjugation, ScCRM1* proteins were mixed with inhibitors and subjected to (i) immediate inhibition assays or either (ii) dialysis or (iii) treatment with 20 mM DTT to remove excess unbound inhibitors followed by CRM1 inhibition assays.
Supplementary Material
Acknowledgments
We thank X. Dong and Z. Zhang for advice and help on CRM1 purification and inhibition assays, J. Humprey and D. Trudgian for help with MS, Karyopharm Therapeutics for KPT-185, and J. Ready, T. Wandless, B. Chait, M. Rout, S. Shacham, J. Kohler, E. Goldsmith, M. Rosen, and M. Phillips for discussions. This work is funded by Cancer Prevention Research Institute of Texas (CPRIT) Grants PR-101496 (to Q.S. and Y.P.C.) and RP120352 (to Y.M.C.), National Institutes of Health Grants R01 CA149833 (to J.M.) and R01 GM069909 (to Y.M.C.), the University of Texas Southwestern Endowed Scholars Program (J.M. and Y.M.C.), Welch Foundation Grant I-1532 (to Y.M.C.), and a Leukemia and Lymphoma Society Scholar Award (to Y.M.C.). Results shown in this report are derived from work performed at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Conflict of interest statement: Y.M.C. is a consultant for Karyopharm Therapeutics.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HAT, 4HAU, 4HAV, 4HAW, 4HAX, 4HAY, 4HAZ, 4HB0, 4HB2, 4HB3, and 4HB4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217203110/-/DCSupplemental.
References
- 1.Kudo N, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96(16):9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24(7):841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 3.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol. 2010;20(6):782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutay U, Güttinger S. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 2005;15(3):121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74(4):648–649. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutka SC, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69(2):510–517. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamamoto T, Gunji S, Tsuji H, Beppu T. Leptomycins A and B, new antifungal antibiotics. I. Taxonomy of the producing strain and their fermentation, purification and characterization. J Antibiot (Tokyo) 1983;36(6):639–645. doi: 10.7164/antibiotics.36.639. [DOI] [PubMed] [Google Scholar]
- 8.Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009;16(5):558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong X, et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458(7242):1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Güttler T, et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol. 2010;17(11):1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- 11.Monecke T, et al. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324(5930):1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 12.Köster M, et al. Ratjadones inhibit nuclear export by blocking CRM1/exportin 1. Exp Cell Res. 2003;286(2):321–331. doi: 10.1016/s0014-4827(03)00100-9. [DOI] [PubMed] [Google Scholar]
- 13.Meissner T, Krause E, Vinkemeier U. Ratjadone and leptomycin B block CRM1-dependent nuclear export by identical mechanisms. FEBS Lett. 2004;576(1–2):27–30. doi: 10.1016/j.febslet.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 14.Bonazzi S, et al. Anguinomycins and derivatives: Total syntheses, modeling, and biological evaluation of the inhibition of nucleocytoplasmic transport. J Am Chem Soc. 2010;132(4):1432–1442. doi: 10.1021/ja9097093. [DOI] [PubMed] [Google Scholar]
- 15.Koyama M, Matsuura Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010;29(12):2002–2013. doi: 10.1038/emboj.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manso JA, Pérez-Prior MT, García-Santos MdelP, Calle E, Casado J. A kinetic approach to the alkylating potential of carcinogenic lactones. Chem Res Toxicol. 2005;18(7):1161–1166. doi: 10.1021/tx050031d. [DOI] [PubMed] [Google Scholar]
- 17.Zhang ZC, Chook YM. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS) Proc Natl Acad Sci USA. 2012;109(30):12017–12021. doi: 10.1073/pnas.1207247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapalombella R, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120(23):4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etchin J, et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia. 2012 doi: 10.1038/leu.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnett EM, Harrelson JA., Jr Ion pairing and reactivity of enolate anions. 7. A spectacular example of the importance of rotational barriers: The ionization of Meldrum's acid. J Am Chem Soc. 1987;109(3):809–812. [Google Scholar]
- 21.Taylor EA, Palmer DR, Gerlt JA. The lesser “burden borne” by o-succinylbenzoate synthase: An “easy” reaction involving a carboxylate carbon acid. J Am Chem Soc. 2001;123(24):5824–5825. doi: 10.1021/ja010882h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.