Abstract
As dominant members of marine mesozooplankton communities, copepods play critical roles in oceanic food webs and biogeochemical cycling. Despite the ecological significance of copepods, little is known regarding the causes of copepod mortality, and up to 35% of total copepod mortality cannot be accounted for by predation alone. Viruses have been established as ecologically important infectious agents in the oceans; however, viral infection has not been investigated in mesozooplankton communities. Here we used molecular and microscopic techniques to document viral infection in natural populations of the calanoid copepods Acartia tonsa (Dana) and Labidocera aestiva (Wheeler) in Tampa Bay, FL. Viral metagenomics revealed previously undocumented viruses in each species, named Acartia tonsa copepod circo-like virus (AtCopCV) and Labidocera aestiva copepod circo-like virus (LaCopCV). LaCopCV was found to be extremely prevalent and abundant in L. aestiva populations, with up to 100% prevalence in some samples and average viral loads of 1.13 × 105 copies per individual. LaCopCV transcription was also detected in the majority of L. aestiva individuals, indicating viral activity. AtCopCV was sporadically detected in A. tonsa populations year-round, suggesting temporal variability in viral infection dynamics. Finally, virus-like particles of unknown identity were observed in the connective tissues of A. tonsa and L. aestiva by transmission electron microscopy, demonstrating that viruses were actively proliferating in copepod connective tissue as opposed to infecting gut contents, parasites, or symbionts. Taken together, these results provide strong independent lines of evidence for active viral infection in dominant copepod species, indicating that viruses may significantly influence mesozooplankton ecology.
Keywords: circovirus, zooplankton, crustacea, ssDNA virus
Given that mesozooplankton provide a key link between primary producers and larger organisms, and play a crucial role in oceanic biogeochemistry (1, 2), there is a strong need to understand the ecology of dominant mesozooplankton constituents. Copepods are one of the most ubiquitous and abundant mesozooplankton groups (3–5), and much work to date has focused on their population dynamics and ecology (6). Mortality is the most poorly constrained parameter in copepod population dynamics (7). It is often assumed that predation is the only significant cause of copepod mortality; however, predation accounts for only 65–75% of copepod mortality (8), suggesting the importance of nonpredatory mortality mechanisms, such as harmful algae, environmental stressors, parasitism, and diseases (9–11), that kill copepods and leave carcasses behind (12). In natural Chesapeake Bay populations, it was found that on average 30% of early naupliar stages and 12–15% of older-stage Acartia tonsa individuals were dead (13). In addition, nonpredatory mortality rates were highest during late spring and summer, coinciding with peaks in population abundance (14). The foregoing studies suggest that nonpredatory copepod mortality plays an important ecological role and imply that population density-dependent factor(s) are involved in nonpredatory mortality, consistent with the characteristics of viral diseases.
Although viruses likely play a role in regulating the abundance and vital rates of organisms at all trophic levels, essentially nothing is currently known about the impact of viruses on the biology and ecology of copepods or marine mesozooplankton communities in general. Copepods have been shown to transmit viruses to fish and penaeid shrimp (15–17), but no previous studies have demonstrated viral infection in copepods. The sole study to date investigating the effects of viral exposure on copepods found no negative impacts on survival or fecundity of A. tonsa (Dana) (18); however, that study did not examine whether the copepods were infected with viruses. To investigate viral infection of mesozooplankton, we used molecular and microscopic methods to explore the types, viral loads, and prevalence of viruses in the ecologically important calanoid copepods A. tonsa (Dana) and Labidocera aestiva (Wheeler) from Tampa Bay, FL.
Results and Discussion
Discovery of Circo-like Virus Genomes in Copepods.
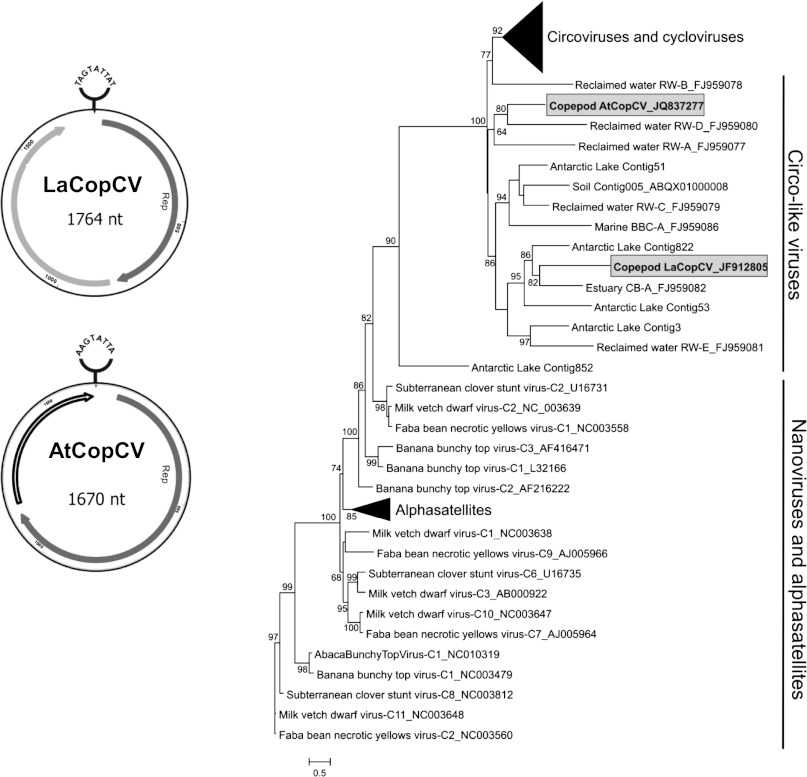
Metagenomic sequencing of viruses purified from each copepod species revealed sequences with weak amino acid similarities to the replication initiator protein (Rep) of viruses with small, circular, single-stranded DNA (ssDNA) genomes, specifically circoviruses (family Circoviridae; A. tonsa metagenome, GenBank Project PRJNA182699; L. aestiva metagenome, GenBank Project PRJNA182700). The use of phi29 rolling circle amplification in metagenome preparation likely biased the metagenome toward circular ssDNA genomes (19); however, this was an important step, allowing sufficient DNA recovery to identify viral targets. Inverse PCR was used to amplify and completely sequence a circo-like virus genome directly from each copepod species: L. aestiva circo-like virus (LaCopCV, 1,764 nt; GenBank accession no. JF912805) and A. tonsa circo-like virus (AtCopCV, 1,670 nt; GenBank accession no. JQ837277). Both genomes share characteristics with known circoviruses, including small genome size, two nonoverlapping ORFs, and a stem-loop with a conserved nonanucleotide motif (Fig. 1) (20). However, in contrast to the ambisense genome organization of known circoviruses, the ORFs of both copepod circo-like virus genomes are oriented in the same direction. This genomic architecture has also been reported in some circo-like virus genomes assembled from environmental and fecal metagenomes (20). Up to 2011, all identified members of the Circoviridae family were known to infect vertebrates; however, considered together with the recent discoveries of circoviruses in dragonflies (21, 22) and an increasing number of circo-like virus sequences identified in environmental viral communities (20, 23), our findings suggest that circo-like viruses may be widespread in both marine and terrestrial invertebrates.
Fig. 1.
Genome organization and phylogenetic placement of the circo-like viruses identified in L. aestiva (LaCopCV) and A. tonsa (AtCopCV). Both genomes have a stem-loop with a conserved nonanucleotide motif at the apex and two nonoverlapping ORFs encoding putative replication initiator (Rep) (gray) and capsid (light gray) proteins oriented in the same direction. ORFs with no hits in the database are shown in white. A maximum likelihood phylogenetic tree of Rep amino acid sequences from these viruses along with viral and associated satellite members of the viral Rep family (PF02407) and other circular Rep-encoding ssDNA viruses from various environmental sources shows that the copepod viruses are only distantly related to known and proposed members of the Circoviridae family (i.e., circoviruses and cycloviruses) and are more closely related to circo-like virus sequences assembled from environmental viral metagenomes. The collapsed circovirus and cyclovirus clade represents Reps from 43 genomes, whereas the alphasatellite clade represents 17 genomes.
Each copepod virus genome contains a Rep-encoding ORF exhibiting the rolling circle replication and SF3 helicase motifs conserved in eukaryotic Rep-encoding circular ssDNA viruses (20, 24). The highest BLASTp similarity of the AtCopCV Rep (376 aa) is to a circo-like virus discovered in bat feces from China (bat circovirus ZS; GenBank accession no. AEL28794.1; E-value, 2e-40) (25). The LaCopCV Rep (255 aa) has BLASTp similarity to a circo-like virus with an unknown host identified from free virioplankton in the Chesapeake Bay (CB-A; GenBank accession no. YP_003084293.1; E-value, 9e-30) (23). Phylogenetic analysis of Reps identified in Circoviridae (circoviruses and cycloviruses) and circo-like viral genomes from environmental samples indicates that both LaCopCV and AtCopCV are highly divergent from known circoviruses, yet cluster with circo-like viruses identified in environmental samples (Fig. 1). Thus, LaCopCV and AtCopCV likely represent unique ssDNA viral lineages infecting marine invertebrates.
Known circoviruses have a stem-loop that acts as the point of rolling circle replication initiation, with a nonanucleotide motif, NANTATTAC, at the apex (20). Both copepod circo-like virus genomes exhibit this stem-loop, with two variations of the nonanucleotide motif (AAGTATTAC in AtCopCV and TATTATTAC in LaCopCV).
The other (non-Rep) ORF (159 aa in AtCopCV and 227 aa in LaCopCV) has no BLAST similarities; however, this ORF is presumed to be a capsid protein (Cap) based on the genome organization of known circoviruses. A Pfam search revealed that the putative capsid of LaCopCV shares weak similarities with the capsid of circoviruses (PF02443; E-value, 0.001). In addition, the N terminus of the putative LaCopCV capsid contains an arginine-rich region predicted to be a nuclear localization signal that is characteristic of circovirus capsid proteins (26, 27). In contrast, the non-Rep ORF in the AtCopCV genome does not have any significant similarities in Pfam and does not exhibit a nuclear localization signal in its N terminus.
Evidence of Active Viral Infection in Copepods.
Viruses have been reported to infect larger marine crustaceans, including crabs (26–30), isopods (31), and penaeid shrimp (32, 33). However, previous studies have focused on virus pathology, and few estimates of viral load or prevalence for marine crustacea have been published. To examine the viral load of LaCopCV within individual animals, we applied a quantitative PCR (qPCR) approach used previously to study viral loads in larvae of aquacultured prawns (34, 35). qPCR of the putative capsid gene of LaCopCV revealed viral loads ranging from 77 to 7.75 × 105 copies per L. aestiva individual, with an average viral load of 1.13 × 105 copies per individual copepod and 100% of L. aestiva individuals infected in three of the four Tampa Bay locations sampled (Table 1). This finding indicates a high prevalence of the virus among wild L. aestiva populations and is consistent with reports of high incidences of viral infections in other marine arthropods, including penaeid shrimp (34–37). Currently, quantitative data on DNA viruses in marine arthropods are sparse. Most of the research on this topic to date has involved white spot syndrome virus (WSSV) in crustaceans. The average viral load for LaCopCV is on the same order of magnitude as estimated WSSV loads in postlarvae of Fenneropenaeus chinensis (34), and approximately one order of magnitude higher than WSSV loads in Portunuus trituberculatus larvae obtained from diverse locations in Korea (35). The range of LaCopCV viral loads observed is similar to ranges reported in surveys of WSSV among P. trituberculatus larvae (35).
Table 1.
LaCopCV viral loads in L. aestiva from four Tampa Bay locations
| Sampling date | Location | No. of specimens | Prevalence of positives, % | Copies/individual |
| March 27, 2009 | Bayboro Harbor | 18 | 100 | 1.24 × 105 ± 2.89 × 104 |
| June 9, 2011 | Alafia River | 6 | 50 | 99 ± 14 |
| August 18, 2011 | Fort Desoto Beach | 10 | 100 | 1,619 ± 950 |
| October 6, 2011 | Eckerd Pier | 5 | 100 | 3.60 × 105 ± 1.05 × 105 |
Location coordinates are as follows: Bayboro Harbor, 27° 45' 38.94” N, 82° 37' 54.12” W; Alafia River, 27° 51' 2.80” N, 82° 24' 59.08” W; Fort Desoto Beach, 27° 36' 54.62” N, 82° 43' 32.21” W; Eckerd Pier, 27° 42' 38.54” N, 82° 41' 27.77” W.
To confirm that LaCopCV was actively replicating, we examined viral transcription in individual L. aestiva collected in Bayboro Harbor in March 2009 using a modified quantitative RT-PCR (qRT-PCR) assay similar to the approach used in previous studies examining porcine circovirus type 2 replication (38, 39). We found low but detectable levels of LaCopCV transcription in 11 of the 14 copepods examined, with an average of 25 ± 5 transcripts per individual. These results should be considered lower estimates of absolute LaCopCV transcript numbers, because some transcripts might have been lost as a result of S1 nuclease activity on weakly bound or unstable DNA-RNA hybrid molecules (Methods).
AtCopCV was detected by conventional PCR in pools of ∼500 A. tonsa from Tampa Bay in all monthly samples from 2011 except January, May, August, September, and December. Although it is difficult to draw seasonality conclusions from a single year of data, these results suggest that AtCopCV is much less prevalent (and possibly not present) during peak winter and summer months, and reemerges during times of population changes in the spring and fall. AtCopCV and LaCopCV were detected only in A. tonsa and L. aestiva, respectively, not in any other zooplankton species. Neither virus was detected in 50-L Tampa Bay seawater virioplankton concentrates, but LaCopCV was frequently detected in Tampa Bay sediments at an average concentration of 5.72 ± 0.55 × 104 copies g−1, suggesting a potential environmental reservoir for this virus.
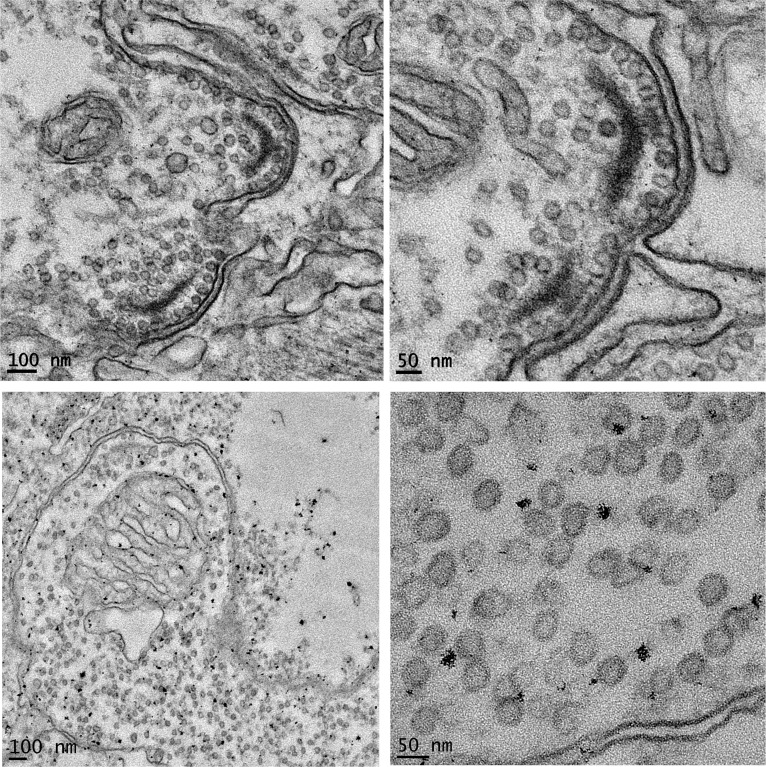
To further document viral infection in copepods, thin sections of A. tonsa and L. aestiva were examined under transmission electron microscopy. Virus-like particles were observed in the connective tissues of both species (Fig. 2), demonstrating that viruses were propagating directly in copepod tissues, as opposed to infecting parasites, symbionts, or gut materials. The average diameter of the virus-like particles was 39.5 ± 7.2 nm in L. aestiva and 37 ± 4.9 nm in A. tonsa, significantly larger than known eukaryotic circoviruses, with diameters of 17–20 nm (40); thus, whether the observed particles represent LaCopCV and AtCopCV remains to be determined. The extremely limited number of metagenomic reads analyzed in this study to initially identify viral sequences was biased toward ssDNA viruses and certainly is not an exhaustive sampling of the copepod-associated virome. More extensive metagenomic sequencing (of both DNA and RNA viruses) likely would yield additional viral sequences, some of which may be the source of the viral particles observed by transmission electron microscopy.
Fig. 2.
Transmission electron micrographs (Left, broad view; Right, close-up) showing virus-like particles in connective tissue of A. tonsa (Upper) and L. aestiva (Lower).
Implications and Future Work.
By documenting unique circo-like virus genomes in two distinct copepod species, as well as the presence of viral particles within copepod tissues, this study provides definitive evidence for viral infection in natural populations of calanoid copepods. LaCopCV and AtCopCV, which are Rep-encoding circular ssDNA viruses, represent hitherto undocumented viral types in marine zooplankton or any marine host. These genomes suggest that mesozooplankton represent a potential source for the unique circular ssDNA genome architectures identified in marine virioplankton through metagenomics (23). The causes of nonpredatory mortality of mesozooplankton are poorly understood, and the potential role of viral infection is a major gap in the current knowledge of zooplankton ecology. This study represents an important breakthrough by demonstrating the presence, high prevalence, and active replication of viruses in two numerically dominant copepod species. Future studies are needed to determine the pathology, route of infection, and ecological implications of viral infection in copepods and other mesozooplankton. This information will ultimately allow us to better understand nonpredatory mortality, enabling inclusion of this parameter in population dynamic and food web models.
Methods
Sample Collection.
Zooplankton were collected using a 335-µm plankton net, and copepods were individually picked from the bulk zooplankton samples, washed three times in 100-kDa filtered seawater, and incubated overnight in a fecatron (41) to allow for gut clearing. After gut clearing, the copepods were rinsed three times in 100-kDa filtered seawater and then frozen at −80 °C until further processing. For viral metagenomic analysis, L. aestiva and A. tonsa were collected from Bayboro Harbor, Tampa Bay, FL (27° 45' 38.94” N, 82° 37' 54.12” W) in April 2009 and May 2010, respectively. For qPCR detection, L. aestiva were collected from multiple locations, including Bayboro Harbor, Eckerd Pier (27° 42' 38.54” N, 82° 41' 27.77” W), the mouth of the Alafia River (27° 51' 2.80” N, 82° 24' 59.08” W), and Fort Desoto Beach (27° 36' 54.62” N, 82° 43' 32.21” W) (Table 1). To determine the temporal dynamics of viral infection in A. tonsa, samples were collected from Bayboro Harbor once a month throughout 2011. To test for environmental reservoirs, surface sediments and ambient seawater samples from Bayboro Harbor were also collected and screened for the specific viruses identified in the copepods. Surface sediments were collected in Bayboro Harbor using a Ponar-type grab sampler, and viruses were concentrated from 50 L of surface seawater using a 100-kDa tangential flow filter.
Metagenomics and Sequencing of Copepod Virus Genomes.
Virus particles were purified from each copepod species based on size, density, and nuclease resistance, and viral metagenomes were sequenced according to standard protocols (42–44). In brief, the copepods were homogenized in sterile SM buffer (50 mM Tris⋅Cl, 10 mM MgSO4, 0.1 M NaCl; pH 7.5), centrifuged at 10,000 × g for 10 min at 4 °C to pellet animal tissues, and passed through a 0.22-µm filter to remove bacteria and animal cells. The A. tonsa filtrate was also loaded onto a cesium chloride step gradient with 1 mL each of 1.2, 1.5, and 1.7 g·mL−1 in SM buffer. After ultracentrifugation at 61,000 × g for 3 h at 4 °C, the viral fraction (between the 1.2- and 1.5-g·mL−1 density layers) was collected, then concentrated and washed twice on a Microcon YM-30 column (Millipore). Both the A. tonsa and L. aestiva viral fractions were treated with 0.2 volumes of chloroform for 10 min, and then incubated with 2.5 U DNase I per µL of sample for 3 h to eliminate free nucleic acids. After the reaction was stopped by incubation at 65 °C for 10 min, viral DNA was extracted with the QIAmp MinElute Virus Spin Kit (Qiagen) and amplified with the strand-displacement method of the Genomiphi V2 DNA Amplification Kit (GE Healthcare). The GenomePlex Whole Genome Amplification Kit (Sigma-Aldrich) was used to fragment and amplify the DNA, which was then cloned into the pCR4 vector using TOPO TA cloning (Invitrogen). A total of 38 transformants were sequenced for each copepod species using dideoxynucleotide sequencing, and the resulting metagenomic sequences were analyzed using tBLASTx against the GenBank nonredundant database (45). In this study, the main purpose of sequencing these small viral metagenomes was to identify putative viral targets for further quantitative ecological study.
Several sequences from the viral metagenomes of both A. tonsa and L. aestiva had tBLASTx similarities to viruses in the Circoviridae family. Given that known circoviruses have small circular genomes, back-to-back PCR primers (L. aestiva primers: 5′-CACCAGCAACTACAGCATCAA-3′ and 5′-GTGACTATGATCCGCTTGGG-3′; A. tonsa primers: 5′-ACGAAGTAGCGCTCGAACTG-3′ and 5′-CGTGAACTACGCTGGTCGTA-3′) were designed from the metagenomic sequences using Primer 3 (46) to amplify the complete circular genome of the copepod circo-like viruses directly from unamplified copepod DNA extracts through inverse PCR. The PCR reactions [containing 1 µM of each primer, 200 µM dNTPs, 1 U RedTaq DNA Polymerase (Sigma-Aldrich), 1× Red Taq Reaction Buffer, and 5 µL of target DNA in a 50-µL reaction] were amplified as follows: 95 °C for 5 min; 45 cycles of 94 °C for 1 min, 58 °C minus 0.2 °C per cycle for 1 min, and 72 °C for 3 min; and a final extension at 72 °C for 10 min. The resulting whole genome PCR products were cloned into the pCR4 vector using TOPO TA cloning and sequenced to 3× coverage. ORFs were predicted and annotated using SeqBuilder (DNASTAR), and stem- loop structures were manually annotated by locating complementary sections. Genomes were analyzed for the presence of a nuclear localization signal using NLStradamus (47). Alignments of the copepod circo-like virus replication initiator protein (Rep) amino acid sequences with viral and associated satellite members of the Pfam viral Rep family PF02407 (48) and other circular Rep-encoding ssDNA viruses from various environmental sources were performed using the PRALINE server (49). A maximum likelihood phylogenetic tree was constructed using the PhyML server (50), with the (LG+I+G) model chosen as the best-fit substitution model according to ProtTest (51). Branch support was assessed with the approximate likelihood ratio test (52), and values >60% are reported.
PCR Detection.
qPCR of the putative capsid gene was used to assay the prevalence and viral load of LaCopCV in DNA extracted from individual Tampa Bay L. aestiva using the Zymo Research Insect and Tissue DNA-5 Kit. DNA extracts were subjected to qPCR, with each run containing 1× TaqMan Master Mix (Applied Biosystems), 80 pmol of primers 5′-CTTCCGCAGGAGAAAGTCAG-3′ and 5′-GCATGGTACCAGGACGAGTT-3′, 80 pmol of probe 5′-CACCAAAGAGGAGGGCACGTGG-3′, and 2 µL of template DNA. The probe was dual-labeled with 5′ FAM and 3′ TAMRA fluorochromes. Triplicate reactions were prepared for each DNA extract and were compared with duplicate synthesized oligonucleotide standards (5′-GCTTCCGCAGGAGAAAGTCAGCACCAAAGAGGAGGGCACGTGGGCGCAAGGCCCGTGCTCGACGTTCACCGTTCAACTCGTCCTGGTACCATGCA-3′) that represented 10-fold dilutions from 108 copies µL−1 to 101 copies µL−1. Standard copy number and cycle threshold values were correlated at R2 >0.94 (Fig. S1 and Table S1). Detected quantities were multiplied by the volume of extracted DNA and further multiplied by 2 to correct for the ssDNA oligonucleotide standard comparison with yield of viral load per individual animal. L. aestiva individuals were considered positive at >25 copies of the LaCopCV Cap gene per individual; this cutoff was calculated from the minimum detection threshold of one copy and corrected for the total volume of extracted DNA.
Active transcription of LaCopCV was examined using qRT-PCR for the putative capsid gene. Individual animals picked from bulk tow material were washed in nuclease-free water and placed into RNase-free 2.0-mm BashingBead Lysis Tubes (Zymo Research), and RNA was extracted using the Insect and Tissue RNA Kit (Zymo Research). Extracted RNA was purified of DNA contamination using the DNA-Free RNA Kit (Zymo Research). The RNA was converted to ssDNA-RNA hybrids using the SuperScript III First-Strand Synthesis System (Invitrogen). For each animal, duplicate reactions containing 8 µL of extracted RNA, 0.5 mM dNTP mix, and 50 ng of random hexamers were incubated at 65 °C for 5 min, then placed on ice for 1 min. After this, both reactions received 1× RT Buffer, 5 mM MgCl2, 10 mM DTT, and 40 U RNAseOUT. To one replicate reaction, 200 U of SuperScript III was added, and to the other reaction, 1 µL of nuclease-free water was added. The reactions were incubated at 25 °C for 10 min, then at 50 °C for 50 min. After incubation, reactions were terminated at 85 °C for 5 min and cooled on ice. The detection threshold for the qRT-PCR assay was eight copies, as calculated from the minimum detection threshold of one copy and corrected for the total volume of extracted RNA.
Initial attempts to compare reverse-transcriptase–treated and untreated RNA samples revealed extensive ssDNA contamination in RNA extracts, likely because genomic ssDNA was not removed efficiently by DNase I (http://www.invitrogen.com/site/us/en/home/References/Ambion-Tech-Support/nuclease-enzymes/general-articles/dnase-i-demystified.html). To eliminate ssDNA, reactions were amended with 1× S1 Nuclease Buffer, 15.5 µL of S1 Nuclease Dilution Buffer, and 750 U of S1 Nuclease (Invitrogen). The reactions were incubated at 37 °C for 10 min, after which 1 µL of Tris-EDTA was added, and reactions were heated to 70 °C for 10 min to inactivate the S1 nuclease. The samples were then subjected to qPCR as described above for viral load estimation. Transcript abundance was calculated by accounting for the total volume of RNA extracted, dilution of extracted RNA in reverse-transcriptase, and S1 nuclease treatment, and multiplied by 2 because an ssDNA oligonucleotide standard was used for comparison. Furthermore, the quantities detected in reactions containing reverse-transcriptase were corrected for ssDNA carry-through by subtracting the values from reactions containing no reverse-transcriptase. Treatment with S1 nuclease after reverse-transcriptase treatment lowered quantities of ssDNA in samples not treated with reverse-transcriptase to less than one copy per reaction. Although S1 nuclease should not digest DNA-RNA hybrids, we cannot discount the possibility that some LaCopCV transcripts were lost as a consequence of nuclease activity on weakly bound and unstable DNA-RNA hybrid molecules. Thus, these transcription estimates are likely underestimates of absolute transcript numbers.
Conventional PCR targeting the AtCopCV Rep gene (primers: 5′ AGTGTCCACATCAAGGCACA-3′ and 5′-CGGAGGAGTTGTCCAAAGAC-3′) was applied to DNA extracted with the Zymo Research Insect and Tissue DNA-25 Kit, using pools of ∼500 A. tonsa collected from Tampa Bay monthly throughout 2011 as a template. The PCR [containing 1 µM of each primer, 200 µM dNTPs, 1 U of Apex Taq DNA polymerase (Genesee Scientific), 1× Taq Reaction Buffer, and 1 µL of target DNA in a 50-µL reaction] was amplified as follows: 95 °C for 5 min; 30 cycles of 94 °C for 1 min, 56 °C minus 0.2 °C per cycle for 1 min, and 72 °C for 3 min; then a final extension at 72 °C for 10 min. The resulting 345-nt PCR product was run on a 1.5% agarose gel and verified by sequencing.
Transmission Electron Microscopy.
Five copepods of each species were fixed in cacodylate-buffered 4% glutaraldehyde with sucrose (0.1 M sodium cacodylate, 0.35 M sucrose, and 4% glutaraldehyde; pH 7.6; Electron Microscopy Sciences) at 4 °C for 2 d. Fixed copepods were rinsed in cacodylate buffer with sucrose, incubated in 1% OsO4 on a rotator for 2 h, then rinsed with deionized water. The copepods were en bloc stained with 1% aqueous uranyl acetate for 2 h in the dark, then rinsed in deionized water, dehydrated in an ethanol series, and transferred to 100% acetone. The copepods were infiltrated with Embed 812 (Electon Microscopy Sciences) without accelerator in increasing concentrations on the rotator, followed by several fresh changes of Embed 812 with accelerator. The copepods were placed in flat embedding molds, oriented longitudinally with the head positioned toward the top for cross-sectioning, and the molds were filled with Embed 812 with accelerator and cured in an oven at 60–70 °C overnight. Then 60-nm sections were sliced with an ultramicrotome and visualized on a Hitachi 7100 transmission electron microscope.
Supplementary Material
Acknowledgments
We thank Lita Proctor, Curtis Suttle, and Kendra Daly for scientific advice; Scott Burghart, Ralph Kitzmiller, and Ernst Peebles for help with sample collection and identification; and Kelly Vasbinder and Sennai Habtes for laboratory assistance. This work was funded by the National Science Foundation (Early Concept Grant for Exploratory Research OCE-1049670).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF912805 and JQ837277; metagenomic sequences have been deposited under GenBank accession nos. JY253391–JY253440).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216595110/-/DCSupplemental.
References
- 1.Honjo S, Manganini SJ, Krishfield RA, Francois R. Particulate organic carbon fluxes to the ocean interior and factors controlling the biological pump: A synthesis of global sediment trap programs since 1983. Prog Oceanogr. 2008;76:217–285. [Google Scholar]
- 2.Small LF, Ellis SG. Fecal carbon production by zooplankton in Santa Monica Basin: The effects of body size and carnivorous feeding. Prog Oceanogr. 1992;30:197–221. [Google Scholar]
- 3.Kleppel GS, Burkart CA, Carter K, Tomas C. Diets of calanoid copepods on the West Florida continental shelf: Relationships between food concentration, food composition and feeding activity. Mar Biol. 1996;127:209–217. [Google Scholar]
- 4.Baird D, Ulanowicz RE. The seasonal dynamics of the Chesapeake Bay ecosystem. Ecol Monogr. 1989;59:329–364. [Google Scholar]
- 5.White J, Roman M. Seasonal study of grazing by metazoan zooplankton in the mesohaline Chesapeake Bay. Mar Ecol Prog Ser. 1992;86:251–261. [Google Scholar]
- 6.Paffenhöfer G-A. On the ecology of marine cyclopoid copepods (Crustacea, Copepoda) J Plankton Res. 1993;15:37–55. [Google Scholar]
- 7.Runge J, et al. Diagnosis and Prediction of Variability in Secondary Production and Fish Recruitment Processes: Developments in Physical-Biological Modeling. Cambridge, MA: Harvard Univ Press; 2004. pp. 413–475. [Google Scholar]
- 8.Hirst A, Kiørboe T. Mortality of marine planktonic copepods: Global rates and patterns. Mar Ecol Prog Ser. 2002;230:195–209. [Google Scholar]
- 9.Kimmerer WJ, McKinnon AD. High mortality in a copepod population caused by a parasitic dinoflagellate. Mar Biol. 1990;107:449–452. [Google Scholar]
- 10.Turner JT, Tester PA. Zooplankton feeding ecology: Nonselective grazing by the copepods Acartia tonsa Dana, Centropages velificatus De Oliveira, and Eucalanus pileatus Giesbrecht in the plume of the Mississippi River. J Exp Mar Biol Ecol. 1989;126:21–43. [Google Scholar]
- 11.Bickel S, Hammon J, Tang K. Boat-generated turbulence as a potential source of mortality among copepods. J Exp Mar Biol Ecol. 2011;401:105–109. [Google Scholar]
- 12.Tang KW, Bickel SL, Dziallas C, Grossart HP. Microbial activities accompanying decomposition of cladoceran and copepod carcasses under different environmental conditions. Aquat Microb Ecol. 2009;57:89–100. [Google Scholar]
- 13.Elliott D, Tang K. Spatial and temporal distributions of live and dead copepods in the lower Chesapeake Bay (Virginia, USA) Estuaries Coasts. 2011;34:1039–1048. [Google Scholar]
- 14.Elliott DT, Tang KW. Influence of carcass abundance on estimates of mortality and assessment of population dynamics in Acartia tonsa. Mar Ecol Prog Ser. 2011;427:1–12. [Google Scholar]
- 15.Nylund A, Wallace C, Hovland T. Pathogens of Wild and Farmed Fish: Sea Lice. 1993. The possible role of Lepeophtheirus salmonis (Kroyer) in the transmission of infectious salmon anaemia. Ellis Horwood Series in Aquaculture and Fisheries Support, eds Boxshall GA, Defaye D (Ellis Horwood, Chichester, UK), pp 367–373. [Google Scholar]
- 16.Overstreet RM, Jovonovich J, Ma HW. Parasitic crustaceans as vectors of viruses, with an emphasis on three penaeid viruses. Integr Comp Biol. 2009;49(2):127–141. doi: 10.1093/icb/icp033. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JS, Dong SL, Dong YW, Tian XL, Hou CQ. Bioassay evidence for the transmission of WSSV by the harpacticoid copepod Nitocra sp. J Invertebr Pathol. 2008;97(1):33–39. doi: 10.1016/j.jip.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Drake LA, Dobbs FC. Do viruses affect fecundity and survival of the copepod Acartia tonsa Dana? J Plankton Res. 2005;27:167–174. [Google Scholar]
- 19.Kim K-H, Bae J-W. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol. 2011;77(21):7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch Virol. 2012;157(10):1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 21.Rosario K, et al. Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata: Anisoptera) J Gen Virol. 2011;92(Pt 6):1302–1308. doi: 10.1099/vir.0.030338-0. [DOI] [PubMed] [Google Scholar]
- 22.Rosario K, et al. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta) J Gen Virol. 2012;93(Pt 12):2668–2681. doi: 10.1099/vir.0.045948-0. [DOI] [PubMed] [Google Scholar]
- 23.Rosario K, Duffy S, Breitbart M. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J Gen Virol. 2009;90(Pt 10):2418–2424. doi: 10.1099/vir.0.012955-0. [DOI] [PubMed] [Google Scholar]
- 24.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20(13):3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge X, et al. Genetic diversity of novel circular ssDNA viruses in bats in China. J Gen Virol. 2011;92(Pt 11):2646–2653. doi: 10.1099/vir.0.034108-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Tikoo SK, Babiuk LA. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology. 2001;285(1):91–99. doi: 10.1006/viro.2001.0922. [DOI] [PubMed] [Google Scholar]
- 27.Heath L, Williamson AL, Rybicki EP. The capsid protein of beak and feather disease virus binds to the viral DNA and is responsible for transporting the replication-associated protein into the nucleus. J Virol. 2006;80(14):7219–7225. doi: 10.1128/JVI.02559-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mari J, Bonami JR. W2 virus infection of the crustacean Carcinus mediterraneus: A reovirus disease. J Gen Virol. 1988;69(Pt 3):561–571. doi: 10.1099/0022-1317-69-3-561. [DOI] [PubMed] [Google Scholar]
- 29.Sprague V, Beckett RL. A disease of blue crabs (Callinectes sapidus) in Maryland and Virginia. J Invertebr Pathol. 1966;8(2):287–289. doi: 10.1016/0022-2011(66)90156-x. [DOI] [PubMed] [Google Scholar]
- 30.Widada JS, Bonami JR. Characteristics of the monocistronic genome of extra small virus, a virus-like particle associated with Macrobrachium rosenbergii nodavirus: Possible candidate for a new species of satellite virus. J Gen Virol. 2004;85(Pt 3):643–646. doi: 10.1099/vir.0.79777-0. [DOI] [PubMed] [Google Scholar]
- 31.Kuris AM, Poinar GO, Hess R, Morris TJ. Virus particles in an internal parasite, Portunion conformis (Crustacea: Isopoda: Entoniscidae), and its marine crab host, Hemigrapsus oregonensis. J Invertebr Pathol. 1979;34:26–31. [Google Scholar]
- 32.Bonami JR, Hasson KW, Mari J, Poulos BT, Lightner DV. Taura syndrome of marine penaeid shrimp: Characterization of the viral agent. J Gen Virol. 1997;78(Pt 2):313–319. doi: 10.1099/0022-1317-78-2-313. [DOI] [PubMed] [Google Scholar]
- 33.Vogt G. Cytopathology of Bay of Piran shrimp virus (BPSV), a new crustacean virus from the Mediterranean Sea. J Invertebr Pathol. 1996;68(3):239–245. doi: 10.1006/jipa.1996.0091. [DOI] [PubMed] [Google Scholar]
- 34.Jang I-K, et al. A TaqMan real-time PCR assay for quantifying white spot syndrome virus (WSSV) infections in wild broodstock and hatchery-reared postlarvae of fleshy shrimp, Fenneropenaeus chinensis. Aquaculture. 2009;287:40–45. [Google Scholar]
- 35.Meng X-H, Jang I-K, Seo H-C, Cho Y-R. White spot syndrome virus quantification in blue crab Portunus trituberculatus hatchery-produced larvae and wild populations by TaqMan real-time PCR, with an emphasis on the relationship between viral infection and crab health. Aquaculture. 2009;291:18–22. [Google Scholar]
- 36.Claydon K, Tahir RAH, Said HM, Lakim MH, Tamat W. Prevalence of shrimp viruses in wild Penaeus monodon from Brunei Darussalam. Aquaculture. 2010;308:71–74. [Google Scholar]
- 37.Tang KF, Lightner DV. Duplex real-time PCR for detection and quantification of monodon baculovirus (MBV) and hepatopancreatic parvovirus (HPV) in Penaeus monodon. Dis Aquat Organ. 2011;93(3):191–198. doi: 10.3354/dao02293. [DOI] [PubMed] [Google Scholar]
- 38.Mankertz J, Buhk HJ, Blaess G, Mankertz A. Transcription analysis of porcine circovirus (PCV) Virus Genes. 1998;16(3):267–276. doi: 10.1023/a:1008022521329. [DOI] [PubMed] [Google Scholar]
- 39.Yu S, Carpenter S, Opriessnig T, Halbur PG, Thacker E. Development of a reverse transcription-PCR assay to detect porcine circovirus type 2 transcription as a measure of replication. J Virol Methods. 2005;123(1):109–112. doi: 10.1016/j.jviromet.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 40.Todd D. Family Circoviridae: Virus taxonomy. In: Fauquet C, Mayo M, Maniloff J, Desselberger U, Ball L, editors. Eighth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Academic; 2005. pp. 327–341. [Google Scholar]
- 41.Tang K. Defecation of dimethylsulfoniopropionate (DMSP) by the copepod Acartia tonsa as functions of ambient food concentration and body DMSP content. J Plankton Res. 2001;23:549–553. [Google Scholar]
- 42.Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques. 2005;39(5):729–736. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- 43.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4(4):470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 44.Ng TFF, et al. Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J Virol. 2009;83(6):2500–2509. doi: 10.1128/JVI.01946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Vol 132. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: A simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simossis VA, Heringa J. The PRALINE online server: Optimising progressive multiple alignment on the web. Comput Biol Chem. 2003;27(4-5):511–519. doi: 10.1016/j.compbiolchem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 51.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 52.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.