Abstract
Actin filament nucleation by actin-related protein (Arp) 2/3 complex is a critical process in cell motility and endocytosis, yet key aspects of its mechanism are unknown due to a lack of real-time observations of Arp2/3 complex through the nucleation process. Triggered by the verprolin homology, central, and acidic (VCA) region of proteins in the Wiskott-Aldrich syndrome protein (WASp) family, Arp2/3 complex produces new (daughter) filaments as branches from the sides of preexisting (mother) filaments. We visualized individual fluorescently labeled Arp2/3 complexes dynamically interacting with and producing branches on growing actin filaments in vitro. Branch formation was strikingly inefficient, even in the presence of VCA: only ∼1% of filament-bound Arp2/3 complexes yielded a daughter filament. VCA acted at multiple steps, increasing both the association rate of Arp2/3 complexes with mother filament and the fraction of filament-bound complexes that nucleated a daughter. The results lead to a quantitative kinetic mechanism for branched actin assembly, revealing the steps that can be stimulated by additional cellular factors.
Keywords: actin nucleation, cytoskeleton, TIRF microscopy
The actin-related protein (Arp) 2/3 complex is a key actin filament nucleation factor (1) that rapidly assembles distinctive branched filament networks (2–6) to drive many forms of cellular motility, endocytosis, and pathogen invasion. Nucleation of actin filament branches involves large-scale conformational changes in the Arp2/3 complex (6–10), binding of one or more verprolin homology, central, and acidic (VCA) domains (10–15), recruitment of one or more actin monomers (2, 16), and formation of a stable association of the complex with a mother filament (6). The ordering, dynamics, and regulation of these individual reaction steps in the overall process of branch nucleation have been subjects of intense interest (13, 15, 17, 18). Elucidating the mechanism of this elaborate process has been challenging using conventional bulk biochemical experiments, because they average together the signals from multiple reaction steps. Here we directly observed the nucleation of single-branched actin filaments by individual molecules of Arp2/3 complex, using dual-wavelength single-molecule fluorescence techniques. By identifying the dynamic interactions between Arp2/3 complex and actin filaments along the pathway to branch formation, we build a quantitative understanding of the mechanism of nucleation and how it is regulated by VCA.
Results
Individual Arp2/3 Complexes Bind the Sides of Preexisting Actin Filaments and Seed New Filament Growth.
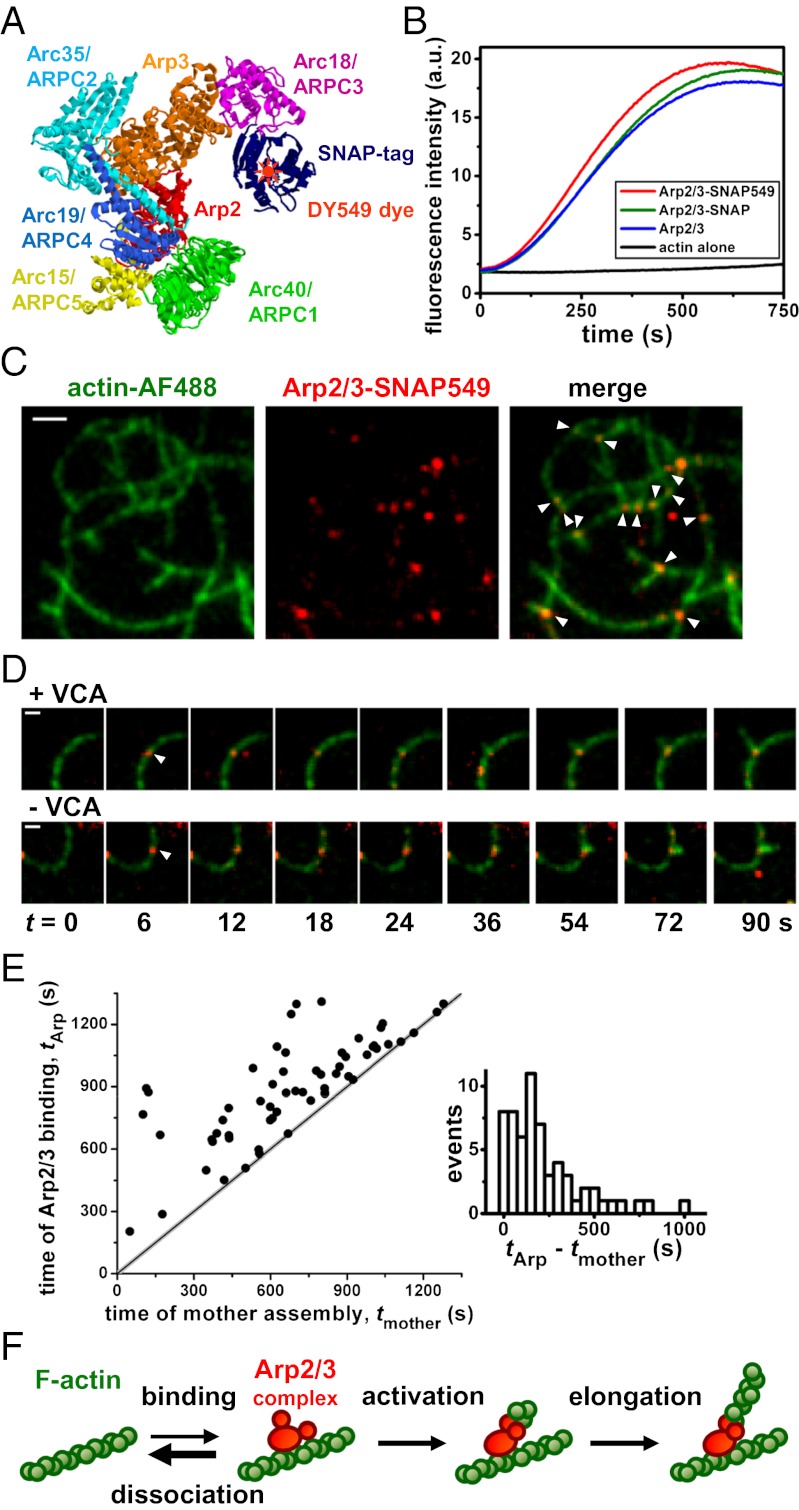
We mixed a fully functional Saccharomyces cerevisiae Arp2/3 complex construct labeled with an orange-emitting dye DY549 (Fig. 1 A and B; Fig. S1 A and B; Movie S1), monomeric actin (sparsely labeled with the green-emitting dye Alexa Fluor 488) and the VCA portion of S. cerevisiae Wiskott-Aldrich syndrom protein (WASp/Las17; unlabeled), and observed filament assembly by total internal reflection fluorescence (TIRF) microscopy. As previously seen using labeled actin and unlabeled Arp2/3 complex, we observed formation of branched filament networks (4, 5, 7, 19–28), and branches rarely if ever dissociated over tens of minutes (26). We saw that 90% (143/160) of branch junctions displayed well-defined spots of orange Arp2/3 complex fluorescence that typically persisted for tens or hundreds of seconds (Fig. 1C). When spots at a persistent branch junction disappeared, almost all of them (112/116) did so in a single step, presumably due to dye photobleaching (Fig. S1 C and D). These data strongly suggest that the spots were individual Arp2/3 complexes (SI Materials and Methods), consistent with cryoelectron microscopy reconstructions that show density corresponding to a single Arp2/3 complex at branch junctions (6).
Fig. 1.
Arp2/3 complex binds to the side of a preexisting actin filament to nucleate a branch. (A) Model structure of Arp2/3-SNAP adapted from refs. 43 and 44, with cartoon indicating the DY549 dye label. (B) Assembly of 2 µM pyrene–actin, with or without 10 nM Arp2/3 complex and 10 nM yeast WASp (Las17), shows that neither the SNAP tag nor the SNAP tag conjugated with DY549 interferes with Arp2/3 complex activity. (C) TIRF images showing that fluorescent Arp2/3-SNAP549 localizes to filament branch junctions (arrowheads). Reaction: 1 µM actin, 20 nM Arp2/3 complex, 300 nM VCA. (Scale bar: 2 μm.) (D) Time-series image sequences showing an Arp2/3 complex binding (arrowhead) to a mother filament followed by growth of a branch from the Arp2/3 complex with or without VCA. (Scale bar: 1 μm.) (E) The subset of Arp2/3 complex filament-binding events that yield branches occur over a broad range of times after the assembly of the mother filament segment. (F) Sequence of events leading to actin nucleation by Arp2/3 complex.
Interleaved visualization of both Arp2/3 complex and actin filament fluorescence enabled direct observation of the sequence of events leading to branch growth. In most cases, Arp2/3 complex bound to the side of a filament, and then dissociated after only a few seconds. However, in a small fraction of binding events, a daughter filament grew from the mother filament-bound Arp2/3 complex, generating a new branch that stably retained Arp2/3 complex (Fig. 1D, Upper). Nearly all (90%, 55/61) of these events occurred sufficiently far (>0.4 μm) from the mother filament ends that it was unambiguous that Arp2/3 complex bound to the side of, rather than the end of, the mother filament. Also, there was a broad range of time delays between polymerization of the mother filament segment to which Arp2/3 complex bound and the time of Arp2/3 complex binding, tArp–tmother (Fig. 1E). A larger number of branches were observed to extend from newer mother filament segments, in part because they are more abundant than older segments in the network, and in part because branches preferentially form on newer vs. older segments (22, 24). Both observations are inconsistent with proposals that Arp2/3 complex is incorporated only at the growing barbed ends of polymerizing filaments (29, 30). Instead, the data strongly support a filament side-binding mechanism of Arp2/3 complex activation (Fig. 1F) (4, 19, 27).
The rate of branch formation, kB, in the presence of 300 nM VCA (9,700 ± 2,900 M−1⋅s−1⋅μm of mother filament) was significantly higher than in the absence of VCA (2,500 ± 700 M−1⋅s−1⋅μm−1; P = 0.015). This four- ± twofold stimulation by VCA was similar to that detected in bulk nucleation assays (6.3- ± 0.8-fold; Fig. S2), suggesting that the extent of VCA stimulation is not substantially affected by surface tethering of the actin filaments. Network assembly by Arp2/3 complex is autocatalytic. Branch formation produces new filaments that in turn act as substrate (mother filament) for Arp2/3 complex activation. Thus, a small increase in branch formation rate can produce a large increase in network assembly rate. To account for this, we derived a method for estimating branching rate per unit length of filament directly from bulk assembly data, which yielded values indistinguishable from our TIRF data (Fig. S2 C and D). Individual branch formation events in the absence of VCA (23) occurred on a similar time-scale and by the same sequence of events (i.e., binding of Arp2/3 complex to the mother filament side followed by nucleation of the daughter filament; Fig. 1D, Lower) as branch formation in the presence of VCA.
Binding of Arp2/3 Complex to Filament Sides Is Accelerated by VCA.
To test the hypothesis that VCA promotes branch nucleation at least in part by increasing the rate at which Arp2/3 complex binds to mother filaments, we immobilized capped filaments on the surface of a flow chamber and observed binding of individual Arp2/3 complexes (Fig. 2A). Filament-specific binding rate constants were measured to be 3.0 ± 0.7 × 103 M−1⋅s−1 (+VCA) and 1.4 ± 0.3 × 103 M−1⋅s−1 (−VCA) per F-actin subunit (mean ± SEM from three independent trials; Fig. 2B and Fig. S3). Thus, VCA accelerates Arp2/3 complex binding to filaments by 2.2- ± 0.7-fold, but this by itself likely cannot explain the overall four- to sevenfold VCA stimulation of daughter filament formation.
Fig. 2.
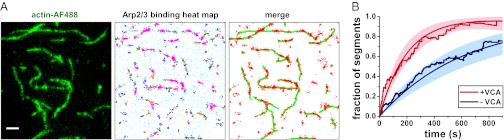
Binding of Arp2/3 complexes to filaments is enhanced by VCA. (A) Preassembled, immobilized, fluorescent actin filaments (Left) bound Arp2/3-SNAP549 molecules (10 nM) in the presence of 1 µM G-actin, 300 nM VCA, and 20 nM capping protein. Locations of all Arp2/3 complex binding events recorded over 15 min are plotted (heat map; marker color reflects local density: cyan <10; blue 10–19; green 20–29; orange 30–39; and magenta >39 events within a 0.4-μm radius). Locations where >9 neighboring events occurred (Right; red) correspond closely with filament locations (green traces). (Scale bar: 2 μm.) (B) Fraction of filament segments (0.4 µm in length) that had bound at least one Arp2/3 complex at or before the indicated time (total of 113 and 105 segments with and without VCA). Data are corrected for background (Materials and Methods). Envelopes: 90% confidence intervals. Smooth curves were calculated from exponential fits (Materials and Methods).
VCA Increases the Efficiency of Daughter Nucleation After Arp2/3 Complexes Associate with Filament Sides.
To identify additional ways that VCA might stimulate daughter filament formation, we measured the activation efficiency fB, the fraction of mother filament–Arp2/3 complexes that produced a daughter filament. This analysis showed that successful daughter production was extraordinarily rare: no branch formation was seen at the vast majority of locations on filaments where Arp2/3 complex fluorescence spots were transiently associated. In the absence of VCA, only 21 of 6,514 observed mother filament–Arp2/3 complexes that persisted for >0.5 s produced a daughter filament, corresponding to fB = 0.4 ± 0.2% (mean ± SEM from three experiments). When a near-maximally activating concentration of VCA was included (300 nM; Fig. S2), efficiency increased significantly (fB,V = 1.3 ± 0.4%; 46 of 3,558 over three experiments). Thus, though the activation of Arp2/3 complex following mother filament binding is highly inefficient, VCA increased the efficiency approximately threefold. Taken together, our data suggest that VCA stimulates two different aspects of the mechanism: (i) the association of Arp2/3 complexes with sides of mother filaments and (ii) the production of branches by mother-bound Arp2/3 complexes.
Daughter Filament Nucleation Rapidly Follows Arp2/3 Complex Binding to Filament Sides.
A merit of single-molecule analysis is that it can be used to specifically characterize the properties of productive branch formation events, excluding from the analysis the much-larger number of Arp2/3 complex binding events that do not produce branches. From all productive events in each reaction we computed the characteristic time delay <ta> between the binding of Arp2/3 complex on the mother filament and the initiation of daughter-filament elongation (Fig. 3). In the presence of VCA, <ta> was small (5 ± 2 s), indicating that for the rare branch-producing events, the daughter typically nucleated shortly after Arp2/3 complex bound to the mother. When VCA was absent, <ta> was 3 ± 2 s, a value similar to the VCA-stimulated value. Thus, activation time is short and is not appreciably lessened by VCA.
Fig. 3.
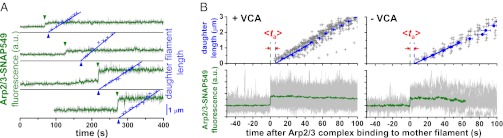
The time delay between mother binding and daughter nucleation is short. (A) Example records of four branch-formation events showing Arp2/3 complex binding (green arrowheads) and the length of the growing daughter filament (+). The length record was extrapolated (line) back to zero length to estimate the daughter nucleation time (blue arrowheads). (B) Averages of all branch formation recordings (n = 61 and 18) from single experiments with and without VCA. Individual daughter filament length measurements (+, as in A) were aligned relative to Arp2/3 complex binding time and combined to produce a consensus length/time record (Materials and Methods), which was extrapolated (line) to zero length to reveal the characteristic activation time <ta>. Error bars indicate SEM.
Evidence for Three Distinct States of Arp2/3–Filament Complexes.
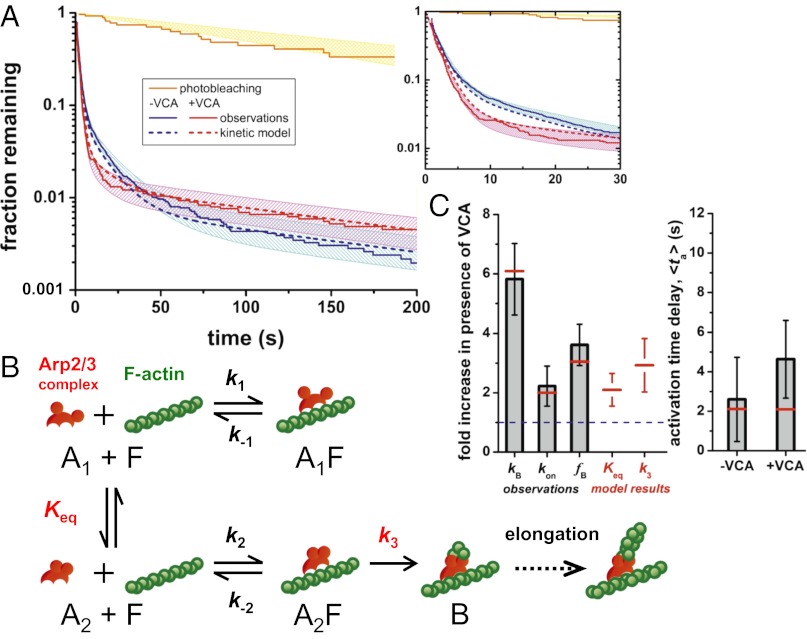
To examine the nature of Arp2/3–filament complexes that form before daughter nucleation, we examined the lifetime distributions of all Arp2/3–filament complexes, whether or not at a branch, in samples with and without VCA (Fig. 4A and Fig. S4). The lifetimes ranged from a fraction of a second to hundreds of seconds (Movie S2) and three exponential terms were required to adequately fit the distributions (Fig. S5; Table S1), demonstrating that at least three distinct species of filament-bound Arp2/3 complex were present. The vast majority (∼98%) of Arp2/3–filament complexes were unsuccessful in nucleation and dissociated from the filament in two shorter lifetime components, with time constants of τ1 = 2 s (∼90% of events) and τ2 = 10–15 s (∼8% of events). These observations suggest that most Arp2/3–filament complexes are >103-fold more kinetically unstable than was inferred from previous experiments (15, 18), and that the simple scheme of Fig. 1F, in which there are only two filament-bound species, cannot adequately explain the experimental data.
Fig. 4.
Complex lifetimes and hypothesized branch nucleation mechanism. (A) Example lifetime distributions of filament-bound Arp2/3-SNAP549 complexes (Materials and Methods; Fig. S4). Curves show lifetimes of all complexes with (red; NV = 2,151) and without (blue; n = 3,654) VCA, and the subset of complexes at branches (yellow; NB = 24). Shading: 90% confidence intervals. (Inset) Magnified view. (B) Kinetic scheme for actin nucleation by Arp2/3 complex. Red circles represent the Arp2 and Arp3 subunits. Only the off-filament conformational equilibrium constant (Keq) and the on-filament activation rate (k3) are altered by the presence of VCA. The presence of three filament-bound states (A1F, A2F, and B) produces the predicted triexponential lifetime distributions (A, dashed lines). (C) Comparison of observed quantities (gray) and predictions from the model in B (red). VCA-stimulation ratios (Left) of the rate of branch formation (kB), the rate of Arp2/3 complex binding to filaments (kon), and the fraction of filament-bound Arp2/3 complexes that nucleate daughter filaments (fB) are shown together with the values of the two model parameters, Keq and k3, that are VCA-dependent. Also shown (Right) are the observed activation times and the corresponding model predictions.
Only ∼2% of the Arp2/3–filament complexes were in the longest-lived component of the lifetime distribution. The τ3 time constants (Fig. S5 B and C) were equal within experimental error to the photobleaching lifetime, which suggests that disappearance of the longest-lived class of fluorescent spots was due to photobleaching, not to dissociation of the Arp2/3–filament complex. Some of these long-lived events were Arp2/3 complexes incorporated into branch junctions, whereas others were “dead-end” complexes that did not show visible branches but were irreversibly bound to the mother filament on the timescale of the experimental observations (Fig. S4). We do not know whether formation of dead-end complexes is intrinsic to the Arp2/3–filament interaction or whether it is the result of photodamage. However, both the branches and dead-end complexes represented only a small minority (∼2%) of the total binding events.
WASp VCA Domain Stimulates Arp2/3 Complex at Two Distinct Steps in the Branch Formation Pathway.
In summary, the data show that (i) VCA stimulates overall Arp2/3 complex-mediated branch production by a factor of four- to sevenfold; (ii) this results from a ∼twofold stimulation of the rate of Arp2/3–mother filament binding and a ∼threefold increase in the efficiency with which bound Arp2/3 complexes produce daughter branches; (iii) even at maximally activating concentrations of VCA, the efficiency is low (1.3%); (iv) the activation delay is short (upper limit ∼7 s); (v) most Arp2/3–mother filament complexes are short-lived (∼90% last ∼2 s); and (vi) there are at least two reversibly bound states in addition to the stably bound branches. All of these observations are consistent with the minimal kinetic scheme and rate constants shown in Fig. 4B and Table S2.
The scheme postulates that Arp2/3 complex free in solution exists in two conformations (Fig. 4B; open [A1] vs. closed [A2]) (7–10) and that only mother filament binding of closed Arp2/3 complex can ultimately lead to branch formation (B). The formation of the initial closed Arp2/3–filament complex (A2F) is reversible, but the Arp–filament interaction is stabilized by subsequent activation processes (13, 17), which may include binding of actin monomers (13, 15), changes in the nucleotide state of Arp2 and/or Arp3 (16, 25), additional Arp2/3 complex conformational changes (6, 10, 17), or alteration of the mother filament structure or curvature (6, 28, 31). Because the experiments were conducted at either zero or maximally activating concentrations of VCA, the scheme does not explicitly show VCA binding steps or define the occupancies of the multiple VCA-binding sites (10, 14, 15). Instead, we make the simplifying assumption that the arrangement of states is the same in both cases and, consequently, the effect of VCA can be incorporated by changing the apparent rate or equilibrium constants.
The predictions of the model agree with all of our observations; we see agreement in the lifetime distributions (Fig. 4A) and the branch nucleation rates, binding rates, and activation efficiencies (Fig. 4C; Table S1); this demonstrates that all of our data can be accounted for by a mechanism with only two VCA-dependent steps. In the scheme shown, VCA is proposed to (i) promote the closed conformation (increased Keq), which increases the apparent binding rate because this conformation binds filaments faster than the open one (k2 > k1), and (ii) increase the rate of the activation step between mother binding and daughter nucleation (k3). This change in k3 causes an increased fraction of A2F complexes to nucleate daughters instead of dissociating (k−2). Both the Keq and the k3 effects result in an increase in the efficiency of branch formation when VCA is present, consistent with our observations.
The model presented in Fig. 4B is intentionally simplified; as already mentioned it does not explicitly include some known features of the system, such as the multiple binding sites for VCA on the Arp2/3 complex. Nevertheless, the above analysis shows that even this simple model with two VCA-regulated steps is fully consistent with our observations. In contrast, we tested versions of the Fig. 4B scheme in which only one, rather than two, constants were modulated by VCA, as well as other models that used different arrangements of states (SI Materials and Methods; Table S3). The data could not be explained by any tested mechanism in which only one step is affected by VCA. Alternative models of equivalent or lower complexity with two VCA-dependent steps were inconsistent with the observations (Tables S1 and S3; Fig. S6). One model of higher complexity was consistent with the observations (Table S3), but was not significantly better than the simpler model of Fig. 4B. In all models reported, a small fraction of Arp2/3 complexes was assumed to bind filaments irreversibly without forming branches (SI Materials and Methods) to account for the observation of dead-end complexes (Fig. S4).
Discussion
The single-molecule fluorescence colocalization techniques used here are powerful tools for directly observing binding and deciphering complex reaction mechanisms (32–36). These techniques allowed us to directly measure key features of Arp2/3 complex-mediated branch formation, including Arp2/3 binding and dissociation rates, branch formation efficiency, and activation time, and to determine how these parameters are affected by VCA. Earlier studies observed actin only (4, 5), or used indirect bulk methods based on fluorescence enhancement of pyrene–actin and/or pyrene–Arp2/3 complex (15, 17, 18). Our activation time-delay results are roughly consistent (within ∼10-fold; SI Materials and Methods) with those derived from kinetic modeling based on earlier observations. However, our observations reveal that binding and most dissociation of Arp2/3 complex from filaments is ∼100 and ∼1,000-fold faster than inferred previously (15). Conversely, branch formation is extremely inefficient (0.5–1.5%), in contrast to the ∼100% efficiency that would be implied if dissociation were slow. The discrepancy between our direct observations of binding and earlier results suggests that the fluorescence enhancement of pyrene–Arp2/3 complex reflects a slow step subsequent to mother filament binding rather than the initial binding itself.
What does this work imply about Arp2/3 complex regulation in vivo? WASp proteins that contain VCA domains function in cells to activate Arp2/3 complex nucleation of actin polymerization at biologically appropriate times and locations (37). The observation that VCA both promotes productive Arp2/3 complex binding to mother filaments and increases the fraction of such binding events that produce daughter filaments suggests two distinct mechanisms by which WASp could promote the temporally and spatially regulated formation of branched actin networks in vivo. First, activated WASp may serve to recruit Arp2/3 complex to sites where nucleation is needed (e.g., adjacent to the necks of endocytic vesicles) and promote its interaction with F-actin. Second, activated WASp may promote branch formation by filament-bound Arp2/3 complex regardless of whether the activator played a role in initial recruitment of Arp2/3 complex. More generally, our observations have provided a quantitative understanding of Arp2/3-mediated actin network assembly dynamics, and revealed an inherent baseline inefficiency and complexity to the branch formation mechanism. There are multiple points in this mechanism at which the efficiency of daughter filament nucleation could be increased by factors that stimulate Arp2/3 complex, including VCA and non-VCA domains in WASp, factors that multimerize WASp, cortactin, and coronin (20, 21, 38–40). Further, their combined effects may allow the integration of multiple stimulatory pathways.
Materials and Methods
Protein Purification and Labeling.
WASp/Las17, its VCA domain, chicken muscle capping protein CapZ, rabbit muscle actin, biotin–actin, and fluorescently labeled actin were prepared by modifications of published methods (SI Materials and Methods). Arp2/3-SNAP was purified from an S. cerevisiae strain in which the ARC18 locus was modified to produce an Arc18-SNAP fusion protein. The complex was labeled in vitro with the orange-emitting fluorescent dye DY549 (SNAP-Surface 549; New England BioLabs). The resulting Arp2/3-SNAP549 retained full activity of wild-type Arp2/3 complex to stimulate actin assembly in vitro (Fig. 1B).
Actin Assembly Kinetics.
Fluorometric measurements of actin network assembly kinetics were made from mixtures of actin monomers (2 μM, 5% of monomers pyrene labeled) and either 10 nM Arp2/3 complex with 10 nM Las17 (Fig. 1B) or 20 nM Arp2/3 complex with the Las17 VCA fragment (as indicated in Fig. S2).
TIRF Microscopy.
Dual-wavelength TIRF microscopy was performed on a custom-built micro-mirror microscope described previously (41). TIRF buffer contained 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole (pH 7.5), 10 mM DTT, 0.2 mM ATP, 15 mM glucose, 0.02 mg/mL catalase, 0.1 mg/mL glucose oxidase, 0.1% bovine serum albumin (BSA), and 0.25% methylcellulose.
To observe actin branch formation, 1% biotin/10% AF488-labeled filaments were briefly preassembled (3 μM actin for ∼5 min), then introduced and allowed to adsorb to the surface of a microscope flow chamber. Monomeric actin (1 μM; 10% AF488- and 1% biotin-labeled), Arp2/3-SNAP549 (20 nM), and VCA (0 or 300 nM) was introduced, and sequences of fluorescence images were recorded at 0.1 or 0.2 s per frame. A single frame of actin fluorescence was recorded every 6–9 s, with continuous recording of Arp2/3 fluorescence between each actin observation.
To measure binding and dissociation of Arp2/3 complex to filaments over long periods without continued filament assembly, filaments were preassembled for an extended period (1–2 h), then capped (20 nM CapZ) before tethering in the flow chamber. CapZ was also included in the reaction mixture, and the intervals between actin fluorescence recordings were extended to 30–40 s.
Data Analysis.
Image processing was performed with custom programs developed in LabVIEW (National Instruments) and in MatLab (MathWorks), and with ImageJ (National Institutes of Health).
Branch formation rates were calculated by dividing the number of observed branches by the product of the length of mother filaments and the observation time, ignoring branches formed on filament segments assembled after the start of the observation. Activation times were computed by first integrating Arp2/3-SNAP549 fluorescence in a small region at each branch site to identify the time of Arp2/3 complex binding to the mother filament (Fig. 3A). Then daughter filaments were traced in each actin image, and length records were aligned relative to Arp2/3 binding time. Average activation times for all branches in each experiment were then calculated from the average times to reach set daughter length intervals in the range ∼0.3–1.5 μm, linearly fit and extrapolated back to zero length to identify the mean nucleation time following Arp2/3–mother binding (Fig. 3B).
Arp2/3–filament binding and dissociation kinetics were quantified by automated detection (adapted from ref. 42) of Arp2/3-SNAP549 fluorescence spots and comparing those detected on regions of the microscope slide containing filaments to those detected on background regions where no filaments were observed. Image sequences were averaged such that the final time resolution was 0.5–1 s. Filament-specific binding rates were calculated by first fitting the times to first-binding for each filament or background segment (0.4 μm in length) with single-exponential probability functions (Fig. S3), then calculating the difference between rates of binding filament and binding background regions. Filament-specific binding data curves (Fig. 2C) were generated by calculating the ratio of the surviving fraction of unbound filament segments to unbound background segments.
Lifetime distributions were also corrected by subtracting the number of background bindings from the number of filament bindings (Fig. S4), per segment and per observation time, for equivalent lifetimes. The resulting filament-specific lifetime distributions with and without VCA (Fig. 4A) were globally fit, together with the lifetime distribution measured at branch sites, using a triple-exponential function for all events and a single-exponential for branch-site events with the longest time-constant constrained to the same value for each (i.e., the photobleaching time; Table S1).
All fitting routines for binding and dissociation data used maximum-likelihood methods and accounted for events missed because they were too short (<0.5–1 s) or too long (>10–15 min). Standard errors were estimated by bootstrapping.
Kinetic Modeling.
For each kinetic scheme tested (Table S3), a set of relations between model parameters (rate and equilibrium constants) and experimental observables (Arp2/3–filament binding rates, time constants, and amplitudes from lifetime distributions, activation times and efficiencies, and overall stimulation of branch formation by VCA) was established. The optimal model parameters that best described all data were then found by reducing the total χ2 difference between model predictions and experimental observations using a Levenberg–Marquardt nonlinear least-squares algorithm in MatLab. See SI Materials and Methods for details of kinetic model development and comparisons.
Supplementary Material
Acknowledgments
We thank L. Friedman and J. Chung for technical assistance with TIRF microscopy and analysis; C. Ydenberg for help with bulk actin assembly kinetics; members of the B.L.G. laboratory for help with CapZ and actin purification and actin labeling; A. Okonechnikov for single-molecule analysis software development; D. Breitsprecher, S. Padrick, and M. Rosen for comments and discussions; and D. Theobald for help with statistical analysis. This work was supported by National Institutes of Health Grants GM43369 (to J.G.) and GM63691 (to B.L.G.) and National Science Foundation Materials Research Science and Engineering Centers Grant 0820492 (to K.D.-C. and B.L.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211164110/-/DCSupplemental.
References
- 1.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7(10):713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 2.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95(11):6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145(5):1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404(6781):1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 5.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci USA. 2001;98(26):15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180(5):887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16(2):269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Rodal AA, et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12(1):26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 9.Martin AC, et al. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168(2):315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X-P, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2012;31(1):236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96(7):3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97(2):221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 13.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3(1):76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 14.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108(33):E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ti S-C, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108(33):E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dayel MJ, Mullins RD. Activation of Arp2/3 complex: Addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004;2(4):E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11(24):1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 18.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283(11):7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 19.Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3(3):306–310. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- 20.Suetsugu S, Miki H, Yamaguchi H, Obinata T, Takenawa T. Enhancement of branching efficiency by the actin filament-binding activity of N-WASP/WAVE2. J Cell Sci. 2001;114(Pt 24):4533–4542. doi: 10.1242/jcs.114.24.4533. [DOI] [PubMed] [Google Scholar]
- 21.Weaver AM, et al. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11(5):370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 22.Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12(1):79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- 23.Wen K-K, Rubenstein PA. Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J Biol Chem. 2005;280(25):24168–24174. doi: 10.1074/jbc.M502024200. [DOI] [PubMed] [Google Scholar]
- 24.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91(9):3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8(8):826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- 26.Gandhi M, et al. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 2010;20(9):861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Achard V, et al. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 2010;20(5):423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 28.Risca VI, et al. Actin filament curvature biases branching direction. Proc Natl Acad Sci USA. 2012;109(8):2913–2918. doi: 10.1073/pnas.1114292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantaloni D, Boujemaa R, Didry D, Gounon P, Carlier MF. The Arp2/3 complex branches filament barbed ends: Functional antagonism with capping proteins. Nat Cell Biol. 2000;2(7):385–391. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- 30.Bugyi B, Carlier M-F. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–470. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 31.Galkin VE, Orlova A, Schröder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17(11):1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191(3):571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara I, Remmert K, Hammer JA., III Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J Biol Chem. 2010;285(4):2707–2720. doi: 10.1074/jbc.M109.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331(6022):1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman LJ, Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148(4):679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breitsprecher D, et al. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336(6085):1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollitt AY, Insall RH. WASP and SCAR/WAVE proteins: The drivers of actin assembly. J Cell Sci. 2009;122(Pt 15):2575–2578. doi: 10.1242/jcs.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padrick SB, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32(3):426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S-L, Needham KM, May JR, Nolen BJ. Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J Biol Chem. 2011;286(19):17039–17046. doi: 10.1074/jbc.M111.219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys J. 2006;91(3):1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 43.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294(5547):1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 44.Mollwitz B, et al. Directed evolution of the suicide protein O6-alkylguanine-DNA alkyltransferase for increased reactivity results in an alkylated protein with exceptional stability. Biochemistry. 2012;51(5):986–994. doi: 10.1021/bi2016537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.