Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common form of lymphoma in adults. The disease exhibits a striking heterogeneity in gene expression profiles and clinical outcomes, but its genetic causes remain to be fully defined. Through whole genome and exome sequencing, we characterized the genetic diversity of DLBCL. In all, we sequenced 73 DLBCL primary tumors (34 with matched normal DNA). Separately, we sequenced the exomes of 21 DLBCL cell lines. We identified 322 DLBCL cancer genes that were recurrently mutated in primary DLBCLs. We identified recurrent mutations implicating a number of known and not previously identified genes and pathways in DLBCL including those related to chromatin modification (ARID1A and MEF2B), NF-κB (CARD11 and TNFAIP3), PI3 kinase (PIK3CD, PIK3R1, and MTOR), B-cell lineage (IRF8, POU2F2, and GNA13), and WNT signaling (WIF1). We also experimentally validated a mutation in PIK3CD, a gene not previously implicated in lymphomas. The patterns of mutation demonstrated a classic long tail distribution with substantial variation of mutated genes from patient to patient and also between published studies. Thus, our study reveals the tremendous genetic heterogeneity that underlies lymphomas and highlights the need for personalized medicine approaches to treating these patients.
Keywords: next-generation sequencing, cancer genetics, cancer heterogeneity
Diffuse large B-cell lymphoma (DLBCL) is the most common form of lymphoma in adults (1). Although nearly half the patients can be cured with standard regimens, the majority of relapsed patients succumb. Thus, there is an urgent need to identify the genetic underpinnings of the disease and to identify novel treatment strategies. Gene expression profiling (2, 3) has uncovered distinct molecular signatures for DLBCL subtypes that have unique biology and prognoses. High-throughput sequencing has provided rich opportunities for the comprehensive identification of the genetic causes of cancer (4–6). Whereas exhaustive portraits of individual cancer genomes are emerging, the degree to which these genomes represent the disease is unclear.
We generated a detailed analysis of a DLBCL genome by sequencing a primary human tumor and paired normal tissue(Dataset S1). We further characterized the genetic diversity of DLBCL by sequencing the exomes of 73 DLBCL primary tumors (34 with matched normal DNA) and 21 DLBCL cell lines for comparative purposes. This in-depth sequencing identified 322 DLBCL cancer genes that were recurrently mutated in DLBCLs. We also experimentally validated the effects of genetic alteration of PIK3CD, an oncogene that we identified in DLBCL. Our work provides one of the largest genetic portraits yet of human DLBCLs and offers insights into the molecular heterogeneity of the disease, especially in the context of other recently published studies in DLBCL (7, 8).
Results
Sequencing of a Lymphoma Genome Uncovers the Spectrum of Somatic Variation in DLBCL.
Lymphoma biopsy tissue and unaffected bone marrow were obtained from the same patient. Using the Illumina platform, we generated at total of 171 Gb of 100 bp paired-end sequences from the tumor and matched normal genomes corresponding to an average per-base sequencing coverage of 37-fold and 20-fold, respectively.
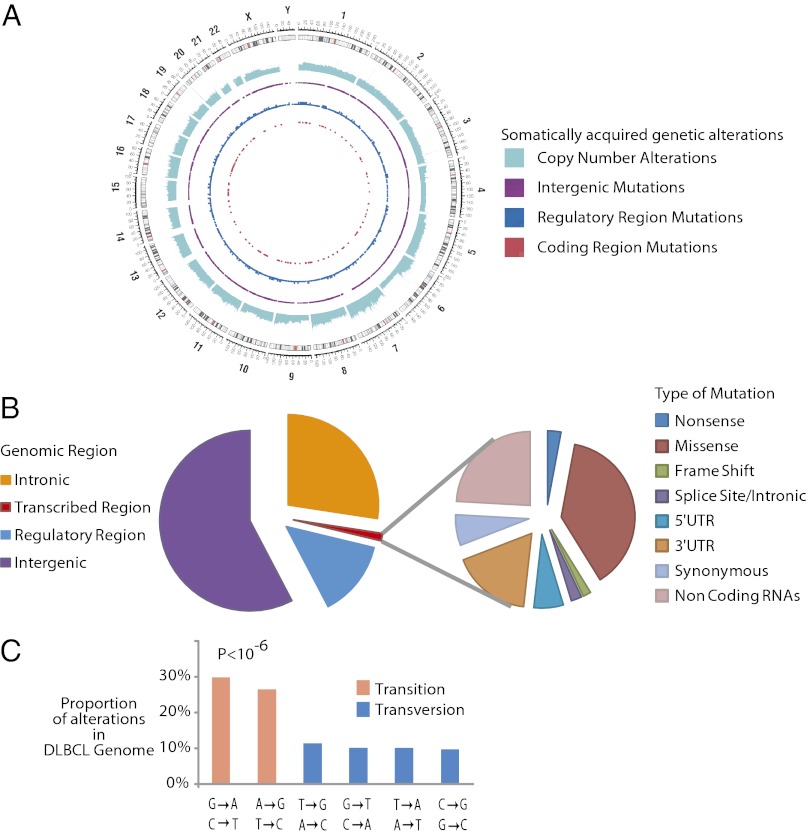
We identified 23,214 somatic sequence alterations occurring throughout the lymphoma genome, summarized in Fig. 1 A and B and SI Appendix, Table S1.
Fig. 1.
Results from sequencing a lymphoma genome. (A) Circos diagram (36) summarizing the somatically acquired genetic variants in a DLBCL genome. The outermost ring depicts the chromosome ideogram oriented clockwise, pter-qter. The next ring indicates copy number alterations in the DLCBL genome. The next three rings indicate somatically acquired mutations in intergenic regions, potential regulatory regions, and the exome respectively. (B) Pie chart depicts the relative number of somatically acquired mutations in the DLBCL genome, which can be classified by their genomic location as intergenic, intronic, potential regulatory, or transcribed regions (Left). (Right) Breakdown of different mutation types observed in the transcribed regions. (C) Histogram depicts the mutation profile of DLBCL. The proportion of mutations in each of the six mutational classes is shown. Transitions represent the majority of the somatically acquired mutations (P < 10−6).
Transitions accounted for about 60% of these events (Fig. 1C), similar to patterns observed in a number of other malignancies (9–11) and suggest that the majority of these DLBCL mutations arise from stochastic endogenous processes rather than environmental exposures, for example, in the context of tobacco exposure and lung cancer (6).
Known oncogenes (12) found to be somatically mutated in this DLBCL patient included ARID1A, SETD2, CARD11, and PIK3R1. Of these genes, only CARD11 (13) has been previously experimentally identified as an oncogene in DLBCL. We also identified structural genetic alterations using approaches described previously (14, 15). In all, we identified seven deletions and three amplifications. Known oncogenes that were implicated by these copy number alterations include PTEN (chromosome 10) and CDKN2A (chromosome 9).
Exome Sequencing Defines the Spectrum of Coding Region Mutations in DLBCL.
To identify recurrently mutated genes in DLBCLs, we obtained a total of 73 cases of primary human samples. We divided the primary human cases into a discovery set (n = 34) and a prevalence set (n = 39). For each of the discovery set cases of primary DLBCLs, we also sequenced paired normal tissue. In addition, we sequenced the exomes of 21 DLBCL cell lines that are widely used to model the disease.
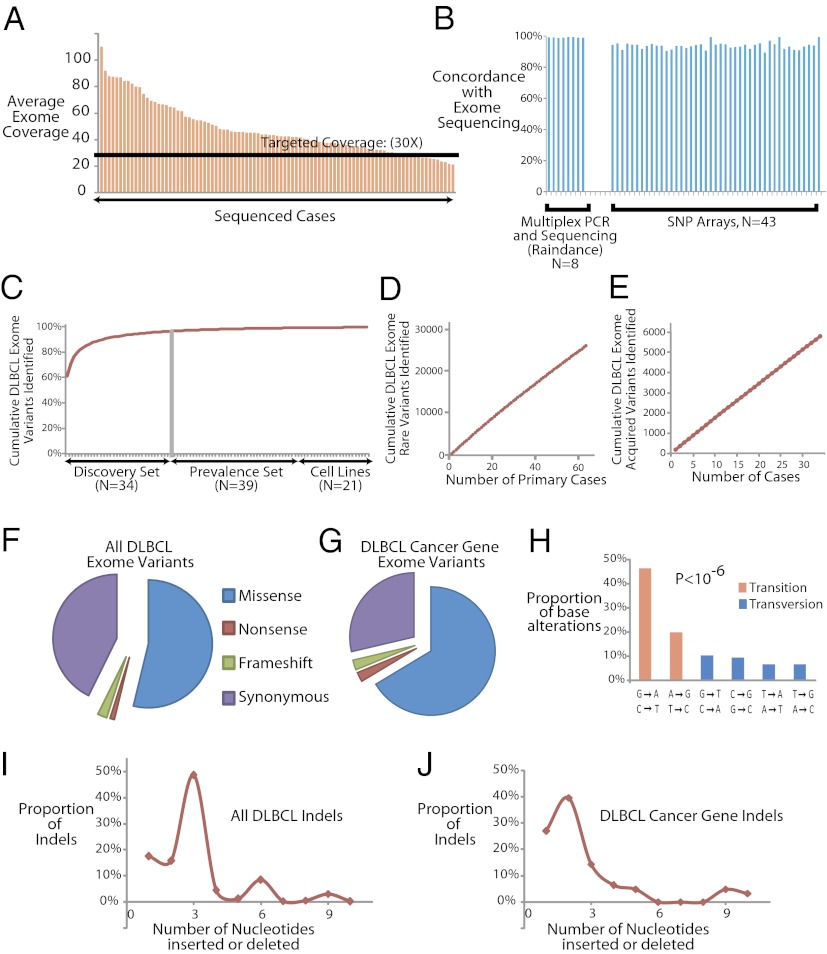
We performed whole-exome sequencing for all of these DLBCL and paired normal cases using the Agilent solution–based system of exon capture, which targets the NCBI Consensus CDS database (CCDS) (16). We generated more than 500 Gb of mappable sequence data and generated sequence data for 94% (median) of the targeted bases in each sample. Our average exome coverage was 47-fold (median, 42.5-fold) per targeted base (Fig. 2A). In all, we identified 121,589 distinct variants in these cases.
Fig. 2.
Exome sequencing in DLBCLs. (A) Bar graph depicts the coverage achieved in each of the cases. The black line indicates our targeted level of 30-fold coverage. (B) Bar graph depicts the overlap between the variants identified by exome sequencing and multiplex PCR followed by deep sequencing (Raindance) in 179 genes in eight cases, as well as exome sequencing and SNP arrays in 43 cases. (C) Plot indicates the average number of additional sequence variants detected in the exomes as a function of adding each additional case. (D) Plot indicates the cumulative estimated number of rare exome variants discovered as a function of sample size. (E) Plot indicates the cumulative estimated number of somatically acquired exome variants discovered as a function of sample size (n = 1 through n = 34). (F) Pie chart indicates the relative distribution of missense, nonsense, frameshift, and synonymous base alterations in the entire dataset. (G) Pie chart indicates the relative distribution of missense, nonsense, frameshift, and synonymous base alterations in the 322 DLBCL cancer genes. (H) Histogram shows the relative distribution of different mutation classes in the 322 DLBCL cancer genes. The difference between the rates of transitions and transversions was highly statistically significant (P < 10−6). (I) Plot shows the relative sizes of all insertions/deletions in the entire dataset. (J) Plot shows the relative sizes of all insertions/deletions among the 322 DLBCL cancer gene variants.
Validation of Genetic Variant Identification.
To verify our methods for exome capture and bioinformatics analyses, we performed exome sequencing on a single Hapmap sample (NA12762) that was previously published (17). We found more than 99% concordance with the published data. We also used three different approaches for further validation.
First, we performed high-throughput, multiplex PCR in microdroplets (Raindance Technologies; ref 18) and sequencing to more than 100-fold coverage for 179 genes (SI Appendix, Table S5) in eight cases. More than 99% of the variants were identical (Fig. 2B; SI Appendix, Table S6). The 100-fold coverage did not result in a substantial increase in variant discovery, confirming our estimates that our coverage of the exome was adequate for the identification of variants in most instances.
Second, we genotyped 43 of these cases using an Illumina SNP array comprising more than 733,000 probes. We found excellent concordance (94.2%) between the microarray calls and the exome sequencing analysis in that sample (Fig. 2B and Dataset S2). Finally, we performed PCR amplification and Sanger sequencing for 25 variants corresponding to 24 genes in 50 cases. Again, we found excellent concordance (93.5%) of the exome capture calls with the calls generated by conventional Sanger sequencing (SI Appendix, Fig. S2). Taken together, these results indicate that our methods for exome-enrichment, sequencing, and bioinformatic analysis produce robust results.
We empirically explored the power of our study to identify novel genetic variants by plotting the expected number of new variants that would be discovered from each additional case of DLBCL (SI Appendix). We found a progressively smaller number of unique variants by sequencing each additional sample (Fig. 2C). By n = 25, the number of additional variants contributed by each additional sample fell to less than 1% of the total (Fig. 2C). These values corresponded well with a regression model for exponential increase (R2 = 0.8768, P < 10−6).
However, the vast majority of these variants depicted in Fig. 2C were common variants and present in our normal controls. The number of rare variants (<1% frequency in the general population) discovered per additional sample remained relatively constant and increased linearly with each additional case (Fig. 2D). Similarly, the number of somatic variants identified in our discovery set (i.e., variants absent from the corresponding paired normal tissue) also increased linearly with the addition of each individual pair of tumor-normal sets (Fig. 2E). The number of individual genes implicated by sequencing additional samples showed a similar linear increase as a function of the number of cases. Because cancer arises predominantly from such somatically acquired rare variants, these findings have implications for the number of samples needed to comprehensively characterize a heterogeneous disease like DLBCL, as discussed below.
Patterns of Exome Variation and Identification of DLBCL Cancer Genes.
We began analysis by aligning the sequencing reads to the genome and determining the distribution of our mutations: 53.8% of the variants were missense, and nonsense, frameshift, and synonymous variants comprised 1.1%, 2.4%, and 42.7% of the total number of variants, respectively (Fig. 2F). These overall patterns of genetic variation in the DLBCL exomes are quite similar to what we would expect in the variation of normal exomes (16). We eliminated common genetic variants that occurred in our set of normal controls identified from dbSNP (19), the 1,000 genomes project (17), and 256 recently published exomes from otherwise healthy individuals (20–22).
Our methods for identifying candidate DLBCL cancer genes are detailed in the SI Appendix. We developed a statistical model for comparing the characteristics of the somatically mutated genes in our discovery set of 34 tumor-normal pairs to previously validated cancer genes. We modeled several variables including frequency of nonsynonymous variation in the gene, frequency of somatic mutation, gene size, rate of nonsynonymous variation in healthy controls, and the predicted effect of the genetic alteration on the encoded protein. DLBCL cancer genes were identified as those that had a score distribution most similar to those of the previously validated cancer genes (P < 10−6).
We identified 322 candidate DLBCL cancer genes as recurrently somatically mutated in DLBCL (Dataset S3). The majority of the 52 known cancer-related genes and the remaining 270 genes have not been previously identified as having a role in lymphomas. Among these 322 DLBCL cancer genes, we identified a total of 1,418 variants in these cases (Fig. 2G; Dataset S4). There was a higher proportion of each category of nonsynonymous mutations including missense (66.4%), nonsense (2.4%), and frameshift (2.5%), and fewer synonymous mutations (28.7%) in these genes compared with the patterns observed in the entire exome.
Once again, we observed a predominance of transitions (P < 10−6, χ2 test; Fig. 2H). Overall, the sizes of the insertions and deletions in these DLBCLs preserved reading frames, with peaks observed at insertion/deletion sizes of three, six, nine, and so on (Fig. 2I). However, in the 322 DLBCL cancer genes, we noted a significant depletion of indel sizes that were multiples of three (Fig. 2J; P < 0.01, χ2 test).
Identification of Protein Coding Sequence Variation in DLBCL.
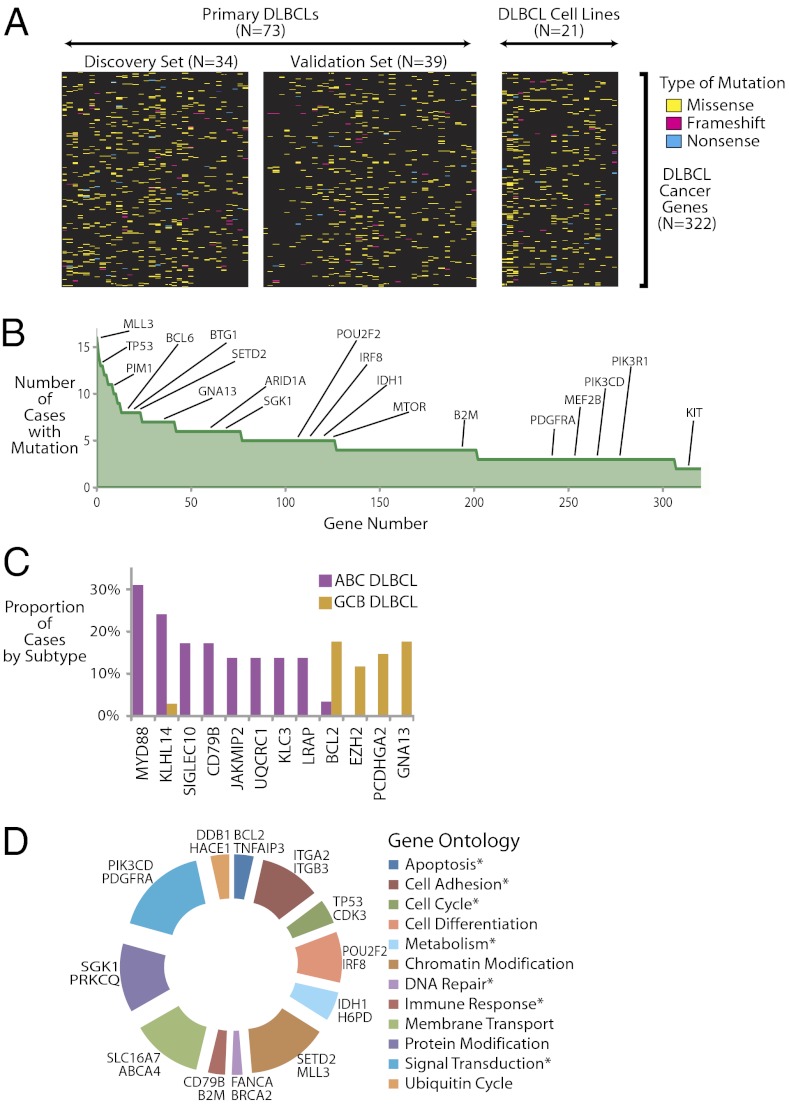
The mutational patterns of these 322 DLBCL cancer genes in the 73 primary lymphomas, as well as 21 cell lines, are depicted in Fig. 3A. The median number of DLBCL cancer gene alterations per patient was 16 (mean, 17).
Fig. 3.
Patterns of exonic mutations in DLBCLs. (A) Heat map indicates the pattern of mutations of the 322 DLBCL cancer genes in 73 primary DLBCLs and 21 DLBCL cell lines. Each column represents a patient or cell line and each row represents a DLBCL cancer gene. Mutation types are indicated in the legend. (B) Frequency of the 322 DLBCL cancer genes are indicated in the graph in descending order. (C) Bar graph shows the frequency of the 12 genes that were found to be differentially mutated in the activated B-cell–like DLBCL subgroup and the germinal center B-cell–like DLBCL subgroup. (D) Relative distribution of genetic mutations by gene ontology categories. The spans of the arcs indicate the relative numbers of different genes annotated with respect to gene ontology (37) terms. Representative genes in each group are shown next to each arc. Terms that match the described hallmarks of cancer (27) are marked with an asterisk.
Fig. 3B shows the frequency of the DLBCL cancer genes, which followed a classic long tail distribution. Our data identify a number of known cancer-related genes in DLBCL that have been previously reported and include TP53 (23), MYD88 (24), PIM1 (25), CARD11 (13), and BCL6 (25). Our data also implicate a number of cancer-related genes that were not previously linked to DLBCL, including PIK3R1, ARID1A, MTOR, and IDH1. These data indicate that the spectrum of mutations and genes involved in lymphomas, and potentially other cancers, may be much larger than has been previously appreciated.
Gene Expression–Based Subgroups of DLBCL Demonstrate Distinct Mutation Patterns.
To better understand potential subgroup-related differences in observed patterns of DLBCL mutations, we performed gene expression profiling using Affymetrix microarrays to distinguish activated B cell-like DLBCLs (n = 29) and germinal center B cell-like DLBCLs (n = 35). We found 12 genes (Fig. 3C) with a frequency of at least 10% in each subgroup that were differentially mutated between the two groups (P < 0.05, Fisher’s exact test). Genes that were more frequently mutated in ABC DLBCLs included MYD88, KLHL14, CD79B, and SIGLEC10, whereas GNA13, BCL2, and EZH2 were more frequently mutated in GCB DLBCL. Of these, we also found GNA13 and EZH2 to be recurrently mutated in Burkitt lymphoma (26), another tumor derived from germinal center B cells.
Functional Categorization of Recurrently Mutated Genes in DLBCL.
Twelve gene ontologies accounted for more than half of the DLBCL cancer genes (n = 203; Fig. 3D). Biological processes comprising signal transduction (e.g., PIK3CD, PDGFRA) and chromatin modification (e.g., MLL3, SETD2) were most commonly implicated as DLBCL cancer genes (SI Appendix, Fig. S3). A number of these biological processes have been directly implicated as hallmarks and enabling characteristics of cancer (27). Thus, a number of these DLBCL cancer genes have directly discernible roles that impact the growth and development of tumors.
The role of signal transduction pathways in tumors is of particular interest because they may be therapeutic targets for small molecule inhibitors. Signaling pathways (28) including JAK-STAT, ubiquitin, WNT, NF-KB, Notch, and PI3 kinase signaling were recurrently mutated in DLBCLs, although mutations in each signaling pathway occurred in only a minority of these cases. Many of these pathways have not been conclusively implicated in lymphomas and might be potential therapeutic targets in DLBCL subsets defined by mutations in them, a notion that we further explored experimentally.
Determination of Genes Enriched with AICDA-Related Mutations.
We further examined the potential role of AICDA (AID) in the acquisition of these somatic mutations in our DLBCL cases. For the 322 DLBCL cancer genes, we determined the number of acquired mutations where the reference sequence was “C,” and of those, the fraction that fell into WRCY motifs (and the reverse complement), which are associated with AICDA activity (29).
We performed a Fisher’s exact test to determine the significance of enrichment for mutations in the WRCY motifs compared with the background rate. We found significant enrichment (P < 0.05) of WRCY motifs in PIM1, BTG1, and CD79B, suggesting that AICDA is a significant contributor to the somatic alterations in these genes.
Gene alterations in PIM1, BTG1, and CD79A have not been described in most solid tumors, suggesting AICDA-related alterations are a lymphoma-specific mechanism, similar to their described role in B-cell biology.
PIK3CD Is Identified as an Oncogene, and PI3 Kinase Inhibition Is a Potential Therapeutic Approach in DLBCL.
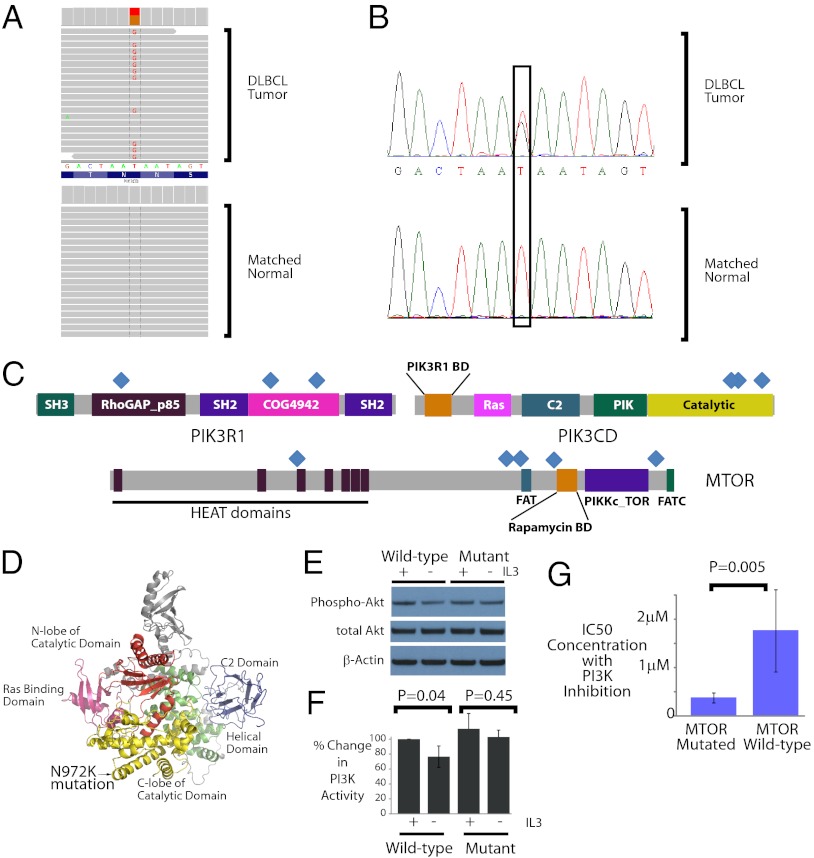
Deregulation of the PI3 kinase pathway is a common feature of many cancers (30). We observed three separate cases with mutations in the PIK3CD gene, which is not thought to be an oncogene. A single point mutation T → G (Fig. 4A), confirmed by Sanger sequencing (Fig. 4B), was identified in the catalytic domain of the PIK3CD gene, which altered the encoded amino acid from one with an uncharged side chain (asparagine) to one with a positively charged side chain (lysine).
Fig. 4.
PI3 kinase pathway in DLBCLs. (A) Deep sequencing reads identify a somatic mutation in PIK3CD in a DLBCL tumor. Sequencing reads matching the genome perfectly are shown in gray. The two samples differ only in a single nucleotide that is G in the tumor, but T (i.e., identical to reference genome) in the matched normal. The data were displayed using the Integrated Genomics Viewer (38). (B) Chromatograms display the results from Sanger sequencing in the same case. The sequenced bases demonstrate a T→G alteration in the tumor but not the matched normal. (C) Distribution of mutations occurring in the PI3 kinase pathway–related genes: PIK3R1, PIK3CD, and MTOR. Each blue diamond marks an individual mutation. Eleven separate events occurred in these three genes. (D) A molecular model of PIK3CD based on threading through the PIK3CG crystal structure. The location of the Asn-972 Lys (N972K) mutation identified in our sequencing data is highlighted within the C-lobe of the catalytic domain. (E) Western blot depicts the alteration in phosphorylated AKT expression initially and 3 h after the withdrawal of IL3. The relative expression of phospho-AKT (S473), with total AKT and β-actin loading controls, are shown. (F) The relative change in PI3 kinase activity is depicted as a function of altered phospho-AKT protein expression normalized to β-actin expression in three separate experiments. The P values were computed using a Student t test comparing the altered PI3 kinase activity in the cells before and after IL3 withdrawal. (+, IL3 exposure; −, IL3 withdrawal). (G) IC50s for 21 cell lines treated with a PI3 kinase inhibitor are shown. The three cell lines with mutated MTOR have approximately fivefold lower IC50s than the 18 cell lines with WT MTOR (P = 0.005).
We found mutations in two additional known oncogenes in the PI3 kinase pathway, PIK3R1 and MTOR, pointing to deregulation of the PI3 kinase pathway as an important oncogenic mechanism in DLBCLs. Other key members of the pathway with known oncogenic roles, including PTEN, FOXO3, and GSK3, were not mutated in our cases. Similar to patterns observed previously in PIK3CA (30), the mutations in PIK3CD, PIK3R1, and MTOR appear to spread across multiple locations of the gene (Fig. 4C) rather than clustering in a single hotspot.
We modeled the PIK3CD protein structure based on that of its paralog PIK3CG, which has been determined through crystallography (31). The identified mutation lies in the catalytic domain in the predicted structure of the protein (Fig. 4D). We overexpressed the WT and the mutant PIK3CD constructs in the FL5.12 lymphoma cell line that is well characterized for its IL3-dependent PI3 kinase signaling (32). In these cells, withdrawal of IL3 is associated with a measurable decrease in PI3 kinase signaling and decreased phophorylated AKT, which is directly downstream of PI3 kinase and provides proliferative signals. In cells expressing the WT form of PIK3CD, we found that withdrawal of IL3 was associated with a measurable decrease in phosphorylated AKT S473 (Fig. 4E). There was no measurable decrease in the phosphorylated AKT in cells expressing the mutant form of PIK3CD, suggesting that the mutation had an activating effect (Fig. 4E). These observations were confirmed in three experimental replicates, all of which showed that IL3 withdrawal was associated with significant down-regulation in phosphorylated AKT in cells expressing WT PIK3CD (P = 0.04), but not in cells expressing mutant PIK3CD (P = 0.44; Fig. 4F). ELISA experiments also demonstrated similar patterns of PI3 kinase activation in cells expressing mutant PIK3CD (SI Appendix, Fig. S8).
Among PIK3CD, PIK3R1, and MTOR, only MTOR was found to be mutated in multiple cell lines (in addition to patient cases). We investigated the effects of a small molecule inhibitor of PI3 kinase, BKM120 (Novartis), on the viability of 21 DLBCL cell lines (Fig. 4G). The three cell lines with MTOR mutations had, on average, a fivefold higher sensitivity to PI3 kinase inhibition than those 18 cell lines that did not harbor these mutations (P = 0.005, Wilcoxon rank test). These results strongly suggest that the presence of mutations in MTOR is associated with sensitivity to PI3 kinase inhibition.
Comparison with Other Genetic Studies Reveals the Striking Genetic Heterogeneity of DLBCL.
Shortly after the completion of our study in June 2011, and during revisions, three separate studies exploring the genetics of DLBCL using similar methodologies and deep sequencing were published (7, 8, 33). Multiple studies applying these methodologies in the same cancer type have generally been lacking thus far, and these publications provided an unusual opportunity for testing the overlapping mutations identified by the different studies.
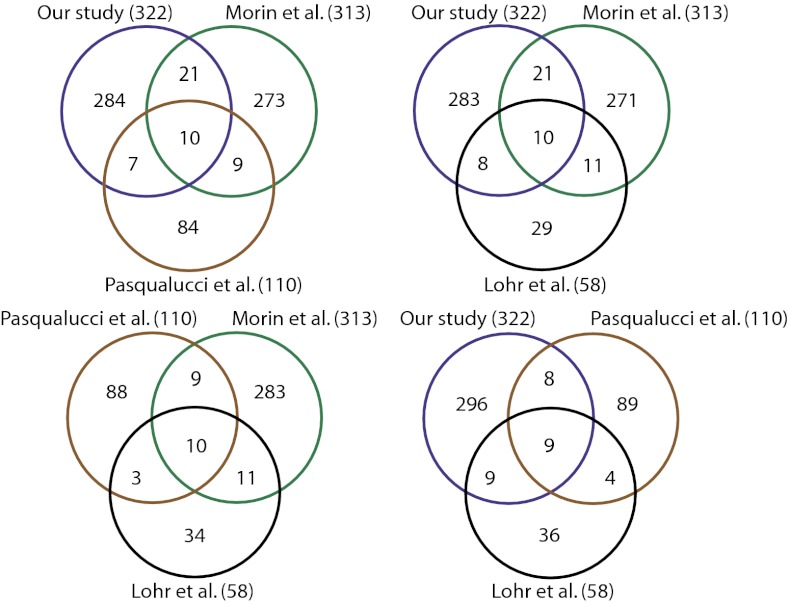
We constructed Venn diagrams depicting the overlap between genes identified in the three studies and our study as shown in Fig. 5. We initially assumed that our study comprising 73 primary DLBCL cases would be sufficient to identify the vast majority of recurrent genetic alterations in the disease. Surprisingly, we noted relatively modest overlaps of roughly 10–20% among the four different studies. Even genes that overlapped between different studies often varied. Similar patterns were observed when we simply compared the overlap between all somatically mutated genes in these studies. The overlap of more frequently mutated genes was still incomplete; when we limited the analysis to those 17 genes that were mutated in more than 10% of the cases reported by Lohr et al. (33), the overlap between the different studies including ours approached 70% (SI Appendix, Fig. S6). Even in that scenario, different studies had overlap with different genes. The remaining 30% of the genes mutated in at least 10% of the cases in that study were not detected by any of the remaining three studies. The overlap is still lower for genes with fewer mutation events. These observations suggest that there is considerable genetic heterogeneity in the disease that contributes the observed patterns of disparate mutations.
Fig. 5.
Overlaps in genes discovered in multiple cancer studies. The Venn diagrams depict the comparison of gene mutations from the four DLBCL studies. The number in parentheses indicates the number of genes identified in each study. The gene lists were as follows: Morin et al. (Table S1 in Ref. 7, genes with confirmed somatic cases), Lohr et al. (Table 1 in Ref. 33, top 58 genes), and Pasqualucci et al. (Table S3 and Fig. 4 in Ref. 8, validated somatic genes).
Although genes that do not overlap among studies might signify some false positives, our analysis indicates that a number of validated oncogenes and tumor suppressor genes were identified in just one study. Examples include NOTCH1 (34), CD74 (34), BCL10, IRF4, MALT1, TET2 (7), BCR, ETV6 (33), PIK3R1, MTOR, KIT, PDGFRA, and ARID1A (our study). The number of identified cancer genes appeared to increase linearly as a function of the size of the study, further indicating that the differences between the individual studies arise from the inherent genetic heterogeneity of the DLBCL tumors, an effect that we also observed in other cancers (SI Appendix, Fig. S7).
Discussion
Through whole-genome sequencing and whole-exome sequencing, we identified the spectrum of sequence variation that occurs in DLBCL. Our data suggest that the majority of genetic variants in DLBCL are stochastically acquired. In all, we identified a total of 322 candidate DLBCL cancer genes that have recurrent somatic mutations in patients with DLBCL. We identified a role for a number of known and previously unknown oncogenes. Many of these genes, including ARID1A, KIT, and IDH1, have not been previously implicated in DLBCL by any previous study.
A central observation of our study is the striking genetic heterogeneity that underlies a relatively common cancer. As we demonstrated, there is a relatively low overlap between four different studies that explore the genetics of DLBCL. Although differences in methodology, the diversity of patient populations, and number of patients might contribute to this low overlap, our data indicate that the major driver of the low overlap is the inherent genetic heterogeneity of the disease. Consistent with this observed heterogeneity, we demonstrate that the number of rare variants and somatic mutations increases linearly with the increased number of cases, suggesting that continued sequencing of tumors will implicate new variants and new cancer-related genes.
Gene expression profiling has previously revealed aspects of the heterogeneity of the disease, particularly with regard to cell of origin of DLBCLs (2). Our data indicate that recurrent mutations in 12 genes were clearly enriched between ABC DLBCLs and GCB DLBCLs. Thus, the two DLBCL subgroups share the mutational patterns of many more genes, suggesting that shared mechanisms underlie their biology. The striking genetic heterogeneity observed in the disease as a whole is also recapitulated in these subgroups.
The recognition of recurrent mutations in the gene coding regions in the disease is an important early step toward understanding its biology and potential therapeutic possibilities. Somatic mutations have previously been observed in multiple genes in the NF-κB pathway, including TNFAIP3(A20) and CARD11 (35). Both of these genes were found to have somatic cases in our study and at least one of the recently published DLBCL studies (7, 8, 33). Histone-modifying genes, such as MLL2 and MEF2B, were found to be frequently mutated in DLBCL (32% and 11.4%, respectively) (7). Although MLL2 was not included in our exome capture library, which was designed using build 36 of the human genome, we also observed somatic mutations in MEF2B and CREBBP, an acetyltransferase gene reported previously (34). MLL3, which forms complexes with MLL2, was the most frequently mutated gene in our cases. Our data also implicate AICDA-related mutations as a major mechanism underlying genetic mutations in the genes PIM1, BTG1, and CD79B. These observations highlight the diverse biological mechanisms underlying the observed genetic diversity.
The genetic heterogeneity of DLBCLs and other cancers implies that no matter what recurrently altered gene or pathway is considered, only a minority of patients are likely to be affected. For that subgroup of patients whose tumors harbor a growing number of recognized genetic lesions that can be targeted therapeutically, the recognition of such alterations can make a crucial difference in their treatment. A number of genetic mutations we identified, including those in PIK3CD, KIT, and PDGFRA, suggest therapeutic possibilities in the affected patients. Our data suggest that such targeted therapeutic approaches in patients will need to be combined with carefully selected assays for those genetic lesions to better understand their role in response to targeted therapies. Our data also have major implications for how we model cancers and the need to ascertain whether extant mouse and other models recapitulate the primary disease. Thus, our study sheds light on the genetic heterogeneity of lymphomas, as well as cancers in general, and underscores the need for individualized approaches for treating patients.
Methods
Detailed methods are provided in the SI Appendix. Whole genome sequencing and exome sequencing were performed on the Illumina platform. Sequencing reads were mapped to the reference genome, and variants were identified, collated, and annotated.
Supplementary Material
Acknowledgments
We thank Susan Sunay for assistance with the sample collection and the Georgia Cancer Coalition for support with sample collection. This study was supported by National Institutes of Health Grants R21CA1561686 and R01CA136895. S.S.D. was also supported by the American Cancer Society. J.C.B. and A.J.J. are supported by the Leukemia and Lymphoma Society. We gratefully acknowledge the generous support of Charles and Daneen Stiefel.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in dbGap, http://www.ncbi.nlm.nih.gov/gap (accession no. phs000573.v1.p1), and the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE22898).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205299110/-/DCSupplemental.
References
- 1.Morton LM, et al. Lymphoma incidence patterns by WHO subtype in the United States, 1992-2001. Blood. 2006;107(1):265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Monti S, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105(5):1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 4.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 12.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 14.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26(3):256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 18.Tewhey R, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27(11):1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry ST, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42(11):969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 23.Young KH, et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood. 2008;112(8):3088–3098. doi: 10.1182/blood-2008-01-129783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 26.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J Biol Chem. 2004;279(8):6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 30.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 31.Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402(6759):313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 32.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18(4):1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.