Abstract
Small, glutamine-rich, tetratricopeptide repeat protein 2 (Sgt2) is the first known port of call for many newly synthesized tail-anchored (TA) proteins released from the ribosome and destined for the GET (Guided Entry of TA proteins) pathway. This leads them to the residential membrane of the endoplasmic reticulum via an alternative to the cotranslational, signal recognition particle-dependent mechanism that their topology denies them. In yeast, the first stage of the GET pathway involves Sgt2 passing TA proteins on to the Get4/Get5 complex through a direct interaction between the N-terminal (NT) domain of Sgt2 and the ubiquitin-like (UBL) domain of Get5. Here we characterize this interaction at a molecular level by solving both a solution structure of Sgt2_NT, which adopts a unique helical fold, and a crystal structure of the Get5_UBL. Furthermore, using reciprocal chemical shift perturbation data and experimental restraints, we solve a structure of the Sgt2_NT/Get5_UBL complex, validate it via site-directed mutagenesis, and empirically determine its stoichiometry using relaxation experiments and isothermal titration calorimetry. Taken together, these data provide detailed structural information about the interaction between two key players in the coordinated delivery of TA protein substrates into the GET pathway.
Keywords: NMR, X-ray crystallography, macromolecular interactions
Tail-anchored (TA) proteins mediate numerous cellular roles, including stress-response, apoptosis, and electron transfer, and are characterized by a single transmembrane domain (TMD) at the extreme C terminus, which tethers them to membranes with the majority of the protein located in the cytoplasm (1). This distinct topology precludes traditional cotranslational membrane insertion via the signal recognition particle as the TMD is obscured by the ribosome until translation terminates (2). An alternative, highly conserved, mechanism named GET (for Guided Entry of TA proteins) was recently delineated in yeast and mammalian systems (reviewed in refs. 3, 4). In Saccharomyces cerevisiae the GET pathway involves at least six proteins including the small glutamine-rich tetratricopeptide repeat (TPR)-containing protein 2 (Sgt2), which is thought to catch TA proteins upon their release from the ribosome (5, 6). The TA-protein substrates are then passed on to a heterotetrameric complex consisting of a dimer each of Get4 and Get5 (7), which then transfers them to an ATPase, the Get3 dimer. An ATP hydrolysis event by Get3 and handover to the transmembrane complex of Get1 and Get2 (8–9) finally facilitates their entry into the membrane. To date, Get3 has been the major focus of high-resolution structural studies, with crystal structures solved in a variety of nucleotide-bound states (3) and, more recently, in complex with the cytoplasmic domains of Get1 and Get2 (8–9). Central TPR domains from human (10) and fungal (11) homologs of Sgt2 have been solved by crystallography, but no high-resolution structures currently exist for the N- and C-terminal domains. The crystal structure of Get4 has been elucidated in isolation (12) and in complex with a short N-terminal (NT) fragment of Get5 (7). In combination with a recently solved structure of the C-terminal dimerization domain of Get5, SAXS data obtained for the full-length complex show an extended conformation with a Get5 dimer linking two Get4 proteins (11). The ubiquitin-like (UBL) domain of Get5, which provides the crucial upstream link to Sgt2, has yet to be solved. Furthermore, little is known about high-resolution intercomplex interactions, which promote or facilitate transfer of TA proteins. The interaction between Get3 and the Get1/Get2 complex (8, 9) is the only one described thus far.
All known cytosolic components of the yeast GET pathway exist as homodimers, and in Sgt2, dimerization is facilitated via the NT domain, which is also known to bind UBL motifs including the Get5 UBL domain (6). In addition, Sgt2 has a central TPR domain known to bind heat-shock proteins and, in the case of its mammalian homolog SGTA, HIV proteins Gag and Vpu (10), growth hormone receptor (13), myostatin (14), and other disease-related proteins. The glutamine-rich C-terminal region of both Sgt2 and SGTA is thought to bind hydrophobic substrates (10), including the TMDs of TA-proteins (15, 16). Here we present the NMR solution structure of the Sgt2_NT dimer in complex with the UBL domain from Get5, the structure of which we first solved by X-ray crystallography and then assigned by NMR for the purposes of mapping the interface with Sgt2 and solving the structure of the complex. Moreover we identify the key residues of the human Get5 homolog, Ubl4a (17), involved in binding the equivalent human SGTA_NT domain, showing that the mode of interaction between these proteins is conserved from fungi to higher eukaryotes. We confirm the stoichiometry of the Sgt2_NT/Get5_UBL complex using isothermal titration calorimetry (ITC) and NMR relaxation experiments.
Results
NMR Solution Structure of Sgt2_NT.
After construct optimization and NMR assignments [BioMagResBank (BMRB) accession no. 18341] as described in Simon et al. (18), we solved the solution structure of Sgt2_NT, residues 1–78, by NMR spectroscopy (Fig. 1 and structural statistics in Table S1) and deposited the coordinates (PDB Accession No. 4ASV). Sgt2_NT forms a tight symmetrical homodimer whose interface was delineated from extensive intermolecular NOEs (244) collected from filtered NOESY experiments on mixed 13C/15N- and unlabeled Sgt2_NT dimers. Each monomer consists of four alpha helices (α1 = K5–K22; α2 = E27–F44; α3 = R48–K57; α4 = L65–S71) connected by short loops and arranged in a fold, as yet unseen in published literature. The dimer interface is highly hydrophobic, resembling the core of a globular protein, and spans an area of 3,222 Å2 as calculated by protein interfaces, surfaces and assemblies (PISA) software (19).
Fig. 1.
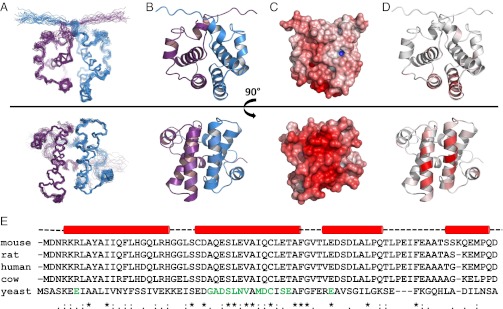
NMR structures of Sgt2_NT dimer rotated 90° around the x-axis. (A) Ensemble views with monomers represented in blue and violet. (B) Ribbon representation with monomers represented in blue and violet. (C) Electrostatic views ranging from –10 negative charge in red to +10 positive charge in blue modeled using the adaptive Poisson-Bolzmann solver (APBS) PyMol plug-in, which calculates the charge distribution displayed on the solvent accessible surface of the protein. (D) Ribbon views colored according to chemical shift perturbation upon binding to Get5_UBL. Residues greater than 80% of maximum chemical shift are colored the darkest red. Between 0% and 80% is divided equally across the seven remaining shades. (E) Sequence alignment of SGT proteins from mouse, rat, human, cow, and yeast with sequence conservation and similarity indicated below. Residues known, from this study, to participate in binding to UBL domains are indicated in green. Helical secondary structure above is derived from our structure solution in yeast. The binding surface is predominantly localized to the second helix (in both yeast and human according to results in this article) and hence the helical dimer interface.
Crystal Structure and NMR Assignment of Get5_UBL.
The structure of Get5_UBL (comprising residues 70–152 of full-length Get5) was solved by molecular replacement (using the UBL domain of Rad23 from S. cerevisiae crystallized in complex with Ufd2: PDB accession no: 3M62) (20) and refined to 1.8 Å resolution in space group P3221 [Protein Data Bank (PDB) accession no. 4A20; see statistics in Table S2]. It has a UBL beta-grasp fold (Figs. 2B, 3, and 4) and comprises one α-helix (α1 = I98–E108) in addition to two parallel sets of antiparallel β-strand pairs (β1 = V74–K80; β2 = F86–F92; β3 = I117–L120; β4 = T143–I148) and two 3/10 (η) helices (η1 = I114–E116; η2 = L132–D134). For the purposes of interaction studies, and calculating the complex structure, a 15N/13C double-labeled Get5_UBL protein sample was used to record the complete battery of standard triple resonance experiments allowing for backbone and side chain assignment of Get5_Ubl via the automated MARS assignment tool in conjunction with manual methods (see details in Simon et al., ref. 18; BMRB accession no. 18342).
Fig. 2.
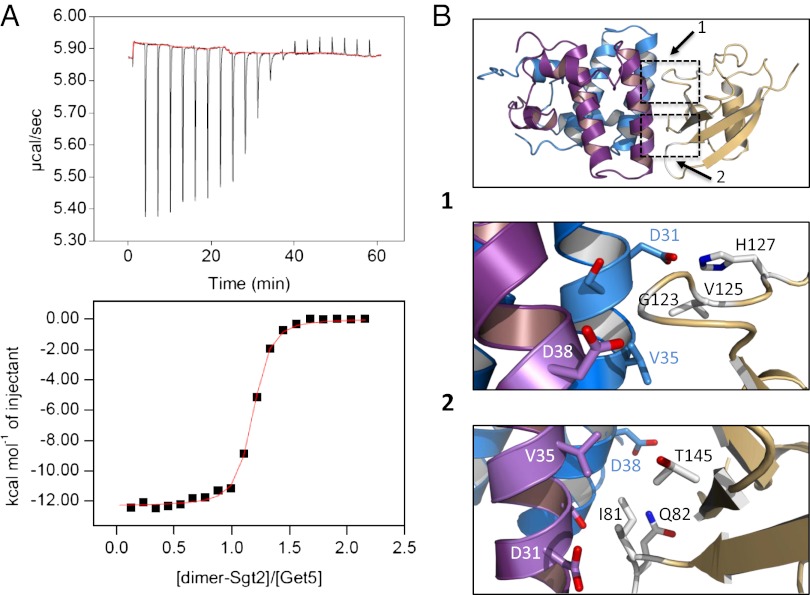
(A) ITC data showing binding of one Get5_UBL domain per dimer of Sgt2_NT. (B) Expansion of the Get5_UBL/Sgt2_NT binding interface with key residues shown as sticks. Although the individual Sgt2_NT monomers contribute symmetrically to binding, Get5_UBL displays an asymmetric binding interface.
Fig. 3.
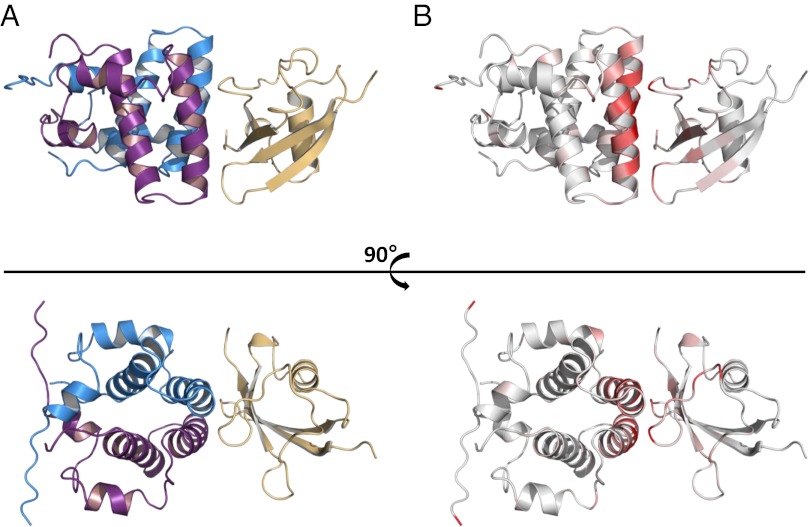
Structure of Sgt2_NT/Get5_UBL complex. (A) Lowest energy structure as calculated by HADDOCK from chemical shift perturbation data and intermolecular NOEs. Sgt2_NT dimer in violet and blue; Get5_UBL in gold. (B) Sgt2_NT and Get5_UBL with mapped chemical shifts aligned to the HADDOCK structure.
Fig. 4.
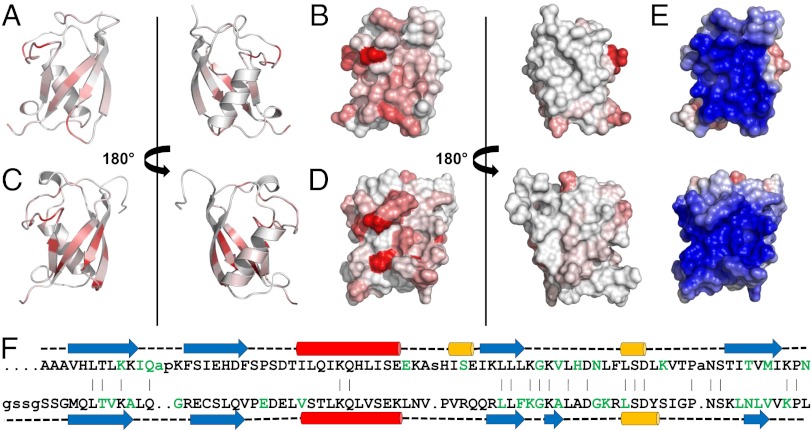
Representations of UBL domains with residues colored white to red in eight shaded increments of increasing normalized 1H/15N chemical shift perturbations observed in the presence of the equivalent SGT. Residues greater than 80% of maximum chemical shift are colored the darkest red. Between 0% and 80% are divided equally across the seven remaining shades. (A) Ribbon and (B) surface views of Get5_UBL binding interface with Sgt2_NT; (C) ribbon and (D) surface views of Ubl4a_UBL binding interface with Sgta_NT; (E) surface views of UBL domains from Get5 (Upper) and Ubl4a (Lower) colored by electrostatics; (F) structure-based sequence alignment of the UBL domains from Get5 (Upper) and Ubl4a (Lower). Red, α-helix; blue, β-strand; yellow, η-helix; green, residues perturbed upon binding to equivalent SGT protein.
Sgt2/Get5 Complex Structure.
Reciprocal chemical shift perturbation experiments were carried out to determine the binding interface between Sgt2_NT and Get5_UBL [see Fig. S1 for heteronuclear single quantum coherence (HSQC) data]. The shifted residues were mapped onto their respective structures (Figs. 1D and 3B) and revealed defined patches of interaction characteristic of a specific binding event. The binding surface on the Sgt2_NT dimer locates to a well-conserved helix (Fig. 1 D and E) and delineates a contiguous negatively charged surface (Fig. 1C). In contrast, the reciprocal site on Get5_UBL reflects the canonical ubiquitin-associated (UBA)-binding patch on UBL proteins (21) and is highly positively charged (Fig. 4E), revealing a strong electrostatic interaction between the two proteins. Get5_UBL presents a β-sheet promoted interface, with β-strands β1 and β4 as well as the loops connecting β1–β2 and β3–β4 providing the main contacts to Sgt2_NT (Fig. 4 A and C). Moreover, filtered NOESY experiments were run on complex samples consisting of labeled Sgt2_NT dimer bound to unlabeled Get5_UBL or unlabeled Sgt2_NT dimer bound to labeled Get5_UBL to identify distance restraints within the binding surface. In addition a filtered NOESY experiment was run on a sample of 50% labeled Sgt2_NT dimer bound to unlabeled Get5_UBL to test whether complex formation with Get5_UBL disrupts the dimer interface. The integrity of the dimer was found to be fully maintained in the presence of Get5_UBL, consistent with its tight hydrophobic nature.
The high ambiguity driven docking program HADDOCK (22) was supplied with all of the experimental data to solve the structure of the Sgt2_NT/Get5_UBL complex (Fig. 3; PDB accession no. 4ASW). The lowest energy ensemble is shown in Fig. S2. A single copy of Get5_UBL binds close to the dimer interface, precluding the binding of two Get5 monomers to the Sgt2 dimer. This is supported by the SAXS model and size exclusion chromatography with multiangle laser light scattering (SEC-MALLS) data previously generated by Chartron et al. on the Sgt2/Get5 complex (11). To examine the stoichiometry of the complex empirically, we carried out ITC and established that one Get5_UBL monomer interacts with each Sgt2_NT dimer (Fig. 2A). We also found that this interaction has a dissociation constant (Kd) of 100 nM, which is consistent with the timescale of the NMR experiments.
NMR relaxation experiments were run on the Sgt2_NT dimer and the full Sgt2_NT/Get5_UBL complex. The estimated rotational correlation times (derived from trimmed mean T1 and T2 relaxation data shown in Fig. S3) were 11 and 15 ns for the free dimer and full complex, respectively, which further supports the presence of a complex comprising one dimer of Sgt2_NT and a single bound copy of Get5_UBL.
Analysis of the Sgt2_NT/Get5_UBL Complex by Site-Directed Mutagenesis.
To verify our model of the complex and confirm the importance of the conserved Sgt2 residues in Get5 binding (Fig. 1), we made single-point mutants of Sgt2_NT and tested the effect on Get5_UBL interaction by ITC (Fig. S4 and Table S3). Mutations were made in the key residues providing electrostatic contacts: D31R, D38R, E42R, and additionally V35A, which is intimately involved in the interaction surface. All mutations resulted in a drop in affinity of at least an order of magnitude (Table S3). The structure of the mutants was retained in all cases, as judged by 1D NMR (Fig. S5), and mutational effects on binding are in qualitative agreement with our structure. Mutation of E42, which interacts with the sidechains of Get5_UBL K95 (in Sgt2 monomer 1) and K70 (monomer 2), resulted in a 15-fold loss of affinity, while mutation of D31, which in each monomer interacts with two basic sidechains (K64 and H73, monomer 1, and K25 and K31, monomer 2) was more severe (∼35× drop). Mutation of V35, which is buried within hydrophobic pockets formed by the aliphatic chains of K64, L66, and M93 (monomer 1) and L66 and T91 methyl group (monomer 2), giving rise to extensive intermolecular NOEs, also results in a severe drop in affinity (∼35×). Mutant D38R reduced affinity to a level not detected by ITC. While this mutation removes only one obvious key interaction per monomer (with the sidechain of K95, monomer 1, and K70, monomer 2), our structure suggests that the increased bulk of the arginine sidechain may clash sterically with the backbone of Get5, resulting in the increased effect.
The extensive hydrophobic dimer interface revealed by our structure of Sgt2 is likely to preclude straightforward dimer-breaking mutations to facilitate analysis of the role played by dimerization in the function of Sgt2. To test this, we sought to take advantage of the dimer symmetry, which results in two key residues, I26 and L83, forming contacts with their equivalent in the opposite monomer. We introduced charged residues at these positions to cause mutual repulsion, disfavoring dimer formation; however, expression of several such mutants yielded no soluble protein. As expected, more conservative mutations such as F16Y, which introduces two buried hydroxyl groups (i.e., one from each monomer) within the hydrophobic core, had little effect on the extensive interface, yielding functional dimer that bound Get5_UBL with wild-type affinity (Table S3).
Equivalent Mammalian System: Sgta/Ubl4a.
The structure of the UBL domain from Ubl4a, the presumptive human homolog of Get5, was solved by Zhao et al. (PDB accession no. 2DZI). Since the chemical shifts were not deposited in the BMRB, we reassigned the backbone using standard methods and titrated the Ubl4a_UBL with an NT dimer of SGTA, the human homolog of Sgt2. Chemical shift mapping results are shown in Fig. 4 C and D along with a structure-based sequence alignment of the two UBL structures in Fig. 4E. The interaction surfaces on the UBL domains are analogous, and SGT sequence alignment data shown in Fig. 1E indicate a high degree of conservation for residues that promote key interactions in the Sgt2/Get5 binding event. These findings show that, despite substantial differences between Get5 and Ubl4a, the UBL domains and their interaction with SGT family members are structurally conserved. This suggests that their functional role(s) with respect to TA-protein biogenesis are also likely to be conserved between the yeast and mammalian GET pathways.
Discussion
Sgt2 plays an important early role in the yeast GET pathway by providing an interface between posttranslational TA-protein binding and efficient entry into the GET pathway for TA-protein delivery to the ER membrane (6). Its actions are facilitated through an interaction of the Sgt2 NT domain with Get5_UBL, occurring within the context of the Get4/Get5 complex (11). In the present study, we structurally elucidate the molecular binding mechanism between Sgt2_NT and Get5_UBL and solve the structures of the complex and both individual components.
The NT dimerization domain of Sgt2 uses a unique configuration of α-helices with a tight hydrophobic interface to present a binding surface for UBL substrates. This structural motif cannot yet be seen in any protein structure deposited in PDB, although it is likely to appear in other unsolved proteins that bind to UBLs, particularly the homologs of Sgt2 from other species. From sequence alignments, Sgt2 is known to be somewhat longer than its mammalian counterparts with the additions localizing to loop regions between the secondary structure elements. Although the structure of full-length Sgt2 has yet to be solved on a molecular level, it is known to have an elongated rather than globular character, based on gel-filtration and SAXS data (11). The NT domain described here probably lies adjacent to the TPR domain from which the C-terminal domain, predicted to be unstructured in the absence of binding partners, emerges.
The dimeric nature of many GET components (Sgt2, Get3, Get4, and Get5 all exist as dimers) has been noted on several occasions (4, 23), but given the current lack of high resolution structures for any large-scale GET complexes, it is unclear exactly how this duplication manifests itself in binding between components. As shown here, Sgt2 dimerizes at the N terminus, leaving the remainder of this elongated molecule with the potential to open like a pair of scissors. In yeast, Get5 dimerization occurs at the C terminus (24), providing its central UBL, as described here, and the NT domain, which binds to Get4, a degree of flexibility. The “open” and “closed” versions of Get3 have been described at length (3), and although Get4, Get5, and Sgt2 all lack the nucleotide binding and hydrolysis capacity of Get3, it is entirely possible that their dimers also adopt variably “open” and “closed” conformations, perhaps depending on which binding partners are present and which stage of the GET pathway is underway. The dimeric state of multiple GET components also provides the potential for many levels of branching within the pathway and could enable multiple distinct processes to occur simultaneously at a single complex. In the case of Sgt2, we speculate that this arrangement likely facilitates a sorting mechanism whereby the C-terminal domains bind hydrophobic substrates and the binding partners of the adjacent TPR and NT domains mediate targeting to the relevant physiological pathways.
Although S. cerevisiae Get5 and its human counterpart, Ubl4a (23), show 20% identity and 43% similarity, much of this localizes to the UBL domain and there are substantial differences between the two proteins, including an additional NT domain in Get5. Although our comparison of the two UBL structures and the binding modes with their equivalent SGT partner proteins (Fig. 4) defines a number of minor differences—for example, the additional 3/10 helix in Get5 and β-strand in Ubl4a—it is the degree of similarity that is the most striking feature. This suggests that the UBL domains of these proteins, and their interactions, are the principal area of commonality between Get5 and Ubl4a.
Chartron et al. have recently characterized a complex between Get5_UBL and an NT Sgt2 construct (residues 1–72) using SEC-MALLS. Based on the apparent molecular weight of the complexes analyzed, and a SAXS envelope, they derive a complex stoichiometry involving the binding of a single copy of Get5_UBL to the Sgt2_NT homodimer (11). Although we had initially anticipated greater disruption to the symmetry of the Sgt2 dimer NMR spectrum upon Get5_UBL binding, our NMR and ITC results also strongly support a stoichiometry of one Get5_UBL to each Sgt2_NT dimer. Furthermore, on the basis of filtered NOESY experiments using partially labeled Sgt2_NT dimers, we determined that the dimer interface is not disrupted upon Get5 binding, which agrees with the strong hydrophobic dimer interface that we delineate and the submicromolar dissociation constant that we measure. In light of our solved complex structure, it is probable that at the elevated temperature required for the NMR experiments (>35 °C), the Get5_UBL swaps rapidly back and forth between the two sides of the Sgt2_NT dimer, thereby averaging out the bound signal. This fast exchange would explain the degeneracy observed in signals from each monomer in the complex and is distinct from the slow exchange behavior arising from the initial binding of Get5 to the Sgt2_NT dimer, which gives rise to the distinct (free and bound state) peaks observable in Fig. S1. The apparent readiness to rapidly exchange between the two binding sites may be enhanced by an apparent pseudosymmetry present in the binding surface of Get_UBL. Considering the key interacting residues, D31, D38, E42, and V35: D31 from one Sgt2 monomer interact with the basic sidechains of K64 and H73 on Get 5, whereas D31 on the other monomer similarly interacts with two basic sidechains on the other side of the UBL interaction surface, K23 and K31. Likewise, D38 in monomer 1 interacts with K95, whereas the equivalent D38 in monomer 2 interacts with a pseudosymmetrical K68. Similarly, E42 from one monomer of Sgt2 interacts with K95, whereas its symmetry-related equivalent contacts K70 on Get5. V35 is at the center of this arrangement, with both sidechains contacting L66 of Get5. Hence Get5_UBL has evolved a pseudosymmetric binding surface for the symmetric arrangement presented by the Sgt2 dimer.
The structure of the Sgt2_NT/Get5_UBL complex provides the first dynamic characterization of the protein interaction that links two key steps of the GET pathway and underlines the structural and functional conservation of GET components involved in targeting TA-proteins to the eukaryotic ER.
Materials and Methods
Protein Production.
Residues 1–78 of Sgt2 (Sgt2_NT) and 70–152 of Get5 (Get5_UBL) from S. cerevisiae and 1–74 of human Ubl4a were amplified from plasmids and inserted via ligation-independent Ek/LIC cloning into pET-46 vectors. These were transformed into Escherichia coli Rosetta cells (DE3), induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at OD600 = 0.8 and expressed overnight at 30 °C. 15N-, 15N/13C-, and 2H/15N/13C-labeled protein samples were prepared according to unlabeled protocols but in M9-based minimal media using correspondingly labeled ammonium chloride (>99% 15N), glucose (>99% U-13C), and deuterium oxide (>99.9% 2H, Sigma Aldrich). Cells were lysed by sonication and protein was purified by affinity chromatography using HisPur Cobalt Resin (Thermo Scientific). Get5_UBL for crystallography was further purified by gel-filtration on a Superdex S200 size-exclusion column (GE Healthcare) in 100 mM Mes, pH 6.0, 150 mM KCl.
NMR Spectroscopy.
All samples were buffer-exchanged by dilution/reconcentration into 100 mM pH 6.0 Mes buffer with 150 mM KCl. NMR experiments were performed on samples of >200 µM uniformly 15N, 13C-labeled protein in either 5 mm (Sigma-Aldrich) Shigemi or standard 5 mm NMR tubes at 35 or 30 °C for Sgt2 and Get5/Ubl4a samples, respectively. Backbone atom assignments were completed with standard experiments (25) and extended into aliphatic sidechains using a combination of HBHA(CBCACO)NH, H(C)CH-TOCSY, (H)CCH-TOCSY, and amide-detected (H)C(CCO)NH- and H(CCCO)NH-TOCSY spectra. Aromatic ring assignments were made from a 13C-NOESY-HMQC spectrum in conjunction with the TROSY-1H, 13C-aromatic HSQC (26). Data were collected on Bruker AvanceIII (600 MHz) and AvanceII (800 MHz) spectrometers equipped with TCI and TXI cryoprobes, respectively, controlled by Topspin3 (Bruker Biospin Ltd). 15N-NOESY spectra were collected on the homebuilt 950 MHz spectrometer equipped with triple-resonance, triple-axis gradient probehead at the University of Oxford. Data were processed using NMRPipe (27) and analyzed in NMRView (One Moon Scientific). Assignment was aided by NMRView modules that provided rapid input for MARS automated assignment (28) and facile handling of sidechain data (29).
NMR Titrations.
Samples of Sgt2_NT and Get5_UBL for titrations were typically 100 µM in 100 mM Mes, pH 6.0, with 150 mM KCl. Spectra were recorded in the absence and presence of a binding partner in a suitable range of molar ratios at 30 °C. Shift changes were monitored by 1D 1H- and 2D 1H-15N HSQC spectra.
X-Ray Crystallography.
Crystals of Get5_UBL were grown by hanging drop vapor diffusion in 0.1 M sodium citrate, pH 5.25, and 3M (NH4)2SO4 at 25 °C following an incubation step of 18 °C for 24 h. Diffraction data were collected at the Diamond Light Source synchrotron beamline I02 without the addition of any cryoprotectant to a maximum resolution of 1.78 Å with the resolution limit truncated to an outer shell I/sigma cutoff of 2.3. Data were integrated using Mosflm (30). The structure of Get5_UBL was solved by molecular replacement with Phaser (ccp4-suite) based on a chainsaw-generated search model of the Ubl domain of Rad23 (20) from S. cerevisiae crystallized in complex with Ufd2 (PDB accession no. 3M62). To avoid phase bias, several nonconserved loop regions as well as the first β-strand of Rad23 were deleted before molecular replacement, increasing the overall sequence identity from 38% to 49%.
After successful molecular replacement, the initial model was built automatically during 20 cycles of arp/warp-based (31) automated model building with preceding DM-based density modification and construction of a new free atoms model. The final structure was completed and rebuilt in Coot (32) and refined with Refmac5 (33).
ITC.
ITC experiments were performed at 30 °C using an ITC-200 microcalorimeter from Microcal (GE Healthcare) following the standard procedure. Proteins were prepared in 100 mM Mes, pH 6.0, 200 mM KCl. In each titration, 20 injections of 2 µL each of Sgt2_NT (dimer), at a concentration of 250 µM, were added to a sample of Get5_UBL at 25 µM (monomer). A spacing of 180 s between each injection was applied to enable the system to reach equilibrium. Integrated heat data obtained for the titrations corrected for heats of dilution were fitted using a nonlinear least-squares minimization algorithm to a theoretical titration curve, using the MicroCal-Origin 7.0 software package. ΔH (reaction enthalpy change in kJ/mol), Kb (equilibrium binding constant in per molar), and n (molar ratio between the proteins in the complex) were the fitting parameters. The reaction entropy, ΔS, was calculated using the relationships ΔG = −RT⋅lnKb (R = 8.314 J/(mol⋅K), T 303 K) and ΔG = ΔH−TΔS.
Supplementary Material
Acknowledgments
The authors thank Dr. Jan Marchant (Imperial College London) for provision of the assignment modules used in the study and continuing valued assistance with software; and Dr. Jonathan Taylor, Caroline Ewens (Imperial College London), and Dr. Luigi Martino (King’s College London) for advice and assistance with ITC. They also thank Prof. Blanche Schwappach (Göttingen University) for helpful discussions and critical reading of the manuscript. The 950 MHz NMR facility at the University of Oxford was funded by the Wellcome Trust Joint Infrastructure Fund and the E. P. Abraham Fund. R.L.I. is supported by a New Investigator Research Grant from the Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4A20, 4ASV, and 4ASW); and the NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu(accession nos. 18341 and 18342).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207518110/-/DCSupplemental
References
- 1.Borgese N, Righi M. Remote origins of tail-anchored proteins. Traffic. 2010;11(7):877–885. doi: 10.1111/j.1600-0854.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- 2.Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta. 2011;1808(3):937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Simpson PJ, Schwappach B, Dohlman HG, Isaacson RL. Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure. 2010;18(8):897–902. doi: 10.1016/j.str.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chartron JW, Clemons WM, Jr, Suloway CJ. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22(2):217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liou ST, Cheng MY, Wang C. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones. 2007;12(1):59–70. doi: 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohl C, et al. Cooperative and independent activities of Sgt2 and Get5 in the targeting of tail-anchored proteins. Biol Chem. 2011;392(7):601–608. doi: 10.1515/BC.2011.066. [DOI] [PubMed] [Google Scholar]
- 7.Chang YW, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285(13):9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477(7362):61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefer S, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333(6043):758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta S, Tan YJ. Structural and functional characterization of human SGT and its interaction with Vpu of the human immunodeficiency virus type 1. Biochemistry. 2008;47(38):10123–10131. doi: 10.1021/bi800758a. [DOI] [PubMed] [Google Scholar]
- 11.Chartron JW, Gonzalez GM, Clemons WM., Jr A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J Biol Chem. 2011;286(39):34325–34334. doi: 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozkurt G, et al. The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Lett. 2010;584(8):1509–1514. doi: 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- 13.Schantl JA, Roza M, De Jong AP, Strous GJ. Small glutamine-rich tetratricopeptide repeat-containing protein (SGT) interacts with the ubiquitin-dependent endocytosis (UbE) motif of the growth hormone receptor. Biochem J. 2003;373(Pt 3):855–863. doi: 10.1042/BJ20021591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan G, et al. Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer Res. 2007;67(20):10087–10096. doi: 10.1158/0008-5472.CAN-07-1646. [DOI] [PubMed] [Google Scholar]
- 15.Leznicki P, Warwicker J, High S. A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem J. 2011;436(3):719–727. doi: 10.1042/BJ20101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Whynot A, Tung M, Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011;43(5):738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42(6):758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon AC, et al. 1H, 13C and 15N assignments of Sgt2 N-terminal dimerisation domain and its binding partner, Get5 Ubiquitin-like domain. Biomol NMR Assign. 2012 doi: 10.1007/s12104-012-9425-7. in press. [DOI] [PubMed] [Google Scholar]
- 19.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Hänzelmann P, Stingele J, Hofmann K, Schindelin H, Raasi S. The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J Biol Chem. 2010;285(26):20390–20398. doi: 10.1074/jbc.M110.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winget JM, Mayor T. The diversity of ubiquitin recognition: Hot spots and varied specificity. Mol Cell. 2010;38(5):627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 23.Chartron JW, Suloway CJ, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA. 2010;107(27):12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chartron JW, VanderVelde DG, Rao M, Clemons WM., Jr Get5 carboxyl-terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J Biol Chem. 2012;287(11):8310–8317. doi: 10.1074/jbc.M111.333252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 1999;34(2):93–158. [Google Scholar]
- 26.Pervushin K, Riek R, Wider G, Wuthrich K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in C-13-labeled proteins. J Am Chem Soc. 1998;120(25):6394–6400. [Google Scholar]
- 27.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Jung YS, Zweckstetter M. Mars—Robust automatic backbone assignment of proteins. J Biomol NMR. 2004;30(1):11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 29.Marchant J, Sawmynaden K, Saouros S, Simpson P, Matthews S. Complete resonance assignment of the first and second apple domains of MIC4 from Toxoplasma gondii, using a new NMRView-based assignment aid. Biomol NMR Assign. 2008;2(2):119–121. doi: 10.1007/s12104-008-9100-1. [DOI] [PubMed] [Google Scholar]
- 30.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.