Abstract
The COP9 (Constitutive photomorphogenesis 9) signalosome (CSN), a large multiprotein complex that resembles the 19S lid of the 26S proteasome, plays a central role in the regulation of the E3-cullin RING ubiquitin ligases (CRLs). The catalytic activity of the CSN complex, carried by subunit 5 (CSN5/Jab1), resides in the deneddylation of the CRLs that is the hydrolysis of the cullin-neural precursor cell expressed developmentally downregulated gene 8 (Nedd8)isopeptide bond. Whereas CSN-dependent CSN5 displays isopeptidase activity, it is intrinsically inactive in other physiologically relevant forms. Here we analyze the crystal structure of CSN5 in its catalytically inactive form to illuminate the molecular basis for its activation state. We show that CSN5 presents a catalytic domain that brings essential elements to understand its activity control. Although the CSN5 active site is catalytically competent and compatible with di-isopeptide binding, the Ins-1 segment obstructs access to its substrate-binding site, and structural rearrangements are necessary for the Nedd8-binding pocket formation. Detailed study of CSN5 by molecular dynamics unveils signs of flexibility and plasticity of the Ins-1 segment. These analyses led to the identification of a molecular trigger implicated in the active/inactive switch that is sufficient to impose on CSN5 an active isopeptidase state. We show that a single mutation in the Ins-1 segment restores biologically relevant deneddylase activity. This study presents detailed insights into CSN5 regulation. Additionally, a dynamic monomer-dimer equilibrium exists both in vitro and in vivo and may be functionally relevant.
Keywords: cullin regulation, protein degradation, MPN, Rpn11
Cell signaling processes mediated by ubiquitinylation, the posttranslational covalent conjugation of ubiquitin molecules, are of prime importance for cellular activity and particularly for protein turnover. Ubiquitin-ligase enzymes (E3s) are responsible for the last step of the ubiquitinylation reaction, and the multisubunit cullin–RING E3 ubiquitin ligases (CRLs) represent the most prominent of E3 enzymes. Among the several factors that regulate CRL activity, cullin neddylation/deneddylation cycles are central (1).
The Cop9 signalosome (CSN), which is an eight-subunit complex largely conserved through evolution, deneddylates CRLs and thereby regulates CRL activity. As a large number of proteins are ubiquitinylated by CRLs, the CSN complex is implicated in the control of a significant proportion of the proteome, including prooncogenes, tumor suppressors, and other important cellular protagonists (1). Not surprisingly, the CSN has been implicated in various cellular functions, ranging from cell cycles to circadian rhythm and to immunity in various organisms. Furthermore, many studies have found a strong link between the CSN and cancers (2).
The CSN, a multi-protein complex of about 320 kDa, contains six proteasome Cop9 eIF3 (PCI)-based subunits and two MPR1-Pad1-N-terminal (MPN)-based subunits. The subunit 5 [CSN5; also known as c-Jun activation domain-binding protein-1 (Jab1)] (3) , one of the two MPN-containing subunits, carries a zinc-dependent isopeptidase catalytic center that contains a Jab1/MPN/Mov34 (JAMM) motif (also known as MPN+ motif) (4). Several detailed studies suggested that the organization of the CSN complex resembles that of the 26S proteasome lid (5), with the deubiquitinase enzyme Rpn11 being the equivalent of the deneddylating subunit CSN5 (4, 6). The physiology of the CSN has been well researched (2). Intriguingly, the CSN cancer implication is attributable mainly to CSN5, which is located on human chromosome 8q.
Smaller forms of the holo-CSN complex, with variable compositions, have been found in vivo (7–11). Although possibly important in cell cycle progression, these sub-CSN complexes have not yet been fully functionally characterized (12). It is interesting that, as alluded to for Rpn11 in several reports (6, 13), CSN5 is found in two forms: a holo-CSN–associated form that is catalytically active and a stand-alone state void of isopeptidase activity (4, 5). The modularity and topology of the CSN complex have been explored in vitro by nondenaturing mass spectrometry (MS), which revealed that CSN5 is a peripheral subunit that can homo-dimerize outside of the CSN complex and interacts mostly with the other MPN-containing subunit, CSN6, in the context of the CSN complex (5). The potential interactions of CSN5 with other CSN subunits, namely CSN1, CSN2, CSN4, and CSN7, have also been highlighted (1, 8, 14, 15).
To elucidate the molecular regulation of CSN5 activity, we structurally and functionally characterized it in its CSN-independent form by X-ray crystallography, molecular dynamics (MD) simulations, and in vitro and in vivo studies. Our structural work uncovered a potential molecular trigger that regulates the active/inactive transition of CSN5. These experiments contributed to the design of a constitutively active form of isolated CSN5, shedding lights on its activation control mechanism at a molecular level.
Results
Overall Structure and Oligomeric Arrangement.
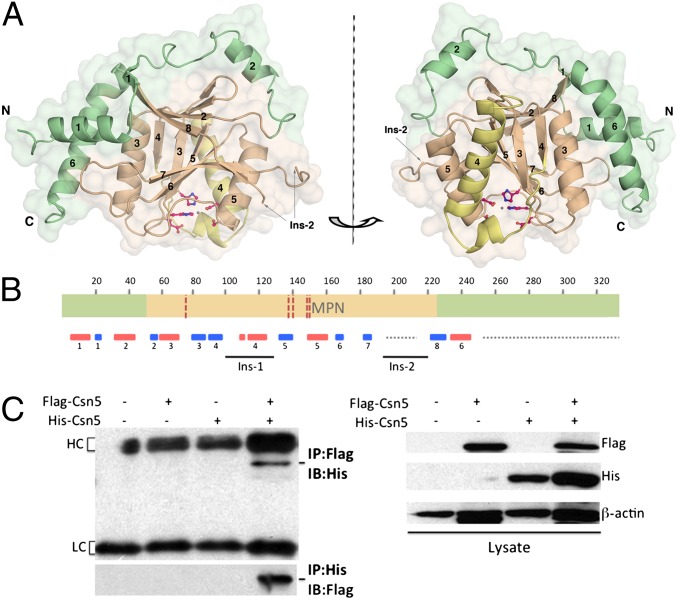
A stable form of human CSN5 comprising residues 1–257 (CSN51–257), identified by MS and N-terminal sequencing, was isolated and crystallized. The crystal structure was solved by selenium– single-wavelength anomalous dispersion (SAD) using diffraction data to 2.6 Å (SI Appendix, Table S1). CSN5, which consists of 334 residues, is a Jab1/MPN superfamily member with a conserved core MPN domain (51–230) and a JAMM motif (Glu76, His138, His140, and Asp151) (Fig. 1 A and B). In addition to the MPN catalytic domain, CSN5 possesses N- and C-terminal regions that tightly pack against the MPN fold and form an extended catalytic domain.
Fig. 1.
CSN5 overall structure and oligomeric arrangement. (A) Overall monomeric structure of CSN51–257 reveals a central MPN core domain (light brown) and peripheral N- and C-terminal extensions (green). The Ins-1 region (residues 97–131) is yellow. Secondary structure elements are numbered, and the catalytic center is shown in ball-and-stick representation. (B) Schematic representation of the CSN5 domain organization. The MPN domain is delineated in orange; the N- and C-terminal extensions are green. The five residues of the JAMM motif, namely E76, H138, H140, S148, and D151, are indicated by red dotted lines. The Ins-1 and Ins-2 insertions are placed with respect to their positions in the sequence. The secondary structure elements are shown in red for α-helices and in blue for β-strands. Regions that are either not ordered in the crystal structure or not included in the crystallized fragment are indicated by a gray dotted line. (C) Biochemical identification of CSN5 dimer in vivo by coimmunoprecipitation experiments. 293T cells were transfected with either Flag- or His-tagged CSN5 or both for 48 h, and then cell lysates were immunoprecipitated (IP) with either anti-Flag (Upper) or anti-His (Lower) tag antibodies and immunoblotted (IB) with anti-His or anti-Flag tag antibodies (Left). The expression of the corresponding pairs of Flag- and His-tagged CSN5 was confirmed by IB using anti-Flag and anti-His antibodies, respectively (Right). IgG heavy chain (HC) and light chain (LC) are indicated.
The asymmetric unit of CSN51–257 crystal contains a dimer, related by a local twofold axis perpendicular to the crystallographic dyad axis, also generating a second dimeric arrangement (SI Appendix; SI Appendix, Fig. S1A). The evidence that CSN5 could adopt a homo-oligomeric arrangement in vivo could be of substantial interest and was further investigated.
CSN5 Can Form Homo-Dimers Both In Vitro and In Vivo.
To investigate the presence of the oligomeric species, our experimental approach was based on chemical cross-linking (SI Appendix, Fig. S1B), dynamic light scattering (DLS; SI Appendix, Table S3), and analytical ultracentrifugation (AUC; SI Appendix, Table S4 and Fig. S1 C and D). The results showed that monomers and dimers were the major species of CSN5 detected in solution. Investigation of the presence of CSN5 oligomers in vivo carried out by coimmunoprecipitation experiments on mammalian cell extracts confirmed that CSN5 assembles in dimer (Fig. 1C). Supported by both in vitro and in vivo data, these observations suggest that a CSN5 dimeric assembly could be present in solution in equilibrium with monomeric species (SI Appendix, Tables S3 and S4). It is noteworthy that other MPN-containing proteins were found to assemble in dimers in the crystals and that each of the described dimers proceeds via totally different interfaces (16, 17), preventing further comparison. Moreover, the question of the physiological relevance of these dimers has not yet been addressed in vivo.
In addition to these experiments and on the basis of the A–B and A–A′ dimer interface analysis, mutations or deletions were designed to selectively weaken these two intersubunit interactions. These data confirmed that CSN5 dimers proceed both in vitro and in vivo predominantly via the A–B interface (SI Appendix; SI Appendix, Fig. S1, E–G) where more than 50% of the contributing residues are highly conserved among the 170 available sequences (SI Appendix, Fig. S2), further suggesting that this assembly may be physiologically relevant. Taken together, these results suggest that CSN5 could form dimers, unveiling a potential new level of regulation in the biology of CSN5. Additional studies will be needed to understand the biological role and the distribution of the CSN5 dimeric form in cells.
Conserved Rigid MPN Domain Is Decorated by CSN5-Specific N- and C-Terminal Extensions.
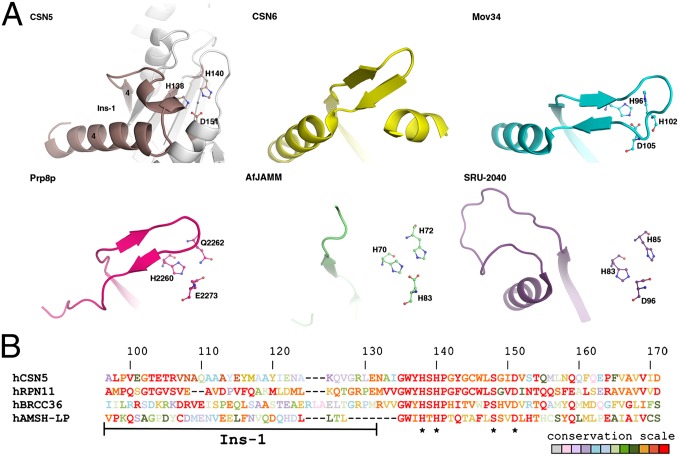
The CSN51–257 structure reveals a fold typical to the Jab1/MPN superfamily that has been described in a number of structures (Fig. 1A; SI Appendix; SI Appendix, Fig. S3) (16–21). Interestingly structural comparison between MPN members revealed that the region spanning from residues 97–131 (referred to as Ins-1) displays an array of conformations in the various MPN members (Fig. 2A). A sequence alignment focused on the Ins-1 segment of four representatives of the MPN+/JAMM family is presented in Fig. 2B. It is noteworthy that the lack of electron density for the CSN5 portion consisting of residues 197–219 [corresponding to Ins-2 in the structure of associated molecule with the SH3 domain of STAM (signal transducing adapter molecule) deubiquitinase (AMSH-LP) (20)] prevented accurate modeling and analysis of this segment.
Fig. 2.
CSN5 MPN domain and specific extensions. (A) Conformational heterogeneity of the Ins-1 region in CSN5 (residues 97–131; brown), CSN6 (yellow; PDB code: 4E0Q), Mov34 (cyan; PDB code: 2O95), Prp8p (magenta; PDB code: 2OG4), AfJAMM (green; PDB code: 1OI0), and SRU-2040 (purple; PDB code: 2KCQ). Secondary structure elements around CSN5 Ins-1 are shown for context in white. (B) Sequence alignment focused on the residues 97–171 (CSN5 numbering) of four human MPN+/JAMM representatives. Residues that correspond to the JAMM motif (with the exception of E76) are indicated with a black *. In addition to each of the proteins, conservation of the residues across the species are illustrated by a color code ranging from red for strictly conserved residues to light gray for residues that are highly variable.
The ensemble of the CSN5-specific N- and C-terminal segments wraps around and makes extensive contacts with the conserved MPN domain core (Fig. 1A;SI Appendix, Fig. S3). Most MPN protein structures solved to date display reduced or are lacking N- and C-terminal additions, with the exception of Prp8p structure that has extensions of similar size to that of CSN5 (19). However, in CSN5 and in the Prp8p scaffolding protein, these regions adopt very different positions and conformations with respect to the core MPN domain.
To extend and complement the structural insights obtained from crystallography, we carried out a series of MD simulations. The CSN5 crystal structure suggests that the central core domain is stable and that some flanking α-helices and loops displaying higher B-factors could be locked into the structure due to the crystal packing. MD simulations of the solvated CSN5 monomer at 300 K for 40 ns confirmed that the core domain is stable (SI Appendix, Fig. S4A) and that the residues forming the Ins-2 segment, the loops, and the N- and C-terminal ends display the maximum fluctuation compared with the central core domain (SI Appendix, Fig. S4B).
CSN5 Zinc-Binding Site Is Catalytically Competent, Similar to Other JAMM-Containing Motifs.
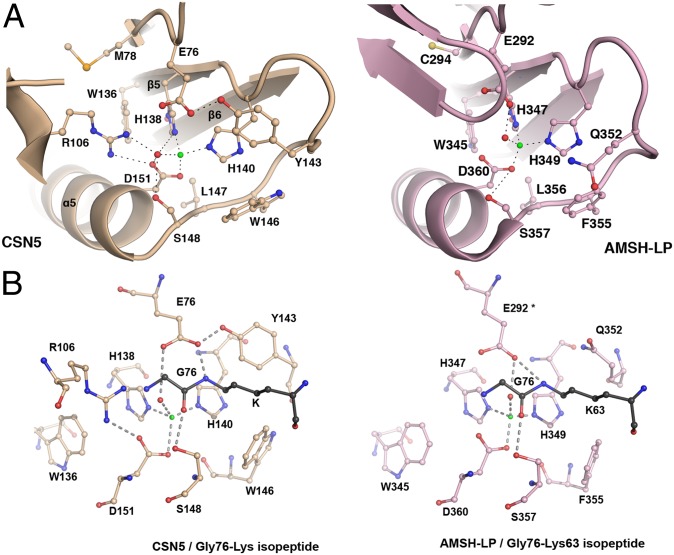
As we anticipated from other MPN+/JAMM proteases, the CSN5 structure contains one zinc atom (Fig. 3A). The strictly conserved zinc coordination site (Fig. 2B) is composed of residues from helix α5 and a subset of the central β-sheet (β5, β5-α5, β6, and β7). The zinc is tetrahedrally coordinated to two His residues (His138 and His140), one Asp residue (Asp151), and a catalytic water molecule hydrogen bonded to Glu76 and Ser148. The importance of the active site zinc-coordinating residues in catalysis was previously tested by mutagenesis (4).
Fig. 3.
Geometry of CSN5 zinc-binding site is compatible with catalysis. Zinc and catalytic water are represented by green and red spheres, respectively. (A) CSN5 active site residues are poised for catalysis. Active site residues and their associated secondary structure are presented for CSN5 (light brown) and AMSH-LP (pink). (B) Model of AMSH-LP Gly-Lys isopeptide in CSN5 confirms that its zinc-binding site geometry is similar to that of AMSH-LP and is therefore competent for isopeptidase activity. The residues of the CSN5 and AMSH-LP zinc-binding sites and of the Gly-Lys isopeptide are shown in ball-and-stick format (light brown, light pink, and dark gray, respectively). The positions of the AMSH-LP residue Glu292, marked by an asterisk, as well as its zinc and catalytic water, were extrapolated from the AMSH-LP structure (PDB code: 2ZNR); the rest is from the structure of the AMSH-LP/K63-Ub2 complex (PDB code: 2ZNV).
To date, AMSH-LP is the only structural example of an active MPN+/JAMM isopeptidase enzyme that exists in an unbound form or in complex with its K63-Ub2 substrate (20). Therefore, it provides for this enzyme family a model for a catalytically competent active site and for substrate interactions. Comparison of the zinc-binding sites of CSN5 and AMSH-LP revealed that the overall topology of their active sites is well conserved (Fig. 3A; SI Appendix, Table S5). In addition, the position and environment of the Gly76-Lys63 isopeptide, inferred from the AMSH-LP/K63-Ub2 complex and straightforwardly placed in the CSN5 active site confirmed that CSN5 adopts a catalytically competent geometry (Fig. 3B). The Gly76-Lys63 isopeptide bond, placed in the CSN5 zinc-binding site, is maintained via a hydrogen bond between the Gly76 carbonyl group and the Ser148 hydroxyl group and between the Lys63 ε-amino group and the Glu76 carboxylate. Moreover, analysis of the crystal packing showed that the C-terminal segment (residues 248–257) of a monomer docks into the active site of the neighboring molecule (SI Appendix, Fig. S4C). Interestingly, a close inspection revealed that this C-terminal portion interacts with the neighboring molecule’s active site in a manner that is reminiscent of the K63-Ub2 isopeptide bond in the AMSH-LP/K63-Ub2 structure (Fig. 3B). Moreover, the CSN5 Ser254 hydroxyl group is hydrogen bonded to Ser148 and occupies a position similar to the distal ubiquitin Gly76 carbonyl in the AMSH-LP active site.
We also investigated the role played by the catalytic zinc ion on the structure and stability of the active site. The side-chain motions of the zinc catalytic site amino acids were analyzed. Their positions were stable over the course of the MD simulations (SI Appendix, Fig. S4D), and their averaged interatomic distances from Zn2+ were in good agreement with those measured from the CSN51–257 and AMSH-LP crystal structures (SI Appendix, Table S5).
Taken together, these observations suggest that, as in AMSH-LP, the zinc-binding site catalytic residues of CSN5 are in a position and are geometry compatible with isopeptidase activity, and therefore the zinc active site conformation of this enzyme in its isolated form is catalytically competent.
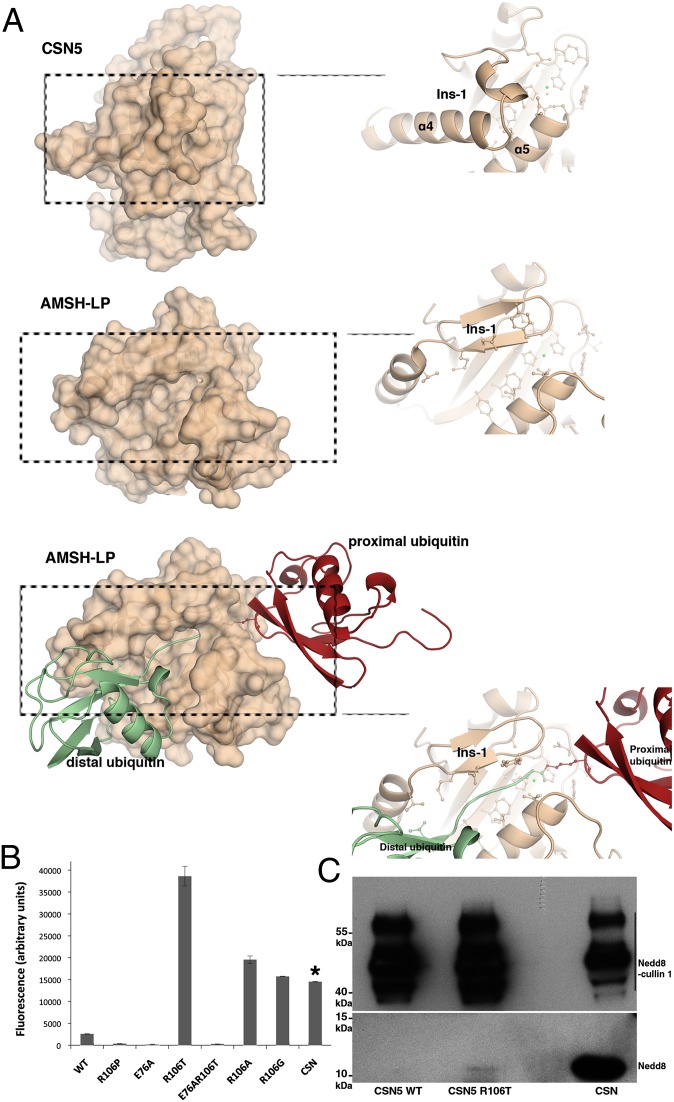
Although the CSN5 zinc-binding site and its catalytic residues are very similar to those of AMSH-LP, their spatial environment and accessibility have several significant differentiating features. In particular, the CSN5 Ins-1 region shielding the active site adopts a radically different topology (loop β4-α4 and α4 helix) compared with AMSH-LP characterized by two antiparallel β-strands followed by a short α-helix (Fig. 4A). An additional crucial feature of CSN5 is the presence of the Arg106 residue projecting out of the Ins-1 segment and establishing a salt bridge with the Asp151 (Fig. 3A).
Fig. 4.
CSN5 adopts a conformation incompatible with Nedd8 recruitment and requires conformational relaxation to perform catalysis. (A) CSN5 Ins-1 masks the putative binding site for Nedd8. (Top) CSN5 surface and a close-up view of the Ins-1 region. (Middle) AMSH-LP surface and a close-up view of the Ins-1 region. (Bottom) Surface of AMSH-LP bound to K63-Ub2 and a close-up view of the groove that accommodates the isopeptide. (B) R106T (or R106A,G) substitution constitutively activates CSN5 against Nedd8 substrate. Isopeptidase activity of CSN5 or of the CSN on Nedd8-AMC was measured by the increase of fluorescence intensity at 460 nm (λexcitation = 380 nm) after 50 min at 28 °C. The concentration of CSN5 used in these experiments is 210-fold more than that of the CSN complex, as indicated by an asterisk. Taking into account this dilution factor, the CSN complex appears 79-fold more active than the most active R106 variant, R106T. (C) The R106T variant form of CSN51–257 is able to deneddylate Nedd8-cullin 1. (Upper) Nedd8 signal of the Nedd8-cullin 1 and (Lower) Nedd8 released on Nedd8-cullin 1 isopeptide bond hydrolysis for CSN5 WT, CSN5 R106T, and the CSN complex (from left to right).
Surroundings of the CSN5 Zinc Catalytic Site Is Not Competent for Nedd8 Recruitment, Without Conformational Rearrangements.
Two different activation states of CSN5 are described in the literature (4, 5): an active deneddylase in the context of the holo-CSN complex and a stand-alone inactive form in the isolated subunit. Because our data suggest that the CSN5 active site is poised for catalysis, it seemed logical to explore substrate binding and recruitment by this enzyme.
In the crystal structure of the AMSH-LP/K63-Ub2 complex, the two ubiquitin molecules, referred to as proximal and distal, interact with AMSH-LP via numerous electrostatic and hydrophobic interactions (SI Appendix, Fig. S2) (20). The directionality of the isopeptide bond implies that neural precursor cell expressed developmentally downregulated gene 8 (Nedd8) would occupy the site corresponding to the distal ubiquitin in the AMSH-LP/K63-Ub2 structure. The distal ubiquitin molecule mediates the largest interaction surface area and contributes the most to the binding affinity of K63-Ub2 for AMSH-LP. Correct positioning of the K63-Ub2 isopeptide bond in the long recognition groove of AMSH-LP is ensured by interactions between AMSH-LP [in particular, the Ins-1 region, the Ins-2 loop (disordered in CSN5), and the segment between these two insertions) and the proximal and distal ubiquitins (SI Appendix, Fig. S2)]. The C-terminal portion of the distal ubiquitin adopts an extended conformation that fits in the substrate-binding groove delimitated by two α-helices and a β-hairpin. Ubiquitin and Nedd8 molecules are 58% identical over 76 residues and adopt the same fold (22, 23). The interactions with the last four residues of ubiquitin/Nedd8 preceding the isopeptide bond are likely to be preserved in CSN5. Only one residue at position 72 (arginine and alanine in ubiquitin and Nedd8, respectively) differentiates ubiquitin from Nedd8 in the last 10 residues. Analysis of the AMSH-LP residues implicated in the distal ubiquitin recognition site revealed that more than 65% of them are highly conserved in CSN5 (SI Appendix, Fig. S2). However, most of the residues for which no equivalent could be found in CSN5 belong to the highly divergent Ins-1 region that has a very different sequence (Fig. 2B) and conformation in CSN5 and AMSH-LP. Consequently, without the structure of CSN5 in its active state, detailed analysis of the substrate-binding site in CSN5 is prevented.
Despite the high conservation of the interaction site in CSN5 (SI Appendix, Fig. S2), the conformation of the Ins-1 segment observed here sterically precludes Nedd8 binding (Fig. 4A). Extensive structural changes of this segment, which probably confers some of the specificity for Nedd8 ligand, would be required to create a fully competent binding site.
Arginine Residue Contributes to the Control of CSN5 Isopeptidase Activation State.
The major difference at the active site level between CSN5 and AMSH-LP corresponds to the conformation of the Ins-1 segment. It is therefore most interesting to note that the Ins-1 segment of CSN5 shows signs of flexibility, as indicated by high crystallographic B-factor values and the fact that it exhibits significant conformational variability within representatives of the MPN family (Fig. 2A). Moreover, MD simulations flagged portions of the Ins-1 region as highly flexible (residues 101–104, 108–112, and 123–131), with a propensity to adopt different conformations during the course of the simulations (SI Appendix, Fig. S4B). These CSN5 segments, bracketing the residue Arg106, display pronounced movements opening onto the solvent in MD simulations, whereas Arg106 contributes significantly to the anchoring of the Ins-1 segment to the zinc-binding site via its salt bridge with Asp151 (SI Appendix, Fig. S5A). Our MD studies confirmed the potential importance of Arg106 with this salt bridge being maintained in the 40-ns trajectory. The observations that Arg106 plays a role in CSN5 plasticity were further probed and confirmed by rotamerically induced perturbation (RIP) simulations (SI Appendix, Fig. S5B) (24). These data suggest that the intrinsic flexibility and plasticity of the Ins-1 region could allow major conformational rearrangements to accommodate Nedd8 binding and that Arg106 could have here a triggering function for structural rearrangement of the Ins-1 segment.
To evaluate the role of Arg106 as a potentially important protagonist in CSN5 activation switch, we tested the effect of Arg106 substitution on CSN5 isopeptidase activity. In agreement with published data in the literature (4, 5), we confirmed that the CSN51–257 WT form is void of isopeptidase activity and showed that the R106 substitution to threonine is sufficient to restore robust constitutive isopeptidase activity against two synthetic substrates: LRGG–7-amido-4-methylcoumarin (AMC) and Nedd8-AMC (Fig. 4B; SI Appendix, Fig. S6). Substitution of the arginine residue by alanine and glycine similarly confers isopeptidase activity to CSN5, albeit to a slightly reduced level compared with the R106T variant, whereas the R106P variant is void of activity (Fig. 4B). Analysis of the zinc binding site topology in CSN5 and AMSH-LP revealed that the glutamate residue corresponding to the position 292 in AMSH-LP is the glutamate 76 in CSN5. The position 76 was consequently mutated to an alanine residue. As expected and in comparison with the WT and R106T variants, the E76A and E76A/R106T variants do not display isopeptidase activity on Nedd8-AMC (Fig. 4B). Comparison of the rescued activity of CSN5 R106T with that of the holo-CSN complex using Nedd8-AMC as a substrate highlighted that the multiprotein complex is about 79-fold more active than the stand-alone protein (Fig. 4B). To complement these activity data, pull-down experiments, using GST-CSN51–257 as the bait and Nedd8 as the target, showed that the R106T form was much more efficient at binding Nedd8 than the WT counterpart (SI Appendix, Fig. S7). Furthermore, MD simulations on the R106T variant confirm that the Ins-1 segment is released from the vicinity of the zinc-binding site; similar results were obtained for R106G and R106P (SI Appendix, Figs. S4B and S5C). These observations confirm that releasing the Ins-1 segment from its anchoring point could favor a conformation that is compatible with Nedd8 binding. Taken together, these data strongly suggest the implication of Arg106 in the active/inactive switch of CSN5. The importance of R106 is further supported by its strict conservation in CSN5 across eukaryote species (Fig. 2B; SI Appendix, Fig. S2). These results would indicate that the conformational relaxation of the Ins-1 region allows substrate binding and additionally corroborates our analysis on the intrinsic topological competence of the zinc binding site for catalysis but also point out that the CSN complex brings further activation and/or activity determinants that makes CSN-associated CSN5 more active than its stand-alone form.
To extend our analysis to a physiological substrate, we asked whether R106T also supports deneddylation of neddylated cullin 1, in complex with Rbx1 in vitro. As expected, the CSN complex is very effective at deneddylating cullin 1, as visualized by the important decrease of neddylated forms of cullin 1 and concomitant release of free forms of Nedd8, detected with Nedd8 antibodies. Importantly, although the stand-alone CSN5 WT was unable to deneddylate cullin 1, a modest but significant level of free Nedd8 was released when using the R106T variant (Fig. 4C). Confirming our results using Nedd8-AMC, these observations show that the WT-isolated form of CSN5 is indeed void of deneddylase activity, that the loss of the R106-D151 salt bridge contributes to unveiling its deneddylase activity, and that the integral CSN is much more efficient at deneddylating cullin 1.
Discussion
Mediated most likely through its deneddylase activity, the function of the CSN complex is important for cellular homeostasis, as highlighted by its implication in proliferative diseases (25). The sequence alignment of the CSN catalytic subunit, CSN5, from different organisms reveals highly conserved features throughout the sequence in agreement with its catalytic function within the CSN complex. The distribution of CSN5 between the holo-CSN, different sub-CSN complexes, and CSN-independent forms observed in a number of studies could play a role in CSN5 regulation, but clearly the critical and best understood function resides in the deneddylation of cullins.
Our work reveals that CSN5 can be found in different oligomeric states in vitro and in cell lysates and may predominantly follow a monomer-dimer equilibrium. The interaction between CSN5 and various partners has been investigated in previous studies, but only in its monomeric form (2). Its assembly in dimers could be relevant in mediating protein-protein interactions and subcellular localization of CSN5.
The main aspect in CSN5 biology that is addressed in this work is its activation state in the CSN-independent context. To glean insights into CSN5 isopeptidase activity regulation, we used structural biology and in silico MD simulations, which together created a detailed picture of CSN5 activity control. The crystal structure of CSN5 in a CSN-independent form displays an extended catalytic domain that revealed a number of features, contributing to our understanding of the enzyme’s activation and substrate recruitment. In analogy to the structure of AMSH-LP (20), the stand-alone form of CSN5 adopts a zinc-binding site geometry that appears compatible with isopeptidase activity and potentially with binding of the Gly76-Lys63 isopeptide, as extrapolated from the cocrystal structure of AMSH-LP/K63-Ub2 to the CSN5 zinc-binding site (Fig. 3B). Unlike AMSH-LP/K63-Ub2, however, investigation of the recruitment of Nedd8 by CSN5 revealed that the substrate-binding site is not formed in CSN5 and that the Ins-1 segment would require substantial structural rearrangement for Nedd8 to bind. These observations were confirmed by analysis of Ins-1 flexibility and plasticity by in silico simulations. Our work also helped understanding one of the molecular events that may trigger these conformational changes in CSN5. MD and RIP calculations pointed to a role for the strictly conserved Arg106 in keeping this segment in a conformation not competent for Nedd8 binding. The implication of this residue, validated by in vitro experiments, led to the confirmation that Arg106 is an important protagonist in CSN5 activation switch. Indeed, substitution of this residue by a threonine and to a lesser extent, alanine and glycine (but not proline), restores a constitutive isopeptidase activity against two synthetic substrates: LRGG-AMC and Nedd8-AMC (Fig. 4B). This work was extended by testing isopeptidase activity of the R106T variant on a physiological substrate, neddylated cullin 1. The CSN5 R106T variant shows modest but significant isopeptidase activity, confirming results obtained on synthetic substrates (Fig. 4C). However, comparison of this activity with that of the CSN complex reveals, as observed for Nedd8-AMC, that the CSN complex is a more efficient deneddylase than CSN5 R106T. It is noteworthy that, although we observed robust isopeptidase activity on Nedd8-AMC by the CSN complex, this is in contrast with the observations reported by the Dubiel’s group (26, 27), in which no activity was found. Even though the CSN isopeptidase activity measured in our experiments corresponds to a weaker level than the activity recorded on a physiological substrate reported in ref. 28, probably reflecting the higher efficiency of the CSN complex on the physiological substrate, this activity was readily measured in our fluorescence-based assay. Additionally Pan’s group reported that the CSN complex was unable to process another linear substrate, pro-Nedd8 (29), using a gel-shift assay. These discrepancies, probably reflecting on the complexity of the studied system, would need further investigation to be resolved.
These observations relative to CSN5 activation shed some light onto different regulatory levels that keep CSN5 under control. CSN5 appears to require an activation step for which one of the switches is likely to correspond to the Arg106–Asp151 salt bridge: loss of this strong interaction promotes the existence of an open form of the enzyme that unveils deneddylase activity on both synthetic and physiological substrates. Coming in addition to the Ins-1 release is its restructuration to form the second ridge of the substrate-binding groove in which the isopeptide would insert. Integration of CSN5 into the CSN complex and the consequent protein–protein interactions with CSN subunits such as CSN6, as highlighted by nondenaturing MS experiments (5), are likely to play a part in both CSN5 activation and substrate recruitment. Given the body of evidence (30–33) that supports the involvement of multiple CSN subunits in substrate recruitment (e.g., the interaction between CSN2 and cullin 1), the observed difference in activity between the CSN complex and the stand-alone R106T CSN5 could be directly linked to the presence of substrate recruitment exosites remote from CSN5 catalytic site, achieving optimal activation and contributing to productive binding of Nedd8-cullin isopeptide bond. Supported by our work, CSN5 incorporation into the CSN complex probably does not lead to global structural reshaping of the enzyme. Instead, the structural changes are likely to be limited to the conserved Ins-1 segment (identified as malleable in our MD calculations), the Ins-2 region (disordered in the crystal), and possibly the C-terminal domain (residues 258–334) to prime the deneddylating molecule for catalysis. Integration of CSN5 in the CSN complex is probably providing the conformational energy necessary for the activation switch.
In addition to addressing CSN5 activity regulation, our work generates insights into the development of CSN5-specific inhibitors. Essentially, the C-terminal tail of one molecule docks in the active site of another one. This association could be exploited to build potent peptidomimetic antagonists that would be of possible interest in the context of breast cancers (2).
Taken together, our study suggests that CSN5 in its CSN-independent form is deficient in substrate recruitment and that a single residue contributes significantly to the activation switch. This discovery provides the framework for conducting biochemical and functional investigations to elucidate the regulatory mechanisms that control CSN5 function.
Materials and Methods
Construct Design.
For detailed construct design, cloning, mutagenesis, protein expression, and purification procedures, see SI Appendix.
Crystallization, Data Collection, Structure Determination, and Molecular Dynamics Simulations.
Purified CSN51–257 formed crystals using the sitting drop vapor diffusion method. A dataset, collected at 2.6-Å resolution from a SeMet-labeled CSN51–257 crystal, was used to determine the structure using the SAD method. The Se substructure was solved using the Charge Flipping method (34) and used for phasing with the PHASER program (35). Details are provided in SI Appendix and SI Appendix, Table S1. The A chain from the CSN51–257 crystal structure was used as the initial structure for MD simulations. Detailed procedures and references are provided in SI Appendix.
Coimmunoprecipitation Experiments and Isopeptidase Assay.
Details regarding cell culture, reagents, transfection, and coimmunoprecipitation experiments are provided in SI Appendix. Different types of AMC-derived substrates, LRGG-AMC, and Nedd8-AMC, as well as neddylated cullin 1, were used to assess CSN5 activity. Details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank N. Zheng for constructive discussion and expression plasmids; M. Cohen-Gonsaud, J. Endicott, O. Coux, and L. Chiche for helpful discussions; and D. Staunton, L. Coquet, and A. Le Roy for experimental assistance. This work was supported by the Université Montpellier 1, the Institut National de la Santé et de la Recherche Médicale, the CNRS [Projets Exploratoires Premier Soutien (PEPS) Grant 2009], and the Groupement des Entreprises Françaises dans la lutte contre le Cancer (GEFLUC) Languedoc Roussillon (A.E.); the National Cancer Institute (RO1-CA90853) and Cancer Prevention and Research Institute of Texas (RP120451) (to F.X.C.); and the CNRS, the Association pour la Recherche sur le Cancer, and the City of Paris (L.P.). We acknowledge the use of the ID14-EH4 beamline [European Synchrotron Radiation Facility (ESRF)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4F7O).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209345110/-/DCSupplemental.
References
- 1.Wei N, Serino G, Deng XW. The COP9 signalosome: More than a protease. Trends Biochem Sci. 2008;33(12):592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Shackleford TJ, Claret FX. JAB1/CSN5: A new player in cell cycle control and cancer. Cell Div. 2010;5:26. doi: 10.1186/1747-1028-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claret FX, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383(6599):453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- 4.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 5.Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17(1):31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freilich S, et al. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9(20):1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- 8.Kwok SF, et al. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10(11):1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mundt KE, Liu C, Carr AM. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell. 2002;13(2):493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oron E, et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development. 2002;129(19):4399–4409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- 11.Tomoda K, et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277(3):2302–2310. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- 12.Fukumoto A, Tomoda K, Kubota M, Kato JY, Yoneda-Kato N. Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett. 2005;579(5):1047–1054. doi: 10.1016/j.febslet.2004.12.076. [DOI] [PubMed] [Google Scholar]
- 13.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 14.Kapelari B, et al. Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J Mol Biol. 2000;300(5):1169–1178. doi: 10.1006/jmbi.2000.3912. [DOI] [PubMed] [Google Scholar]
- 15.Serino G, et al. Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell. 1999;11(10):1967–1980. doi: 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanches M, Alves BS, Zanchin NI, Guimarães BG. The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. J Mol Biol. 2007;370(5):846–855. doi: 10.1016/j.jmb.2007.04.084. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, et al. The crystal structure of the MPN domain from the COP9 signalosome subunit CSN6. FEBS Lett. 2012;586(8):1147–1153. doi: 10.1016/j.febslet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 18.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2(1):E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell. 2007;25(4):615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455(7211):358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 21.Tran HJ, Allen MD, Löwe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42(39):11460–11465. doi: 10.1021/bi035033g. [DOI] [PubMed] [Google Scholar]
- 22.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273(52):34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 23.Ramage R, et al. Synthetic, structural and biological studies of the ubiquitin system: The total chemical synthesis of ubiquitin. Biochem J. 1994;299(Pt 1):151–158. doi: 10.1042/bj2990151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho BK, Agard DA. Probing the flexibility of large conformational changes in protein structures through local perturbations. PLOS Comput Biol. 2009;5(4):e1000343. doi: 10.1371/journal.pcbi.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14(11):1209–1225. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- 26.Schmaler T, Dubiel W. Control of deneddylation by the COP9 signalosome. Subcell Biochem. 2010;54:57–68. doi: 10.1007/978-1-4419-6676-6_5. [DOI] [PubMed] [Google Scholar]
- 27.Hannss R, Dubiel W. COP9 signalosome function in the DDR. FEBS Lett. 2011;585(18):2845–2852. doi: 10.1016/j.febslet.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Emberley ED, Mosadeghi R, Deshaies RJ. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J Biol Chem. 2012;287(35):29679–29689. doi: 10.1074/jbc.M112.352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278(31):28882–28891. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, et al. Consequences of COP9 signalosome and 26S proteasome interaction. FEBS J. 2005;272(15):3909–3917. doi: 10.1111/j.1742-4658.2005.04807.x. [DOI] [PubMed] [Google Scholar]
- 31.Min KW, et al. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem Biophys Res Commun. 2005;334(3):867–874. doi: 10.1016/j.bbrc.2005.06.188. [DOI] [PubMed] [Google Scholar]
- 32.Olma MH, et al. An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J Cell Sci. 2009;122(Pt 7):1035–1044. doi: 10.1242/jcs.043539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, et al. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol. 2002;12(8):667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 34.Dumas C, van der Lee A. Macromolecular structure solution by charge flipping. Acta Crystallogr D Biol Crystallogr. 2008;D64(Pt 8):864–873. doi: 10.1107/S0907444908017381. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.