Abstract
The human heart is believed to grow by enlargement but not proliferation of cardiomyocytes (heart muscle cells) during postnatal development. However, recent studies have shown that cardiomyocyte proliferation is a mechanism of cardiac growth and regeneration in animals. Combined with evidence for cardiomyocyte turnover in adult humans, this suggests that cardiomyocyte proliferation may play an unrecognized role during the period of developmental heart growth between birth and adolescence. We tested this hypothesis by examining the cellular growth mechanisms of the left ventricle on a set of healthy hearts from humans aged 0–59 y (n = 36). The percentages of cardiomyocytes in mitosis and cytokinesis were highest in infants, decreasing to low levels by 20 y. Although cardiomyocyte mitosis was detectable throughout life, cardiomyocyte cytokinesis was not evident after 20 y. Between the first year and 20 y of life, the number of cardiomyocytes in the left ventricle increased 3.4-fold, which was consistent with our predictions based on measured cardiomyocyte cell cycle activity. Our findings show that cardiomyocyte proliferation contributes to developmental heart growth in young humans. This suggests that children and adolescents may be able to regenerate myocardium, that abnormal cardiomyocyte proliferation may be involved in myocardial diseases that affect this population, and that these diseases might be treatable through stimulation of cardiomyocyte proliferation.
Keywords: heart failure, pediatrics
Heart failure, a leading public health problem worldwide (1), is linked to the loss of cardiomyocytes (2–4). The only currently available, definitive therapy—heart transplantation—is limited by donor availability. New approaches, such as cell transplantation, have shown encouraging results in clinical trials (5, 6). However, a third, complementary strategy has emerged, based on stimulating endogenous regenerative mechanisms. One approach for developing such regeneration strategies is to examine the cellular mechanisms of myocardial growth, since mechanisms of regeneration should be similar to the mechanisms of development.
Although stem and progenitor cells are important for morphogenesis of the myocardium, developmental growth in a number of nonhuman species is largely driven by cardiomyocyte proliferation (7–9). In biological models that, unlike adult humans, regenerate myocardium, cardiomyocyte proliferation is important for regeneration as well as postnatal heart growth (10, 11). For example, in mice, developmental cardiomyocyte proliferation continues for up to day 7 after birth, which coincides with the loss of regenerative capacity (11, 12). The close temporal relationship between cardiomyocyte proliferation and heart regeneration in animals raises the question of whether and to what age and extent cardiomyocyte proliferation plays a role in humans. The answer may help us understand the endogenous regenerative potential of the human heart and possibly indicate strategies for stimulating cardiomyocyte proliferation to regenerate myocardium.
Our current understanding of human myocardial growth is limited by an overall lack of reliable data about the underlying cellular mechanisms (reviewed in refs. 3, 4). Cardiomyocyte nuclei have been quantified in human fetuses using hematoxylin-eosin staining (13–15). Radiocarbon birth dating has shown that a small portion of cardiomyocytes is replaced in humans older than 20 y (16), but this technique is unreliable for the analysis of recent samples from individuals younger than 20 y of age (17). The detection of thymidine analogs, used to quantify cardiomyocyte turnover in adult cancer patients (18), is also not feasible in children. Thus, due to multiple limitations, little is known about the cellular growth mechanisms in the human heart in the most dynamic time window between birth and 20 y of age.
Technical limitations have hampered progress in addressing these questions. To overcome these limitations, we have developed and implemented image-based assays. In the present study, we examined a set of 21 healthy young (age range from 0–20 y) hearts and an additional set of 15 adult hearts (SI Appendix, Table S1). Our aim was to determine the extent and timing of cardiomyocyte cell cycling, proliferation, and hypertrophy and to relate the activity of these mechanisms to the growth of the human heart.
Results
Validation of Sampling and Methods.
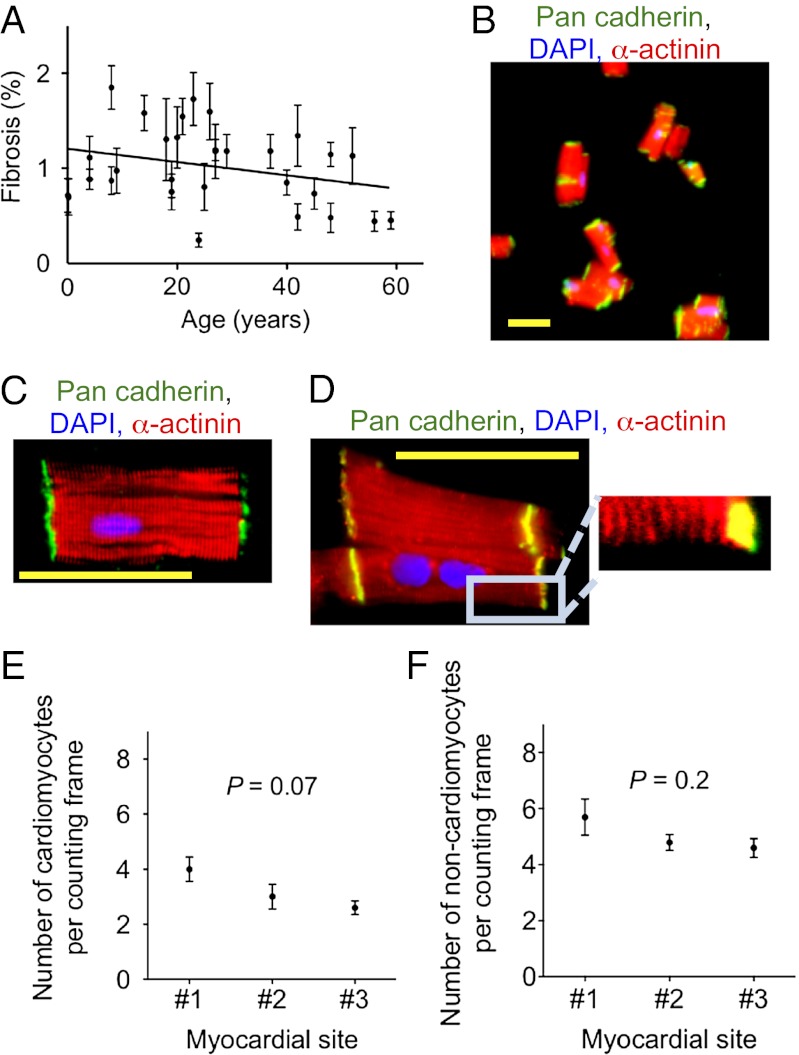
Using digital color thresholding on acid Fuchsin Orange G-stained myocardial samples, we quantified fibrosis, which was 1.02 ± 0.4%, (Fig. 1A). In addition, we performed histopathologic evaluation of these slides, which led to the elimination of four hearts. Thus, evaluation of the selected hearts showed that they are a representative sample (SI Appendix, Table S1 and Figs. S1 and S2).
Fig. 1.
Sampling and isolation methods yield representative probes of healthy human hearts. (A) Analysis of AFOG-stained tissue sections reveals no increased fibrosis; data fitted with linear regression, slope: –0.007 ± 0.002, P = 0.007. (B) Isolation from a 9-y-old donor heart using the fixation-digestion method yields intact cardiomyocytes. (C) Staining of desmosomes with an antibody against pan-cadherin shows intact cardiomyocytes. (D) α-actinin shows intact sarcomeres. (E) Optical dissector method quantifying cardiomyocyte and (F) noncardiomyocyte nuclei on three random myocardial sections from different sites of the same LV shows no significant differences in the nuclear density in different compartments of the same LV. Statistical significance was tested with ANOVA. The results are mean ± SD. Scale bar: 50 μm (B–D).
To be able to use flash-frozen human myocardial samples, we developed a unique isolation method, which involves fixing the myocardium before digestion with collagenase. The resulting yield of cardiomyocytes was 91.3 ± 3% [weight%, determined by (original weight – residual)/original weight]. The percentage of cardiomyocytes with desmosome-containing ends was 92.4 ± 0.4% (Fig. 1 B and C), and sarcomereres were present (Fig. 1D), indicating that most of the isolated heart muscle cells were intact.
To determine whether immunofluorescence microscopy could reliably differentiate between cardiomyocytes and noncardiomyocytes, we used two different structural markers: a sarcomeric marker (α-actinin and/or troponin I) as well as a membrane marker (caveolin-3). Confocal microscopy and visual quantification showed agreement between these markers in the identification of cardiomyocyte nuclei (SI Appendix, Fig. S3). We noted that H&E staining overestimated the number of cardiomyocyte nuclei compared with caveolin-3 and troponin I staining (SI Appendix, Fig. S3 G and H). The optical dissector method for quantifying cardiomyocyte (Fig. 1E) and noncardiomyocyte nuclei (Fig. 1F) on three random myocardial sections from different sites of the same left ventricle (LV) showed no significant differences in the nuclear density in different compartments of the same LV, indicating that our sampling method yielded a probe representative of the whole LV. We determined fixation-related tissue shrinkage to be 21 ± 5.8% and corrected the results accordingly (SI Appendix, Fig. S6). In summary, the applied sampling methods yield representative probes of human hearts.
Human Hearts Show Evidence of Cardiomyocyte Cell Cycle Activity.
Having established the validity of our methods, we visualized mitotic cardiomyocytes with antibodies against phosphorylated histone H3 (a marker for M-phase) and sarcomeric markers (α-actinin, Fig. 2 A and B). Using laser-scanning cytometry (LSC) as an automated and unbiased method for quantification on isolated cardiomyocytes, we determined that during the first year of life the percentage of cardiomyocytes in M-phase was 0.04 ± 0.01% (n = 6, Fig. 2C). Between 10 and 20 y, this decreased to 0.009 ± 0.006% (n = 5, P < 0.05, Fig. 2C) and remained detectable in hearts from subjects older than 40 y.
Fig. 2.
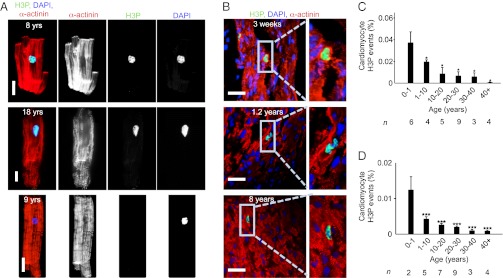
Human hearts show evidence for cardiomyocyte cycling after birth. (A) Isolated cardiomyocytes in M-phase were visualized by immunofluorescent microscopy using antibodies against phosphorylated histone H3 (H3P, green) and α-actinin (red). H3P-negative cardiomyocyte is shown as a control (Lower). (B) H3P-positive cardiomyocytes on tissue sections. (C and D) Quantification of isolated cardiomyocytes by LSC (C) and on tissue sections (D) shows that the percentage of cycling cardiomyocytes decreases with age. Columns represent mean values of the number of hearts investigated per age group (n) that are indicated under each column. One-way ANOVA revealed a significant difference between 0–1 y and the other age groups. *P < 0.05, ***P < 0.001. Scale bar: 25 µm.
To cross-check these results, we quantified cardiomyocytes in M-phase with a different approach, using confocal microscopy of tissue sections (Fig. 2B, SI Appendix, and Movies S1 and S2). The quantification showed that in the first year of life, the percentage of M-phase cardiomyocytes was 0.012 ± 0.003% (Fig. 2D). The percentage of M-phase cardiomyocytes decreased significantly over the first two decades of life but was still detectable above 40 y. The percentages of M-phase cardiomyocytes determined on stained sections correlated well with the results from the LSC analysis of isolated cardiomyocytes, indicating that cardiomyocyte cell cycle activity is higher in young than in adult humans.
Analysis of isolated cardiomyocytes showed that M-phase cardiomyocytes were predominantly mononucleated (P < 0.001), which is in agreement with previous reports of cycling mononucleated cardiomyocytes in growing cats (19), rats (20), and mice (21).
Young Humans Show Evidence of Cardiomyocyte Division.
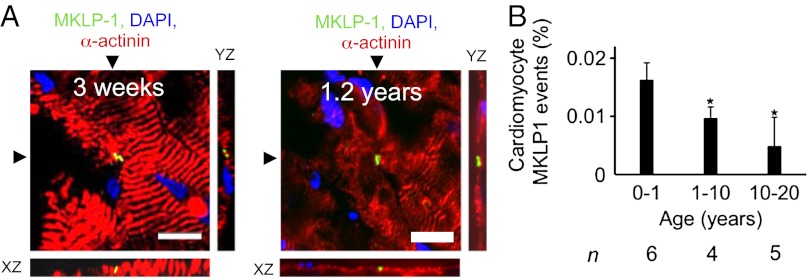
Detection of the contractile ring provides evidence of cardiomyocyte cytokinesis. The contractile ring consists of regulatory and motor proteins (22), including the mitotic kinesin-like protein (MKLP-1), a component of the centralspindlin complex, which is required for completion of cytokinesis (23). We developed a cardiomyocyte cytokinesis assay by staining thick (30 μm) myocardial sections with antibodies against MKLP-1 and α-actinin (Fig. 3A). Confocal stacks and 3D reconstructions showed contractile rings traversing cardiomyocytes (Movies S3, S4, S5, and S6). For quantification, we identified MKLP-1–positive cardiomyocytes on myocardial sections by confocal microscopy. In the age range of 0–1 y, 0.016 ± 0.003% of cardiomyocytes were in cytokinesis (n = 6). Between 2 and 10 y, it was 0.01 ± 0.002% (n = 4), and in the second decade of life, this value decreased to 0.005 ± 0.005% (n = 5, P < 0.05, Fig. 3B). We did not detect cardiomyocytes in cytokinesis in subjects older than 20 y (n = 9). In summary, human infants show evidence of cardiomyocyte cytokinesis, which decreases during childhood and adolescence to nondetectable levels in adults.
Fig. 3.
Humans show evidence for cardiomyocyte cytokinesis in the first two decades of life. (A) Cardiomyocyte cytokinesis was detected with an antibody against MKLP-1; XY and XZ reconstruction planes are indicated with arrowheads. (B) Frequency of MKLP-1–positive cardiomyocyte-specific events declines over first two decades of life. Scale bar: 25 µm. *P < 0.05 compared with 0–1 y (ANOVA); n, number of hearts analyzed. Cardiomyocyte-specific MKLP-1 activity was not found in samples >20 y of age (n = 9). For 3D reconstructions of the photomicrographs in A, see Movies S3, S4, S5, and S6.
Human Cardiomyocytes Show an Increase of Nuclear Ploidy with Age.
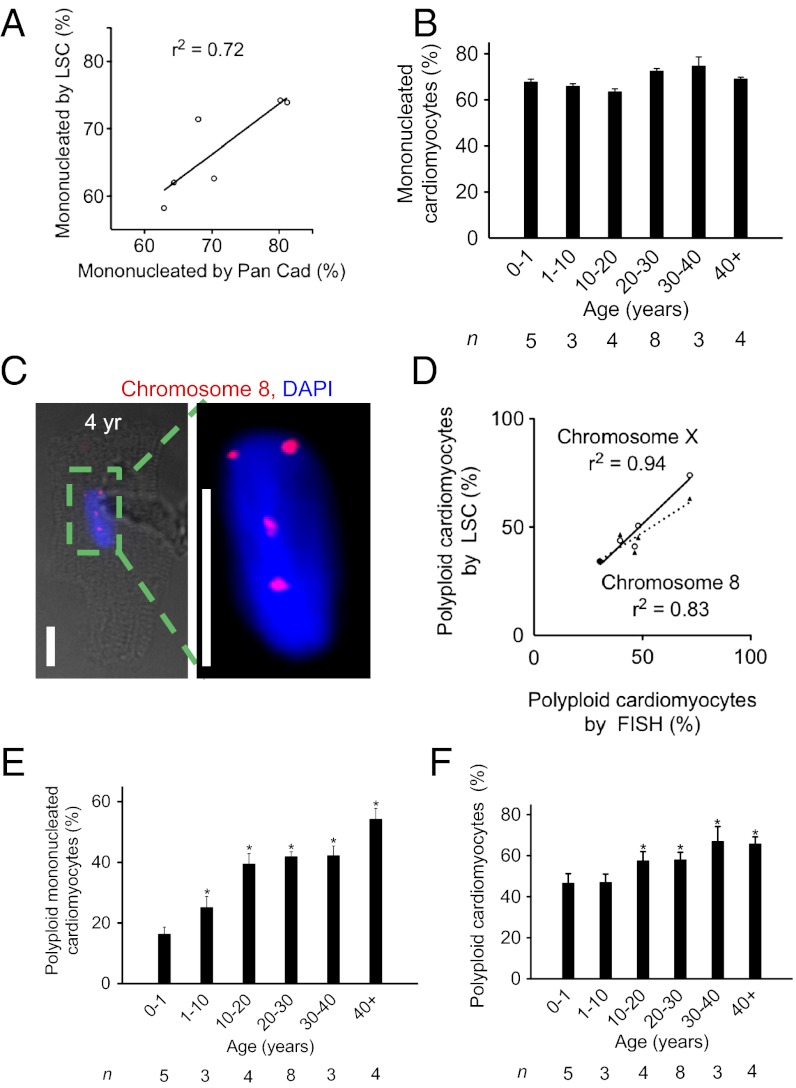
Cell cycle activity in cardiomyocytes may lead not only to division but also to formation of binucleated and polyploid daughter cells (3). Therefore, to determine the association between cardiomyocyte cell cycle activity and cellular proliferation, it is necessary to account for these nonproductive cell cycles. To determine the percentage of mononucleated cardiomyocytes in our samples, we quantified the number of nuclei in isolated human cardiomyocytes by visual count as well as automatically using LSC (SI Appendix, Fig. S4). The results obtained with both methods were highly correlated (Fig. 4A), thus validating the LSC method. Using LSC, we determined that the percentage of mononucleated cardiomyocytes did not change significantly between the first year (67.8 ± 3%) and 10 and 20 y of life (63.5 ± 2.2%, P > 0.05, Fig. 4B) and remained unchanged throughout life. The percentage of mononucleated cardiomyocytes in children has not been published previously, but the percentage in adults is in agreement with a prior study (24). In conclusion, humans do not display the transition to predominantly binucleated cardiomyocytes, which is known to happen in murine models in the first week after birth (12, 25).
Fig. 4.
Human cardiomyocytes show increased formation of polyploid nuclei between birth and 20 y. (A) Validation of LSC method for quantification of mononucleated cardiomyocytes. (B) In humans, the percentage of mononucleated cardiomyocytes remains unchanged after birth. (C) Tetraploid cardiomyocyte nucleus (DAPI, blue) with FISH probe against chromosome 8. Scale bar: 25 μm. (D) Comparison of quantification of nuclear DNA content using FISH and LSC shows significant correlation. (E) The percentage of polyploid mononucleated cardiomyocytes increases with age. (F) Percentage of cardiomyocytes with DNA content >2N determined by LSC increases with age. Statistical difference between 0–1 y and other age groups was tested with one-way ANOVA: *P < 0.05. n, number of hearts analyzed.
We examined ploidy changes with LSC and validated them by fluorescence in situ hybridization (FISH) using probes against chromosomes 8 and X (Fig. 4 C and D and SI Appendix, Fig. S4). In the first year of life, 16.3 ± 5.2% of mononucleated cardiomyocytes were hyperdiploid (>2N), which increased to 39.5 ± 6.9% between 10 and 20 y (P < 0.05), but then remained unchanged (Fig. 4E). Mononucleated polyploid cardiomyocytes increased to 54.2 ± 5.8% above 40 y, which was not significantly different compared with 30–40 y. Taking mono- and binucleated cardiomyocytes together, the percentage of those with a total DNA content >2N increased significantly to 57.5 ± 4.5% by 10–20 y, which remained unchanged in later life (P < 0.05, Fig. 4F). In summary, these results indicate that human cardiomyocytes undergo significant changes in their nuclear ploidy after birth. Together with the result that cycling cardiomyocytes are predominantly mononucleated, this suggests that they are the direct or indirect precursors of cardiomyocytes with a total DNA content of >2N.
Human Cardiomyocytes Proliferate and Enlarge After Birth.
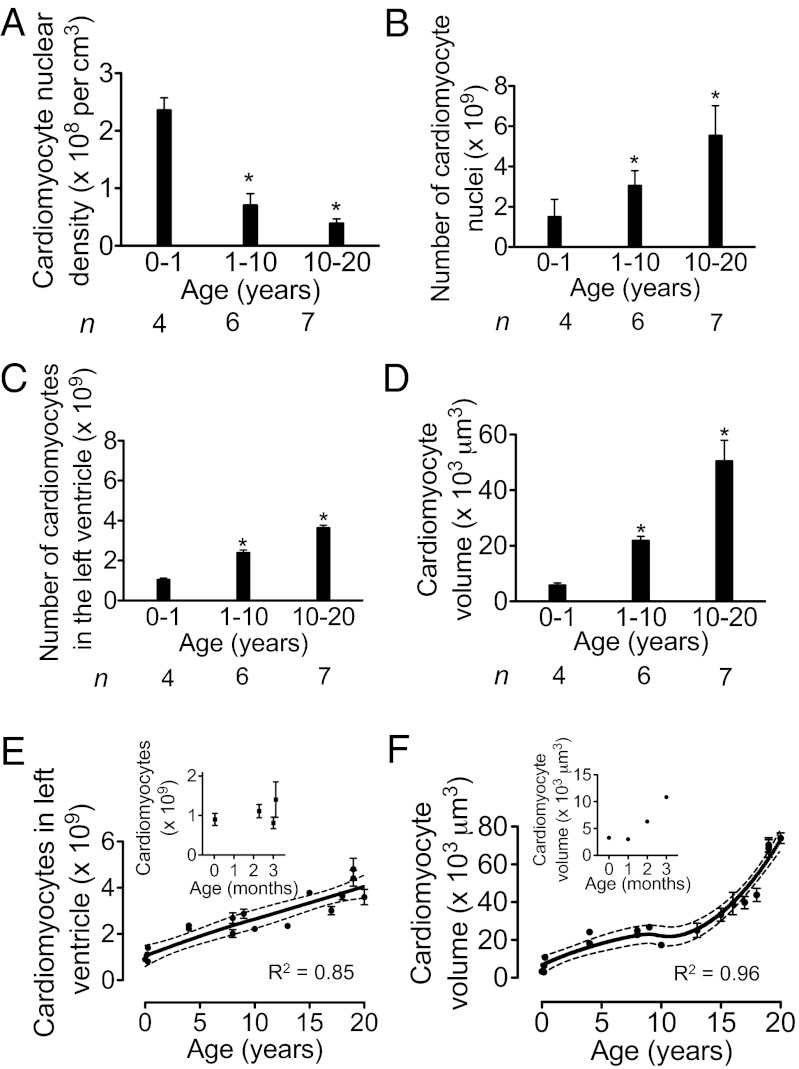
We used stereology techniques to quantify the number of cardiomyocytes per LV (SI Appendix). Using the optical dissector principle, we determined that in the first year of life, the volume density of cardiomyocyte nuclei was 2.4 ± 0.2 × 108 / cm3 (Fig. 5A). Between 10 and 20 y, the volume density decreased to 3.6 ± 0.7 × 107 / cm3 (P < 0.05), in agreement with a previous report from adult hearts (26). Using the specific reference volume for each LV (27), we calculated from the volume density of cardiomyocyte nuclei (Fig. 5A) that newborns had 1.5 ± 0.9 × 109 and young adults had 5.6 ± 1.5 × 109 cardiomyocyte nuclei in the LV (P < 0.05, Fig. 5B). By correcting the number of cardiomyocyte nuclei by accounting for the corresponding percentages of mono- and binucleated cardiomyocytes (from Fig. 4B), we calculated that in the age range of 0–1 y, 1.1 ± 0.1 × 109 cardiomyocytes were present in the LV (n = 4), compared with 3.7 ± 0.3 × 109 at the age of 20 y (n = 7, Fig. 5C). This represents a 3.4-fold increase (P < 0.05, Fig. 5C). The mean cellular volume of cardiomyocytes in the age range of 0–1 y was 5,854 ± 818 µm3 (n = 4), and between 10–20 y, it reached 50,564 ± 7,398 µm3 (n = 7, Fig. 5D), representing an 8.6-fold increase (P < 0.05). Taken together, these results indicate that both cardiomyocyte proliferation and enlargement contribute to postnatal heart growth in humans.
Fig. 5.
Human cardiomyocytes proliferate and enlarge after birth. (A) Cardiomyocyte nuclear density, determined by the optical dissector method, decreases with age. (B) Number of cardiomyocyte nuclei per LV increases with age. (C) Number of cardiomyocytes per LV, calculated from number of cardiomyocyte nuclei (B) and percentages of mono-, bi-, and multinucleated cardiomyocytes. (D) Mean volume of cardiomyocytes increases with age. (E and F) The number (E) and mean volume (F) of cardiomyocytes from individual LVs were graphed over age and modeled using locally weighted scatter plot smoothing (LOWESS). Blow-up graphs of results from the first 3.5 mo of life are shown. Dotted lines indicate 95% confidence intervals. R2 values are indicated. *P < 0.05. n, number of hearts analyzed.
We graphed the number of cardiomyocytes determined in individual LVs (Fig. 5E) and the mean cardiomyocyte volume (Fig. 5F) and fitted the data with LOWESS, validating the fits using generalized cross-validation. The patterns of cardiomyocyte enlargement and proliferation indicated that both contribute simultaneously to myocardial growth between birth and 20 y of age.
Discussion
It has long been thought that the number of cardiomyocytes does not change significantly during physiologic postnatal growth in humans (reviewed in ref 3). This notion arose after a 1950 study of human hearts, some of which were diseased. Cardiomyocyte silhouettes were measured on perpendicular sections of the papillary muscles (28), and the results led researchers to conclude that cardiomyocyte enlargement is the sole mechanism for postnatal growth. By today’s standards, this study was limited by selection, anisotropy, and counting biases (28, 29). Although other reports suggested that cardiomyocytes in human infants show mitotic activity (30–32) and that the number of cardiomyocytes may double in the first year of life (33), the cell-static view has dominated most of our thinking about physiologic heart growth in humans.
Our study demonstrates that myocardial growth in humans is based on two cellular mechanisms: cardiomyocyte enlargement and proliferation. Two lines of evidence support the conclusion that cardiomyocyte proliferation contributes to postnatal myocardial growth. First, we show evidence for cardiomyocyte cytokinesis. Second, we demonstrate that the number of cardiomyocytes increases between birth and 20 y of age. We formally tested the hypothesis that cardiomyocyte cell cycle activity contributes to postnatal cardiomyocyte proliferation by comparing the number of cardiomyocytes predicted to be generated by cell cycle events with the number present in the LV. Calculating the number of cardiomyocytes generated based on cell cycle activity requires input of the incidence of a cell cycle marker (1), the duration of its presence (2), and the probability that the marker indicates division (3). We used the percentage of cardiomyocytes in M-phase, which shows conservation of regulatory mechanisms and duration (∼1.5 h) between species, tissues, and ages (34–36). To calculate the number of cardiomyocytes generated from M-phase activity, determined by H3P analysis, we used the previously determined duration of 1.8 ± 0.3 h in adult ventricular cardiomyocytes (21, 37). To correct our results for nonproductive mitotic events (binucleation, polyploidization), we multiplied by the age-specific percentage of mononucleated and diploid cardiomyocytes (from Fig. 4F), which results in the number of actually generated cardiomyocytes. Thus, for the first year of life, the calculation 0.04% H3P-positive of 0.9 × 109 cardiomyocytes (present at birth) × 24 h × 365 d / 1.8 h per mitosis × 56% productive mitoses would yield 0.9 × 109 de novo cardiomyocytes in the LV. Using the range of duration of mitosis (1.5–2.1 h) (34) rather than the mean, we calculated a range of 0.8–1.1 × 109 de novo generated cardiomyocyte during the first year of life. In the first decade of life, the same calculation would yield ∼0.9 × 109 new cardiomyocytes and in the second decade 0.6 × 109. The stereological quantification showed that in the age range of 0–1 y, 1.1 ± 0.1 × 109 (n = 4) cardiomyocytes were present and, at 20 y, 3.7 ± 0.3 × 109 (n = 7), an increase by 2.6 × 109 (P < 0.05). Therefore, this cross-check demonstrates that the measured M-phase activity can account for the increase of the number of cardiomyocytes. Considering that a large myocardial infarction can wipe out ∼25% of the adult heart (3), which is equivalent to 1 billion cardiomyocytes, the generation of 2.6 billion new cardiomyocytes during heart growth is significant.
We detected cardiomyocyte proliferation in normal human hearts up to 20 y of age, which is longer than would be anticipated by extrapolating the results from mice (12) and rats (25). In addition, in humans, cardiomyocyte proliferation and enlargement contribute simultaneously to myocardial growth, which contrasts with the rapid switch from proliferation to hypertrophy in mice and rats in the first week of life (12, 25).
Recent studies by Bergmann et al. (16) and Kajstura et al. (38) on generation of cardiomyocytes in human hearts offer an opportunity for comparison with our findings (SI Appendix, Table S5). Using the 14C-based birth-dating technique, Bergmann et al. (16) reported a turnover rate of 1.9% in one 19 y-old heart and 1% in five hearts between 21–40 y. We calculated that at 20 y 6.9 × 107 new cardiomyocytes are generated per year—the equivalent to 1.9% of cardiomyocytes present at this age—based on quantifying cardiomyocyte mitotic activity at 20 y (0.00092% H3P, n = 4, age range 19–23 y) and adjusting for nonproductive cell cycles. Thus, our results in young adults are within the same range as the corresponding results by Bergmann et al. (16).
Kajstura et al. (38) reported 0.01% H3P-positive cardiomyocytes between 1–10 y, 0.004% between 10–20 y, 0.003% between 21–40 y, and 0.006% above 40 y. Our corresponding results are 2–3.5-fold higher up to 40 y of age and sixfold lower above 40 y (SI Appendix, Table S5). However, Kajstura et al. (38) used their birth-dating results to arrive at cardiomyocyte generation rates that are 5- to 10-fold higher up to 40 y of age and 80-fold higher above 40 y. In contrast, we did not detect cardiomyocyte cytokinesis in hearts from subjects older than 20 y. This may be because, unlike using the mean birthdate of cardiomyocytes, as done by Bergmann et al. (16) and Kajstura et al. (38), our cytokinesis assay is based upon direct visualization and quantification of individual dividing cardiomyocytes. An alternative interpretation of the lack of detection of cardiomyocyte cytokinesis above 20 y is that the corresponding cell cycle activity that we detected using the H3P assay is exclusively associated with nonproductive cell cycle events. In summary, the pattern of cardiomyocyte cell cycle activity in our study, which included more and younger hearts than prior studies, confirms the suggestion that young humans show more cardiomyocyte cycling and division than adults.
The number of generated cardiomyocytes in our stereologic estimates (Fig. 5 C and E) may include those that originate from cardiogenic stem and progenitor cells, as well as those generated by division of preexisting, differentiated cardiomyocytes. Both mechanisms are not mutually exclusive and may occur simultaneously. Current knowledge of cardiac development would place cardiogenic stem and progenitor cells upstream of dividing cardiomyocytes, but available methods in humans cannot differentiate these two processes (3, 18, 39, 40).
Regardless of the origin of the cycling cardiomyocytes, the findings from our study and others (SI Appendix, Table S5) establish the importance of cardiomyocyte proliferation in the growth and development of postnatal human hearts. In addition to providing a new cell-based growth model for the human heart, our findings point to a potential opportunity for stimulating myocardial growth and regeneration in humans—through the manipulation of endogenous mechanisms of cardiomyocyte proliferation.
Materials and Methods
Study Population and Tissue Sampling.
The Muscle Research Unit at the University of Sydney (Australia) provided LV myocardial samples from 28 unused donor hearts that were procured for transplantation. The National Institute of Child Health and Human Development Brain and Tissue Bank at the University of Maryland provided eight samples from cadaveric hearts with short postmortem intervals (SI Appendix, Table S1 and Figs. S1 and S2). Sample ascertainment was approved by the Institutional Review Boards of St. Vincent's Hospital (H03/118) and the University of Sydney (09-2009-12146).
Isolation of Cardiomyocytes from Flash-Frozen Tissue.
Myocardial samples were flash frozen and stored in liquid nitrogen. For isolation, 1 mm3 blocks of tissue were fixed in 3.7% (vol/vol) normal buffered formaldehyde (Sigma) and incubated on a slow bench top rocker at room temperature for 2 h. The tissue was then washed in PBS for 5 min and digested with collagenase B (1.8 mg/mL, Roche) and collagenase D (2.4 mg/mL, Roche) on a rotator at 37 °C overnight.
Quantification of Fibrosis.
Myocardial tissue samples were stained with AFOG to visualize cardiomyocytes (red) and collagen (blue), and 15 random images per slide were assessed with a Zeiss Axioplan2 microscope and 20× lens. We quantified the proportion of fibrosis by digital color thresholding (Metamorph). The same slides were used for histopathologic evaluation.
Immunofluorescence and Optical Dissector Method.
For each heart, 210 consecutive cryosections were prepared on 70 slides. We selected the initial slide with a random number generator and picked every fifteenth slide after that for staining and microscopy in a random-systematic fashion. Six researchers, blinded with respect to the samples’ corresponding ages and identities, quantified cellular events, either by manual count or by digital thresholding (image segmentation and creation of a binary image from a gray scale). Software analysis of the converted binary images was performed with Image Processing and Analysis in Java (Image J). To quantify the number of cardiomyocyte nuclei per cubed centimeter, we used the optical dissector method (29) and validated it with two cardiomyocyte-specific structural markers (SI Appendix, Fig. S3). To calculate the number of cardiomyocyte nuclei per LV, we multiplied the number of cardiomyocyte nuclei per cubed centimeter with the LV volume (see below and ref. 27). We quantified tissue shrinkage and corrected all results accordingly (SI Appendix, Fig. S6).
Determination of LV Myocardial Mass.
The LV reference mass of each heart for which heart weight was not available as the mean normal value for body surface area was calculated according to normal z-score values described in ref. 41. Our calculations were based on data from 576 healthy humans obtained at Boston Children’s Hospital within an institutional review board–approved study. Quantification of the number of cardiomyocytes per LV using the predicted heart weights matched very closely with estimations based on actual heart weights for those seven hearts where this information was available (SI Appendix, Fig. S8).
Cardiomyocyte Volume Determination.
To determine the cellular volumes of isolated cardiomyocytes, we visualized the cytoplasm with CellMask (Invitrogen, 5 μg/mL, 5 min at room temperature). To select cardiomyocytes for volume analysis, we scanned the stained slide with a ×60 water objective and selected one random cardiomyocyte from every fourth field of view. We acquired confocal stacks with a step size of 1.2 µm (Olympus FV 1000; SI Appendix, Fig. S5A). We used digital thresholding to determine the area of each optical section (SI Appendix, Fig. S5B), and by knowing the distance of 1.2 µm between them, we calculated the cellular volume using Image J. The mean of 60–100 isolated cardiomyocytes from each heart was calculated. We validated this design-based method using a model-based method that assumed a cylindrical shape of the cardiomyocyte (24, 25). Bland–Altman analysis (SI Appendix, Fig. S5 C and D) revealed a significant agreement between the model-based (24, 25) and our design-based method for volume determination.
Cell Cycle Activity.
Cardiomyocyte karyokinesis was visualized with an antibody against phosphorylated histone H3 (Ser10, Upstate, 1:500), and cytokinesis was visualized with an antibody against the centralspindlin component MKLP-1 (Abcam, 1:100). Primary antibodies were detected with AlexaFluor conjugated secondary antibodies (Invitrogen; SI Appendix, Table S4). Nuclei were visualized with 4',6-diamidino-2-phenylindole (DAPI, Invitrogen, 50 nM).
Ploidy Determination.
Isolated cardiomyocytes were spread on slides and examined by LSC for observer-independent quantitative analyses of nuclear DNA content (SI Appendix, Fig. S4). We validated the LSC-based method of quantifying the amount of DNA in cardiomyocyte nuclei using synchronized human umbilical vein endothelial cells. At least 15,000 cardiomyocytes per sample were analyzed. Ploidy data were further confirmed with FISH probes against chromosomes 8 and X (Vysis), which were visually analyzed.
Statistical Analysis.
The numbers of analyzed samples and cells are listed in SI Appendix, Table S3. Numerical results are represented as means ± SEM, unless otherwise indicated. The age bins were 0–1 y (0–365 d), 1–10 y (366 d–11th birthday), and 10–20 y (after 11th to < 21st birthday) and so forth. Linear regression was used to model the relationship between the proportion of fibrosis vs. age and noncardiomyocyte space vs. age. Continuous outcomes by age group were compared with analysis of variance (ANOVA) followed by Bartlett’s post hoc testing. The relationships (mean and 95% confidence interval for the mean) between cardiomyocyte volume and the number of cardiomyocytes per LV vs. age were modeled using locally weighted scatterplot smoothing (LOWESS) (42) to account for nonlinearities in the associations. Generalized cross-validation was used to determine the value of the smoothing parameter that minimized mean squared error. Statistical significance was achieved with a two-sided P value < 0.05. Statistical analyses were performed with GraphPad Prism, version 5 for Windows (GraphPad Software) and S-Plus, Version 8.0 (TIBCO Software).
Supplementary Material
Acknowledgments
We thank R. Mitchell (Brigham and Womens Hospital) for histopathologic evaluation; S. Lal, S. Joseph, and R. Faisal Rasouli (University of Sydney); E. Long, S. Arab, and J. Rosen (Children’s Hospital Boston) for technical contributions; and B. Polizzotti, S. Suresh, D. Zebrowski, W. Pu, C. Edwards, and L. Jackson-Grusby for critical suggestions. We acknowledge support from the Department of Cardiology and the Translational Investigator Program (Boston Children’s Hospital), the Children’s Cardiomyopathy and Geneen Foundations (B.K.), the Biomedical Research Exchange Program and the Helmut-Drexler Foundation (M.M.), and the American Heart Association (S.W.). Human heart tissue was obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214608110/-/DCSupplemental.
References
- 1.Roger VL, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics—2012 update: A report from the American Heart Association. Circulation. 2012;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linzbach AJ. Heart failure from the point of view of quantitative anatomy. Am J Cardiol. 1960;5:370–382. doi: 10.1016/0002-9149(60)90084-9. [DOI] [PubMed] [Google Scholar]
- 3.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vela D, Buja LM. Quest for the cardiovascular holy grail: Mammalian myocardial regeneration. Cardiovasc Pathol. 2008;17(1):1–5. doi: 10.1016/j.carpath.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Bolli R, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): Initial results of a randomised phase 1 trial. Lancet. 2011;378(9806):1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Siu CW, Tse HF. Cardiac regeneration: Messages from CADUCEUS. Lancet. 2012;379(9819):870–871. doi: 10.1016/S0140-6736(12)60236-0. [DOI] [PubMed] [Google Scholar]
- 7.Sedmera D, Thompson RP. Myocyte proliferation in the developing heart. Dev Dyn. 2011;240(6):1322–1334. doi: 10.1002/dvdy.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: Lessons from development. Genes Dev. 2011;25(4):299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi BA, Wernet O, Chien KR. Pregenerative medicine: Developmental paradigms in the biology of cardiovascular regeneration. J Clin Invest. 2010;120(1):20–28. doi: 10.1172/JCI40820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 11.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271(5 Pt 2):H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 13.Austin A, Fagan DG, Mayhew TM. A stereological method for estimating the total number of ventricular myocyte nuclei in fetal and postnatal hearts. J Anat. 1995;187(Pt 3):641–647. [PMC free article] [PubMed] [Google Scholar]
- 14.Mayhew TM, Pharaoh A, Austin A, Fagan DG. Stereological estimates of nuclear number in human ventricular cardiomyocytes before and after birth obtained using physical disectors. J Anat. 1997;191(Pt 1):107–115. doi: 10.1046/j.1469-7580.1997.19110107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayhew TM, Gregson C, Pharaoh A, Fagan DG. Numbers of nuclei in different tissue compartments of fetal ventricular myocardium from 16 to 35 weeks of gestation. Virchows Arch. 1998;433(2):167–172. doi: 10.1007/s004280050232. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spalding KL, Buchholz BA, Bergman LE, Druid H, Frisén J. Forensics: Age written in teeth by nuclear tests. Nature. 2005;437(7057):333–334. doi: 10.1038/437333a. [DOI] [PubMed] [Google Scholar]
- 18.Kajstura J, et al. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107(2):305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Chen X, et al. Adolescent feline heart contains a population of small, proliferative ventricular myocytes with immature physiological properties. Circ Res. 2007;100(4):536–544. doi: 10.1161/01.RES.0000259560.39234.99. [DOI] [PubMed] [Google Scholar]
- 20.Kühn B, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13(8):962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 21.Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 22.Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14(5):440–447. doi: 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- 23.Lewellyn L, Carvalho A, Desai A, Maddox AS, Oegema K. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J Cell Biol. 2011;193(1):155–169. doi: 10.1083/jcb.201008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivetti G, et al. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol. 1996;28(7):1463–1477. doi: 10.1006/jmcc.1996.0137. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28(8):1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y, Nyengaard JR, Andersen JB, Baandrup U, Gundersen HJ. The application of stereological methods for estimating structural parameters in the human heart. Anat Rec (Hoboken) 2009;292(10):1630–1647. doi: 10.1002/ar.20952. [DOI] [PubMed] [Google Scholar]
- 27.Sluysmans T, Colan SD. Theoretical and empirical derivation of cardiovascular allometric relationships in children. J Appl Physiol. 2005;99(2):445–457. doi: 10.1152/japplphysiol.01144.2004. [DOI] [PubMed] [Google Scholar]
- 28.Linzbach AJ. [The muscle fiber constant and the law of growth of the human ventricles] Virchows Arch. 1950;318(5):575–618. doi: 10.1007/BF00947940. [DOI] [PubMed] [Google Scholar]
- 29.Mühlfeld C, Nyengaard JR, Mayhew TM. A review of state-of-the-art stereology for better quantitative 3D morphology in cardiac research. Cardiovasc Pathol. 2010;19(2):65–82. doi: 10.1016/j.carpath.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Macmahon HE. Hyperplasia and regeneration of the myocardium in infants and in children. Am J Pathol. 1937;13(5):845–854. [PMC free article] [PubMed] [Google Scholar]
- 31.Chang KT, Taylor GP, Meschino WS, Kantor PF, Cutz E. Mitogenic cardiomyopathy: A lethal neonatal familial dilated cardiomyopathy characterized by myocyte hyperplasia and proliferation. Hum Pathol. 2010;41(7):1002–1008. doi: 10.1016/j.humpath.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Zerbini C, Weinberg DS, Perez-Atayde AR. DNA ploidy analysis of myocardial hyperplasia. Hum Pathol. 1992;23(12):1427–1430. doi: 10.1016/0046-8177(92)90064-a. [DOI] [PubMed] [Google Scholar]
- 33.Adler CP, Costabel U. Cell number in human heart in atrophy, hypertrophy, and under the influence of cytostatics. Recent Adv Stud Cardiac Struct Metab. 1975;6:343–355. [PubMed] [Google Scholar]
- 34.Hahn AT, Jones JT, Meyer T. Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle. 2009;8(7):1044–1052. doi: 10.4161/cc.8.7.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437(7061):1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 36.Carney BK, Caruso Silva V, Cassimeris L. The microtubule cytoskeleton is required for a G2 cell cycle delay in cancer cells lacking stathmin and p53. Cytoskeleton (Hoboken) 2012;69(5):278–289. doi: 10.1002/cm.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettencourt-Dias M, Mittnacht S, Brockes JP. Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. J Cell Sci. 2003;116(Pt 19):4001–4009. doi: 10.1242/jcs.00698. [DOI] [PubMed] [Google Scholar]
- 38.Kajstura J, et al. Cardiomyogenesis in the aging and failing human heart. Circulation. 2012;126(15):1869–1881. doi: 10.1161/CIRCULATIONAHA.112.118380. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Steinhauser ML, Lee RT. Regeneration of the heart. EMBO Mol Med. 2011;3(12):701–712. doi: 10.1002/emmm.201100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira-Martins J, et al. Cardiomyogenesis in the developing heart is regulated by c-kit-positive cardiac stem cells. Circ Res. 2012;110(5):701–715. doi: 10.1161/CIRCRESAHA.111.259507. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Foster BJ, et al. A novel method of expressing left ventricular mass relative to body size in children. Circulation. 2008;117(21):2769–2775. doi: 10.1161/CIRCULATIONAHA.107.741157. [DOI] [PubMed] [Google Scholar]
- 42.Cleveland WS, Dvelin SJ. Locally-weighted regression: An approach to regression analysis by local fitting. J Am Stat Assoc. 1988;83(403):596–610. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.