Abstract
Despite its highly inflammatory nature, LPS is a molecule with remarkable therapeutic potential. Lipid A is a glycolipid that serves as the hydrophobic anchor of LPS and constitutes a potent ligand of the Toll-like receptor (TLR)4/myeloid differentiation factor 2 receptor of the innate immune system. A less toxic mixture of monophosphorylated lipid A species (MPL) recently became the first new Food and Drug Administration-approved adjuvant in over 70 y. Whereas wild-type Escherichia coli LPS provokes strong inflammatory MyD88 (myeloid differentiation primary response gene 88)-mediated TLR4 signaling, MPL preferentially induces less inflammatory TRIF (TIR-domain–containing adaptor-inducing IFN-β)-mediated responses. Here, we developed a system for combinatorial structural diversification of E. coli lipid A, yielding a spectrum of bioactive variants that display distinct TLR4 agonist activities and cytokine induction. Mice immunized with engineered lipid A/antigen emulsions exhibited robust IgG titers, indicating the efficacy of these molecules as adjuvants. This approach demonstrates how combinatorial engineering of lipid A can be exploited to generate a spectrum of immunostimulatory molecules for vaccine and therapeutics development.
Keywords: outer membrane, cell envelope, gram negative
In 1892, Richard Pfeiffer introduced the revolutionary concept of bacterial endotoxin in his description of a nonproteinaceous, nonsecreted toxin bound to the surface of Vibrio cholerae (1). This toxin, lipopolysaccharide (LPS), is the major surface molecule of Gram-negative bacteria that triggers the host immune response during infection through recognition of the bioactive lipid A domain (2). Lipid A, or endotoxin, is recognized by the innate immune system through the conserved pattern recognition receptor, Toll-like receptor 4/myeloid differentiation factor 2 (TLR4/MD2) complex, which initiates a robust signal cascade leading to cytokine production that is crucial for clearance of infection but can be potent enough to result in lethal endotoxic shock (2).
The structural nature of Escherichia coli lipid A, with six acyl chains and two phosphate groups, is critical for complete activation of human TLR4/MD2 (3). Many bacteria have evolved enzymes that alter their lipid A structure by modifying its number of acyl chains, phosphate groups, or polar functional groups (4). Such modifications can alter the strength of the TLR4 response. For instance, hexaacylated lipid A is maximally stimulatory, whereas tetraacylated lipid A is antagonistic (3). Lipid A modifications can also lead to stimulation of select TLR4 signaling pathways by mediating the recruitment of distinct sets of adaptor proteins: MyD88 (myeloid differentiation primary response gene 88), TRIF (TIR-domain-containing adaptor-inducing IFN-β), or both (5). Whereas strong induction of the MyD88 pathway is harmful because of high production of proinflammatory cytokines, including TNF-α and IL-1β, a low level of induction is beneficial for long-lasting immunity in vaccines (5). TRIF-mediated responses produce lower quantities of inflammatory cytokines and yet are still effective in triggering production of cytokines important for vaccine adjuvants such as interferon regulatory factor-3 inducible genes (6).
Efforts to understand the lipid A–TLR4 interaction have yielded improved vaccines (7). Notably, one LPS derivative with reduced toxicity, termed monophosphorylated lipid A (MPL), has been approved to supplement an adjuvant system in vaccines worldwide (5). MPL is a heterogeneous mixture of lipid A species from Salmonella minnesota R595 that has been chemically detoxified by successive acid and base hydrolysis (8). The primary lipid A species of the MPL mixture is 3-O-deacyl-4′-monophosphoryl lipid A (SI Appendix, Fig. S1), which induces a stronger T-helper cell type 1 response than classical adjuvants such as aluminum salt precipitates (alum) (5) but, unlike LPS, induces a signaling bias toward the TRIF pathway, resulting in the safe stimulation of adaptive immune responses without excessive production of inflammatory cytokines (9). Efforts to engineer E. coli strains for simplified, biological production of MPL have not been successful thus far. E. coli strains that are dephosphorylated at the 1 position have been developed (10); however, the acyl chain arrangement in lipid A from these strains varies structurally from the predominant Salmonella 3-O-deacyl-4′-monophosphoryl lipid A in MPL (8).
Immunomodulation by LPS variants has been recognized as a potentially advantageous therapeutic strategy for many circumstances (7, 11, 12). Additionally, the lipid A-modifying enzymes used in the present study have been individually expressed in wild-type E. coli and/or Salmonella for proof of function (13–18), and two strains have been generated in which two coexpressed enzymes (LpxE/LpxF and PagP/PagL, respectively) resulted in lowered antagonistic properties (19, 20). However, lipid A heterogeneity, its essentiality to bacterial growth, the complexity of lipid A biosynthesis, and the analytical challenges posed by lipidomics have previously impeded the engineering of a single species of bacteria capable of producing nearly any lipid A species. Here, we report a system for the combinatorial engineering of E. coli strains that produce lipid A variants with distinct TLR4 agonist functions and a broad range of effect on the mammalian immune response (Fig. 1). Our method shows that coexpression of individual enzymes is not effective compared with combinatorial expression. Additionally, in many instances, the engineered strains produce highly homogeneous lipid A, a feature that is particularly attractive for therapeutics development. These strains have provided a means to directly compare whole cells, intact LPS, and lipid A in the same background, eliminating confounding factors present when using different bacteria to compare lipid A structures (some not naturally found in nature). Chemical synthesis of many of these lipid A structures is complicated because of varied length of primary acyl chains and asymmetric acyl chain arrangement (21). Also, chemical synthesis cannot be used in settings where whole bacterial cells or purified, intact LPS is required. We report on the construction and characterization of a library of 61 distinctive, engineered strains that mediate graded TLR4-dependent cytokine responses in vitro and result in robust IgG titers following immunization of mice with antigen/lipid A-variant emulsions.
Fig. 1.
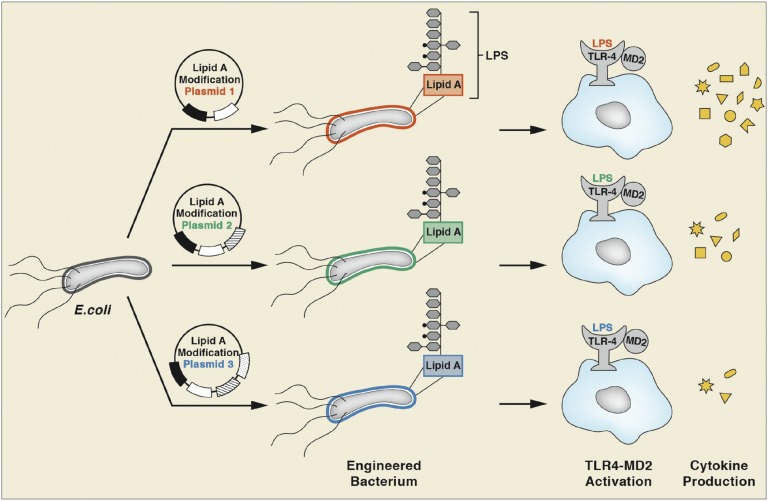
Combinatorial engineering of lipid A anchors to generate diverse immune responses. A schematic is shown illustrating how the outer surface of E. coli strains varies in LPS structure (indicated by different colors) when plasmids are expressed that contain combinations of up to five lipid A-modifying enzymes. The altered LPS molecules bind and activate the TLR4/MD2 complex differentially, altering the nature of downstream cytokine production, represented by shapes that indicate different types and quantity of cytokines released.
Results
Combinatorial Engineering of Lipid A.
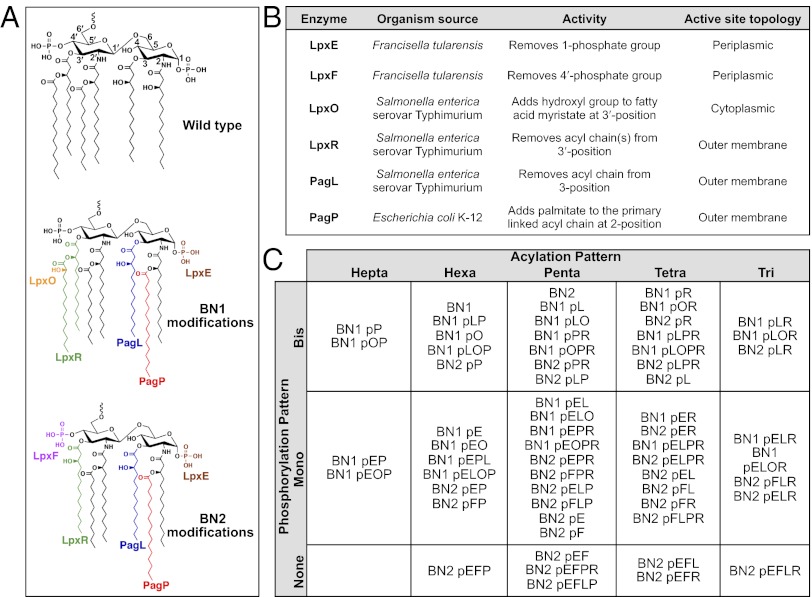
First, to generate an E. coli library with defined lipid A domains of LPS, we generated two background strains that synthesize homogeneous lipid A by deletion of the genes that modify E. coli lipid A under normal growth conditions (Materials and Methods and SI Appendix). Specifically, strain BN1 was generated from strain BN0, an lpxT and eptA double mutant. The LpxT enzyme functions to add a third phosphate group to lipid A, and when mutated, the resulting lipid A is strictly bis-phosphorylated (22). However, LpxT inhibition activates EptA, which adds a phosphoethanolamine to the 1-position of lipid A (23). Mutation of lpxT and eptA increases the activity of PagP to palmitoylate the 2-acyl chain of lipid A (23), so the pagP gene was also deleted. As a result, strain BN1 produces hexaacylated, bis-phosphorylated lipid A, the highly endotoxic, major species synthesized by E. coli (4). Deletion of lpxM in BN1 generated strain BN2, which synthesizes only pentaacylated lipid A. TLC and MALDI-TOF mass spectrometry (MS) confirmed the lipid A profiles (SI Appendix, Fig. S2). BN1 and BN2 provide two distinct templates suitable for alteration by endotoxin-modifying enzymes, and expression of any individual or combination of lipid A modification enzymes in these backgrounds generates previously unreported strains, distinctive in their homogeneous lipid A template. Additionally, 36 of these include combinations of enzymes that have not been coexpressed previously in any strain, which provides 61 strains when introduced into the two backgrounds.
BN1 and BN2 were transformed with a single vector, which allows the expression of an unrestricted number of proteins (24), harboring combinations of genes encoding lipid A-modification enzymes from different bacterial species, specifically PagP from E. coli; PagL, LpxR, and LpxO from Salmonella enterica serovar Typhimurium; and LpxE and LpxF from Francisella tularensis (Fig. 2B and SI Appendix, Tables S1 and S2) (4). Because LpxF from F. tularensis is known to not function on hexaacylated lipid A substrate (4), LpxF was not introduced into BN1. Additionally, LpxO from S. enterica hydroxylates the 3′-acyloxyacyl chain (4) that is absent in BN2, precluding its use in this strain background.
Fig. 2.
Modification machinery used for generation of diversified lipid A molecules in whole bacteria. Lipid A structures of wild-type E. coli K12, BN1, and BN2 are shown with the names of the six lipid A-modifying enzymes represented in color next to the group that each enzyme modifies: LpxR (green), PagL (blue), LpxE (brown), and LpxF (purple) all remove the corresponding group. LpxO (orange) and PagP (red) transfer the group onto the molecule (4). The attachment site for the remaining polysaccharide is indicated at the 6′ position of each molecule (A). The organism source, activity, and active site topology of each of the six enzymes (B) and the 61 combinatorial strains (C) are presented. Combinatorial strains were generated by transformation of BN1 and BN2 with a pQLinkN plasmid expressing combinations of the six lipid A-modifying enzymes. Each enzyme is abbreviated by its final letter and ordered alphabetically in the plasmid name (i.e., LpxE is abbreviated E, LpxF is F, LpxR is R, PagP is P, PagL is L, and LpxO is O).
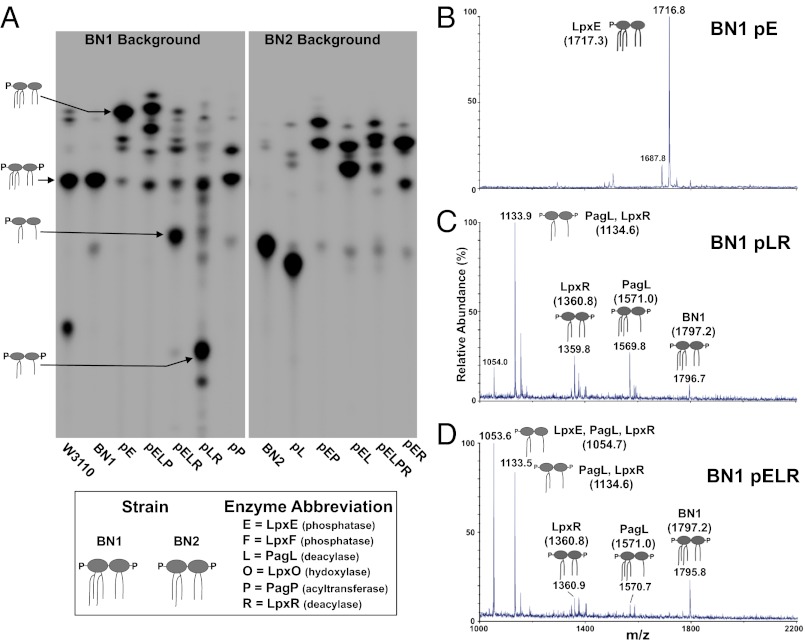
To confirm activity of the lipid modifying enzymes, 32P-labeled lipid A isolated from the 61 engineered E. coli strains was analyzed by TLC, revealing 61 distinct lipid profiles, as expected. Fig. 3A demonstrates the variety of endotoxin species produced in BN1 and BN2 expressing combinations of lipid A-modifying enzymes (Fig. 3A). Eleven strains synthesized nearly homogeneous lipid A (SI Appendix, Fig. S3), such as BN2 expressing PagL (strain BN2 pL) that produces 99.2% tetraacylated lipid A (Fig. 3A). This is rare in nature, because a heterogenous mixture of lipid A species is found on the surface of most Gram-negative bacteria (4). An additional eight strains produce at least 75% homogeneous lipid A (SI Appendix, Fig. S3), whereas other strains (e.g., BN2 pELPR) produce a more heterogeneous mixture as a consequence of the substrate specificity and limited expression level of the transmembrane lipid A-modifying enzymes (Fig. 3A). Although this heterogeneity was not our initial goal, the fact that the Food and Drug Administration-approved adjuvant MPL is a mixture, as well as the fact that all current whole-cell vaccines synthesize heterogeneous lipid A, indicated that a reproducible mixture could be just as valuable as a single uniform lipid A species. Each strain was grown and analyzed at least three times to confirm reproducibility in the lipid A profile for these mixtures.
Fig. 3.
Analysis of engineered lipid A molecules. TLC of 32P-labeled isolated lipid A from combinatorial strains is shown to illustrate the diversity within the collection (A). This method allows species separation, identification, and quantification based upon hydrophobicity-mediated migration. MS of isolated lipid A from selected strains allows further identification of lipid A species (B–D). BN1 pE produces a major peak at m/z 1,716.8, consistent with the expected removal of one phosphate group (B). BN2 pLR produces a major peak at m/z 1,133.9, corresponding to the mass of a triacylated lipid A molecule (C). This is contrasted with BN1 pELR (D), which produces a predominant peak at m/z 1,053.6, corresponding to the dephosphorylation of the major peak seen in BN1 pLR. Minor peaks in both of these strains are similar. Peaks at m/z ∼1,360 and ∼1,570 correspond to masses of lipid A resulting from a single deacylation by either LpxR or PagL, respectively. The peak at m/z ∼1,796 corresponds to residual unmodified BN1 lipid A. In BN1 pLR, there is a slight loss of the labile 1-phosphate group from the major species, yielding a peak at m/z 1,054.0.
In addition to TLC analysis, all lipid A species in the library were analyzed by MS, which allowed structural characterization based on mass and expected enzyme activities (SI Appendix, Fig. S3). Fig. 3 highlights examples of MS results for three categories of lipid A modifications: phosphate-modified (Fig. 3B), acyl-chain–modified (Fig. 3C), or a combination of both (Fig. 3D). The mass spectrum of BN1 pE revealed a major peak at m/z 1,716.8 corresponding to the removal of one phosphate group. Strain BN1 pLR yielded a major peak at m/z 1,133.9 corresponding to a triacylated lipid A, resulting from deacylation by PagL and LpxR. Cells expressing LpxE, PagL, and LpxR (BN1 pELR) yield lipid A, producing a major peak at m/z 1,053.6 corresponding to triacylated monophosphorylated lipid A. These results indicate that through logarithmic growth, E. coli can tolerate drastic changes to its normally tightly regulated lipid A profile (SI Appendix, Fig. S10).
Coexpression of these enzymes allowed further characterization of the substrate specificity of the enzymes themselves. For example, the crystal structure of LpxR and modeling of the structure with lipid A did not indicate any important interactions of the enzyme with the 1-phosphate group of lipid A (25). The enzyme is Ca2+-dependent, and the 4'-phosphate group interacts with this cation. However, analysis of our strains that express LpxR in combination with the phosphatases LpxE and/or LpxF has shown that in the BN1 hexaacylated background, the deacylase has increased activity (almost 100%) in the presence of LpxE, which removes the 1-phosphate group in the inner membrane before the lipid A molecule reaches LpxR in the outer membrane during synthesis and transport. In the pentaacylated BN2 background, removal of the 4′-phosphate group by LpxF completely abolishes LpxR activity, but LpxE activity does not. This is interesting because in Francisella LpxF mutants the 3′-acyl chain is retained, indicating the LpxF activity actually enhances LpxR activity in this organism (26).
Differential TLR4 Stimulation by Whole Bacteria and LPS with Modified Lipid A.
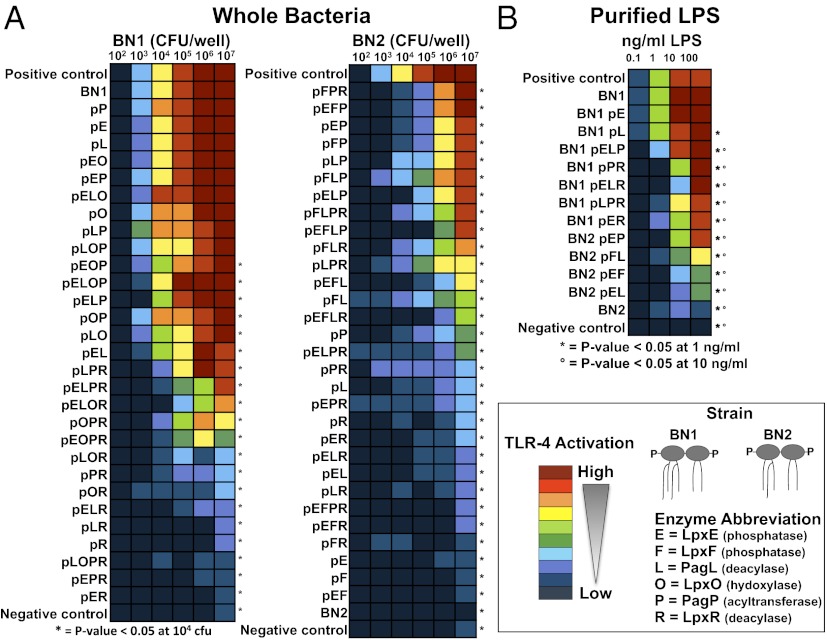
To examine the range of TLR4 activation induced by whole bacterial cells producing diverse lipid A structures, we screened our library using HEK-Blue hTLR4 cells. These cells express TLR4, MD2, CD14, and the NF-κB– and activator protein-1–dependent reporter secreted embryonic alkaline phosphatase (SEAP). The reporter cell line was challenged with a range of colony forming units (cfu) of each of the 61 lipid A library strains. At 104 cfu per well, 51 of the strains displayed significantly different TLR4 responses relative to the BN1 control strain that produces wild-type lipid A (P < 0.05). Additional controls used were the bis-phosphorylated, hexaacylated W3110 parent strain and the bis-phosphorylated, tetraacylated strain CMR300, which acts as a TLR4 antagonist. For direct comparison between strains, the data are presented colorimetrically (Fig. 4 and SI Appendix, Fig. S4), and all graphical data are supplied in SI Appendix, Fig. S5. Similar TLR4 assays were also performed using 0.1–100 ng/mL isolated LPS from 13 of the strains (Fig. 4B and SI Appendix, Fig. S5C). In all experiments, LPS and lipid A samples are quantified and normalized according to number of molecules. All LPS samples except BN1 pE induced significantly lower TLR4 activation than BN1 LPS at 1 ng/mL (P < 0.05), and a similar trend was observed between cells and purified LPS (Fig. 4). To show full induction, pure hexaacylated LPS was used, and Rhodobacter sphaeroides LPS, a known TLR4 antagonist, was used as a negative control. Although the effects of some lipid A modifications on LPS immunogenicity have been reported (20, 27), our combinatorial approach generated a surprisingly diverse range in agonistic TLR4 stimulation and illustrated the need for further characterization of the TLR4-lipid A interaction because of the unpredictable results for many of the strains. This technique also allows investigation into reproducible mixtures of agonistic and antagonistic lipid A species within a bacterial cell that could prove optimal for adjuvant activity because of a reduction in TLR4 activation compared with wild-type cells and LPS.
Fig. 4.
TLR4 stimulation by whole bacterial cells and LPS. Stimulation of TLR4 following incubation of whole bacterial cells with HEK-Blue cells expressing TLR4, MD2, and CD14 is depicted (A). The TLR4 responses to whole cells are shown for all strains. Colors were assigned based on the TLR4 stimulation results in the BN1 strain. The rationale for colorimetric designations is displayed in SI Appendix, Fig. S4. The positive control is E. coli K12 strain W3110, the parent strain of the mutants used in this study. The negative control for this assay is strain CMR300, an E. coli strain that produces only lipid IVA, a tetraacylated TLR4 antagonist (35). *P < 0.05 at 104 cfu per well. HEK-Blue-TLR4-MD2-CD14 cells were also incubated with increasing concentrations of LPS from 13 of the 61 engineered strains (B). E. coli K-12 LPS was used as a positive control and R. sphaeroides LPS, a known TLR4 antagonist, served as a negative control. *P < 0.05 at 1.0 ng/mL; °P < 0.05 10 ng/mL.
In this TLR4-specific assay, stimulation by BN1 bacterial cells expressing a single lipid A-modifying enzyme generally did not differ from the BN1 parental strain, even in BN1 pL, in which PagL cleaves the majority of the lipid A at the 3 position to yield a pentaacylated form predicted to be less inflammatory (28) (Fig. 4 and SI Appendix, Fig. S3). This was surprising, considering the reports from others that purified lipid A modified individually by PagL, PagP, or LpxE shows a reduction of TLR4 antagonism (5, 29). The limited alteration of the TLR4 response to whole bacterial cells expressing only one lipid A-modifying enzyme validates the combinatorial approach using multiple enzymes to generate molecules with varied TLR4 activity. For example, in strain BN1 pLPR, single expression of PagL or PagP or LpxR results in very high or very low TLR4 stimulation, respectively. However, when all three enzymes are coexpressed, an intermediate activity is displayed (Fig. 4A).
We found that the TLR4 response of strains ranging from completely inactive to slightly active could be enhanced when expressed in combination. For example, in the pentaacylated BN2 background strain, expression of PagP with either or both phosphatases, LpxE and LpxF, (strains BN2 pFP, BN2 pEP, and BN2 pEFP) stimulate TLR4 almost at wild-type levels at high cell counts, and yet BN2 alone and each of the enzymes expressed individually in BN2 (BN2 pP, BN2 pE, and BN2 pF) are all much less stimulatory, and some of them might even act antagonistically. Removal of either phosphate group from lipid A was expected to decrease the interaction with TLR4 (3), but, instead, we see that in combination with PagP, the phosphatases act to make the strains more endotoxic. We observe a similar phenomenon in other strains, for example, BN2 pLP and BN2 pFL, which each induce a higher TLR4 response than any potential subspecies within the strain (Fig. 4A). These results clearly demonstrate that TLR4 stimulation is substantially affected by the combinatorial expression of lipid A-modifying enzymes in a manner that does not necessarily reflect the expected results from individual expression of each enzyme.
The importance of acyl chain position on the lipid A molecule during TLR4 activation has been difficult to study in the past because comparing different bacteria that synthesize lipid A with varied acyl-chain position is not a controlled system. Our strains enable a direct comparison. As an example, BN2 is pentaacylated and does not stimulate TLR4 at any of the tested cell concentrations. As expected, expression of PagP (strain BN2 pP) increases activity, because it converts some of the lipid A to hexaacylated but not nearly as much as the highly endotoxic, wild-type hexaacylated E. coli lipid A. However, it is surprising that when PagL (strain BN1 pLP) is expressed (another acyl chain is removed and the strain then synthesizes more pentaacylated lipid A) instead of decreasing, the TLR4 stimulation increases (SI Appendix, Fig. S6).
Lipid A Modification Is Sufficient to Reduce Stimulation of Monocytes Expressing Multiple Pattern-Recognition Receptors.
Next, we investigated whether alteration of the TLR4 response through the lipid A modifications in the library are sufficient to alter the overall innate immune response, even in THP1 (human acute monocytic leukemia) cells that also recognize many conserved bacterial patterns, such as flagellar proteins, lipoproteins, and peptidoglycan in addition to LPS. To do so, bacterial cells were used to stimulate THP1-XBlue-MD2-CD14 cells expressing all TLRs, NOD1 and NOD2 (nucleotide–oligomerization domains responsible for peptidoglycan recognition) proteins, along with the SEAP reporter system that responds to stimulation of any TLR or NOD pathway. Engineered E. coli strains that resulted in reduced TLR4-specific activation in HEK-Blue hTLR4 cells (Fig. 4) also resulted in a reduced overall THP1-XBlue-MD2-CD14 response (Fig. 5A), revealing that the innate immune recognition of 103 to 105 cfu per well of engineered E. coli is dominated by TLR4 activation by the lipid A moiety of LPS and not by signaling through other Toll-like receptors.
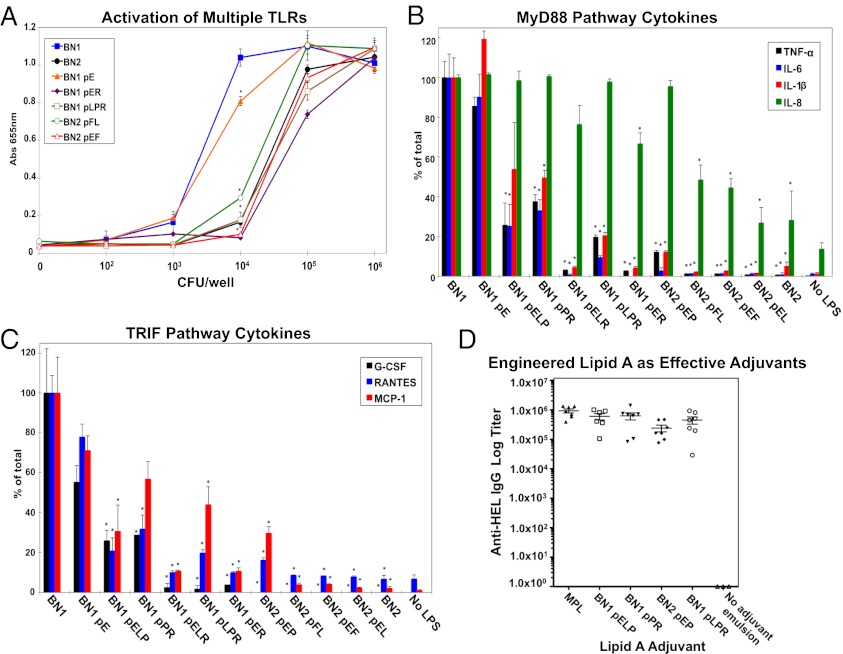
Fig. 5.
Engineered strains induce diverse stimulation and cytokine production in vitro and high IgG titers in vivo. THP1-XBlue monocytes expressing all TLRs, Nod1, Nod2, MD2, and CD14 were incubated with whole bacterial cells, and overall innate immune receptor activation was measured (A). The graph of representative samples illustrates that in the range of 103 to 105 cfu per well, the activation of the THP1 cells was reduced, and all samples were significantly different from BN1 at 104 cfu per well (P < 0.001) (A). Production of TRIF pathway cytokines (G-CSF, RANTES, and MCP-1) and MyD88 pathway cytokines (TNF-α, IL-6, IL-1β, and IL-8) by wild-type THP1 monocytes differentiated into macrophage-like cells when incubated with 100 ng/mL LPS (B and C). Cytokine levels are presented as percentages of the BN1 level. BALB/cJ mice were immunized with 50 µL of an emulsion of 30 µg of lysozyme from chicken egg white (HEL) with 6 pM purified lipid A, and serum was analyzed by ELISA (D). All lipid A adjuvants tested (BN1 pELP, BN1 pPR, BN2 pEP, and BN1 pLPR) induced a high IgG response, and only BN2 pEP was significantly lower than the MPL control (P = 0.0009).
Cytokine Profile of Human Monocytes Stimulated by LPS.
To evaluate the cytokine profile elicited by LPS containing various lipid A modifications, we determined the concentrations of the MyD88-dependent cytokines TNF-α, IL-6, IL-1β, and IL-8 and the TRIF-dependent cytokines G-CSF, regulated and normal T cell expressed and secreted (RANTES) (chemokine ligand 5), and monocyte chemotactic protein-1 (MCP-1) (chemokine ligand 2), after 24 h of incubation with 100 ng/mL LPS (Fig. 5 B and C and SI Appendix, Fig. S7). A full spectrum of cytokine levels was observed, ranging from strong stimulation by LPS from the BN1 parental strain to minimal stimulation elicited by BN2 LPS. Interestingly, LPS from some strains retained the capacity to preferentially stimulate certain cytokines. For example, BN1 pLPR maintains 40% of the MCP-1 level compared with BN1, and yet G-CSF production is almost completely abolished (Fig. 5C). Another instance of variable production of cytokines was observed in IL-8 levels, as shown by BN2 pEP that stimulates IL-8 production equal to BN1, whereas all other cytokine levels were greatly diminished (Fig. 5B). Because MCP-1 and IL-8 are important for T-cell protective immune responses, retention of their stimulation could be valuable to immune modulation, especially with the combined reduction of other inflammatory cytokines (30, 31). These results highlight how chemical modifications in LPS can impart a broad range of agonist properties to modulate cytokine production.
Engineered Lipid A Samples Induce a Strong Acquired-Immune Response in Mice.
To investigate the adjuvant potential of strains in this library, BALB/cJ mice were immunized with emulsions of hen egg white lysozyme (HEL) and purified lipid A from four strains: BN1 expressing LpxE, PagL, and PagP (BN1 pELP); BN1 expressing PagP and LpxR (BN1 pPR); BN2 expressing LpxE and PagP (BN1 pEP); and BN1 expressing PagL, PagP, and LpxR (BN1 pLPR). In this experiment, we used purified lipid A for direct comparison with MPL, which consists of only the lipid A domain of LPS. All tested lipid formulations resulted in high anti-HEL IgG titers, and only the BN2 pEP titer was statistically significantly lower than MPL (Fig. 5D). Cytokine analysis of the serum revealed a low inflammatory response measured by equivalent TNF-α levels between adjuvants and modest variation in some cytokines, such as G-CSF (SI Appendix, Fig. S8). These responses in mice indicate that the combinatorial strains could facilitate biological production of safe adjuvants that are as effective as those currently used, such as MPL. Although animal models are quite useful in testing potential adjuvants, human and murine TLR4-MD2 display differential recognition of LPS. For example, tetraacylated lipid A is an antagonist of human TLR4-MD2 but an agonist of murine TLR4-MD2 (32). This could explain the surprisingly uniform response in mice observed between lipid A samples, which can be partially attributed to lower specificity in ligand binding in the murine TLR4 compared with human TLR4, such that a wider diversity in human response to lipid A adjuvants from the library might be expected (33). Unfortunately, a humanized mouse is not yet available to compare the murine cytokine response to the adjuvant directly with human TLR4 response.
The high IgG response to lipid A from strain BN1 pELP is noteworthy because this strain produces lipid A species characteristic of the MPL mixture derived from S. minnesota LPS. Nonetheless, lipid A from BN1 pELP and the similar strain BN1 pELOP produce a higher percentage of the predominant species, 3-O-deacyl-4′-monophosphoryl lipid A, found in commercial MPL preparation from S. minnesota (SI Appendix, Fig. S9). This finding indicates that engineered E. coli can potentially be used to produce high-purity MPL, thus eliminating the need for extensive chemical treatment of S. minnesota LPS required to generate the adjuvant (8). To ensure that purification of the 3-O-deacyl-4′-monophosphoryl lipid A species is feasible, we performed liquid chromatography and analyzed the purified species by MS, confirming the isolation of the species of interest (SI Appendix, Fig. S9).
Discussion
We report that combinatorial expression of lipid A-modifying enzymes in E. coli results in an array of strains with structurally diverse lipid A species that induce differential TLR responses and cytokine profiles, many of which could not be predicted based on previous data of the TLR4–lipid A interaction. Modified LPS from these engineered strains can be used as (i) the major immunogenic surface component of whole bacteria, (ii) a purified LPS molecule, or (iii) free lipid A molecules following LPS hydrolysis. The effect of some lipid A-modifying enzymes on TLR4 agonist activity were previously unreported. For example, we show that LpxR activity diminishes TLR4 stimulation, as expected because of reported effects of reduced acylation (28). The results of some strains could not have been predicted based on previous work on the importance of acylation and phosphorylation of lipid A (3) [e.g., BN2 pFL, which generates tetraacylated and 4′-dephosphorylated lipid A and yet consistently shows higher TLR4 stimulation than its pentaacylated, bis-phosphorylated parent (Fig. 4A)]. Other strains, such as the extensively modified monophosphorylated, triacylated lipid A of strain BN1 pELR, show that E. coli tolerates severe alterations in lipid A structure through removal of three of its six lipid A acyl chains and one phosphate group without apparent growth defects (Fig. 3 A and C). This library provides a versatile tool and illustrates the utility of E. coli for production of modified lipid A molecules.
The diversification of modified LPS molecules that induce distinct cytokine responses offers the potential to produce new adjuvants that may be able to modulate humoral immune responses for more effective and longer-lasting protection. Although intact LPS or lipid A molecules from the library could allow pairing of adjuvants suitable for the desired response of a particular antigen, lipid A is insoluble and requires adsorption onto alum to enable delivery (5). Thus, detoxified whole LPS may be a more attractive soluble adjuvant.
This combinatorial library also provides tools to overcome hurdles in other aspects of industry, such as DNA and protein expression, gene therapy, and drugs for antisepsis or cancer. E. coli is ideal as an inexpensive, high-level expression strain, but LPS is a major contaminant in such preparations. Using E. coli strains with a diminished threat of endotoxic impurity could improve the safety of these preparations. Altered immunogenicity of whole E. coli cells through combinatorial diversification of lipid A would also improve gene therapy strains. Bacteria can be engineered to present antigen in tumors or invade particular tissues to transfer genetic material to a host, and a strain that selectively modulates the immune response could provide an optimal vector for such mechanisms (11, 12). In addition to lowered toxicity important for expression and gene therapy strains, some lipid A species from the library could possess antagonistic properties that inhibit TLR4 signaling. As antisepsis drugs, they could serve to block the strong inflammatory response during to septic shock (34). In this work, we have followed a synthetic lipid biology approach to generate a combinatorial library of lipid A molecules to satisfy a wide array of biotechnological and therapeutic needs, the scope of which will continue to broaden as further investigation into the potential of these strains is completed.
Materials and Methods
A description of all bacterial strains, plasmids, and the construction of lipid A template strains BN1 and BN2 can be found in the SI Appendix. Methods for the purification of lipid A, mass spectrometry, TLR4 signaling assays, the isolation and quantification of LPS, and quantification of cytokines and IgG are all described in SI Appendix. Statistical analysis of all data is also given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Catherine Schilffarth at Luminex Corporation for help with mouse cytokine data. This work was supported by National Institutes of Health Grants AI064184 and AI76322 (to M.S.T.) and AI075068 (to M.W.); a grant from the Norman Hackerman Advanced Research Program of the Texas Higher Education Coordinating Board (to G.G.); and Army Research Office Grant 61789-MA-MUR (to M.S.T.). M.W. is a Burroughs Wellcome investigator in the pathogenesis of infectious disease.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218080110/-/DCSupplemental.
References
- 1.Pfeiffer R. Untersuchungen uber das Choleragift. Z Hygeine. 1892;11:393–412. German. [Google Scholar]
- 2.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casella CR, Mitchell TC. Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell Mol Life Sci. 2008;65(20):3231–3240. doi: 10.1007/s00018-008-8228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto M, Akira S. Lipid A receptor TLR4-mediated signaling pathways. Adv Exp Med Biol. 2009;667:59–68. doi: 10.1007/978-1-4419-1603-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Keiser PB, et al. A phase 1 study of a meningococcal native outer membrane vesicle vaccine made from a group B strain with deleted lpxL1 and synX, over-expressed factor H binding protein, two PorAs and stabilized OpcA expression. Vaccine. 2011;29(7):1413–1420. doi: 10.1016/j.vaccine.2010.12.039. [DOI] [PubMed] [Google Scholar]
- 8.Garçon N. Preclinical development of AS04. Methods Mol Biol. 2010;626:15–27. doi: 10.1007/978-1-60761-585-9_2. [DOI] [PubMed] [Google Scholar]
- 9.Mata-Haro V, et al. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science. 2007;316(5831):1628–1632. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Tao G, Wang X. Construction of an Escherichia coli mutant producing monophosphoryl lipid A. Biotechnol Lett. 2011;33(5):1013–1019. doi: 10.1007/s10529-011-0521-z. [DOI] [PubMed] [Google Scholar]
- 11.Chen G, et al. Oral delivery of tumor-targeting Salmonella exhibits promising therapeutic efficacy and low toxicity. Cancer Sci. 2009;100(12):2437–2443. doi: 10.1111/j.1349-7006.2009.01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss S, Chakraborty T. Transfer of eukaryotic expression plasmids to mammalian host cells by bacterial carriers. Curr Opin Biotechnol. 2001;12(5):467–472. doi: 10.1016/s0958-1669(00)00247-0. [DOI] [PubMed] [Google Scholar]
- 13.Trent MS, Pabich W, Raetz CR, Miller SI. A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem. 2001;276(12):9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 14.Bishop RE, et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19(19):5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CR. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: Topography of francisella novicida LpxE expressed in Escherichia coli. J Biol Chem. 2004;279(47):49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, McGrath SC, Cotter RJ, Raetz CR. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J Biol Chem. 2006;281(14):9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds CM, et al. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281(31):21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbons HS, Lin S, Cotter RJ, Raetz CR. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem. 2000;275(42):32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- 19.Ingram BO, Masoudi A, Raetz CR. Escherichia coli mutants that synthesize dephosphorylated lipid A molecules. Biochemistry. 2010;49(38):8325–8337. doi: 10.1021/bi101253s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki K, Ernst RK, Miller SI. Purification and characterization of deacylated and/or palmitoylated lipid A species unique to Salmonella enterica serovar Typhimurium. J Endotoxin Res. 2005;11(1):57–61. doi: 10.1179/096805105225006696. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto Y, Shimoyama A, Suda Y, Fukase K. Synthesis and immunomodulatory activities of Helicobacter pylori lipophilic terminus of lipopolysaccharide including lipid A. Carbohydr Res. 2012;356:37–43. doi: 10.1016/j.carres.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Touzé T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol. 2008;67(2):264–277. doi: 10.1111/j.1365-2958.2007.06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrera CM, Hankins JV, Trent MS. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol. 2010;76(6):1444–1460. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35(6):e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutten L, et al. Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium. Proc Natl Acad Sci USA. 2009;106(6):1960–1964. doi: 10.1073/pnas.0813064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CR. Attenuated virulence of a Francisella mutant lacking the lipid A 4′-phosphatase. Proc Natl Acad Sci USA. 2007;104(10):4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong Q, et al. Salmonella synthesizing 1-monophosphorylated lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J Immunol. 2011;187(1):412–423. doi: 10.4049/jimmunol.1100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teghanemt A, Zhang D, Levis EN, Weiss JP, Gioannini TL. Molecular basis of reduced potency of underacylated endotoxins. J Immunol. 2005;175(7):4669–4676. doi: 10.4049/jimmunol.175.7.4669. [DOI] [PubMed] [Google Scholar]
- 29.Kawasaki K, Ernst RK, Miller SI. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J Biol Chem. 2004;279(19):20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 30.Ragnarsdóttir B, et al. TLR- and CXCR1-dependent innate immunity: Insights into the genetics of urinary tract infections. Eur J Clin Invest. 2008;38(Suppl 2):12–20. doi: 10.1111/j.1365-2362.2008.02004.x. [DOI] [PubMed] [Google Scholar]
- 31.Sin J, Kim JJ, Pachuk C, Satishchandran C, Weiner DB. DNA vaccines encoding interleukin-8 and RANTES enhance antigen-specific Th1-type CD4(+) T-cell-mediated protective immunity against herpes simplex virus type 2 in vivo. J Virol. 2000;74(23):11173–11180. doi: 10.1128/jvi.74.23.11173-11180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohto U, Fukase K, Miyake K, Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc Natl Acad Sci USA. 2012;109(19):7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3(4):354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 34.Peri F, Piazza M. 2012. Therapeutic targeting of innate immunity with Toll-like receptor 4 (TLR4) antagonists. Biotechnol Adv 30(1):251–260.
- 35.Reynolds CM, Raetz CR. Replacement of lipopolysaccharide with free lipid A molecules in Escherichia coli mutants lacking all core sugars. Biochemistry. 2009;48(40):9627–9640. doi: 10.1021/bi901391g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.