Significance Statement
Rhythmic mRNA expression is a hallmark of circadian rhythms. To address this regulation in Drosophila, we used high throughput sequencing to compare nascent RNA and mRNA levels in fly heads at different times of day. This indicated that some genes undergo rhythmic transcription with no impact on mRNA, whereas a few strongly rhythmic mRNAs have weak transcriptional oscillations. A substantial number of rhythmic genes manifest more robust mRNA cycling than transcriptional cycling, indicating that post-transcriptional regulation plays a widespread role in boosting circadian gene expression amplitude. Our study highlights the importance of directly assaying transcription to understand gene regulation.
Keywords: Nas-seq, chromatin dynamics
Abstract
Rhythmic mRNA expression is a hallmark of circadian biology and has been described in numerous experimental systems including mammals. A small number of core clock gene mRNAs and a much larger number of output mRNAs are under circadian control. The rhythmic expression of core clock genes is regulated at the transcriptional level, and this regulation is important for the timekeeping mechanism. However, the relative contribution of transcriptional and posttranscriptional regulation to global circadian mRNA oscillations is unknown. To address this issue in Drosophila, we isolated nascent RNA from adult fly heads collected at different time points and subjected it to high-throughput sequencing. mRNA was isolated and sequenced in parallel. Some genes had cycling nascent RNAs with no cycling mRNA, caused, most likely, by light-mediated read-through transcription. Most genes with cycling mRNAs had significant nascent RNA cycling amplitudes, indicating a prominent role for circadian transcriptional regulation. However, a considerable fraction had higher mRNA amplitudes than nascent RNA amplitudes. The same comparison for core clock gene mRNAs gives rise to a qualitatively similar conclusion. The data therefore indicate a significant quantitative contribution of posttranscriptional regulation to mRNA cycling.
The circadian clock controls the rhythmic expression of thousands of genes. These mRNAs mediate the oscillation of numerous biochemical, physiological, and behavioral functions. Although inconsistency among microarray datasets has made it difficult to identify a definitive set of mRNA cyclers in Drosophila, there is good agreement on a small number of robust cycling mRNAs (see below). Moreover, it appears that a much larger number of cycling mRNAs exist within circadian neurons (1), and thousands of mRNAs have been shown to oscillate within individual tissues in mammals (2–5).
Cycling mRNAs are dependent on a functional pacemaker (6, 7), but most probably are regulated only indirectly by the core circadian clock (8). In flies, this mechanism consists of the CLOCK and CYCLE heterodimer (CLK/CYC), which drives the rhythmic transcription of its repressor protein genes period and timeless (9). After translation, the proteins PER and TIM decrease their own transcription by inhibiting the activity of CLK/CYC (10–12). A set of largely orthologous proteins (CLK, BMAL1, PER, and CRYPTOCHROME) functions similarly in mammals. Although posttranslational regulation of these transcription factors makes a major contribution to core clock function (13–19), there is evidence that transcriptional regulation also is important for circadian timing. For example, flies with enhanced CLK/CYC-mediated transcription have unusually high levels of per mRNA and significantly shorter periods (20).
The core clock model drives the assumption that the large population of cycling mRNAs—the hundreds or thousands of output mRNAs under circadian regulation—is also under transcriptional regulation. However, it is not known whether transcriptional regulation accounts for all, most, or only a small fraction of the large number of cycling output mRNAs. Moreover, an individual mRNA can be regulated in several ways: For some oscillating mRNAs, regulation might be mostly transcriptional, whereas for others it might be mostly posttranscriptional. The latter could reflect the circadian regulation of mRNA turnover or of translation.
To address this issue of circadian transcriptional regulation at a global level in flies, we subjected nascent RNA from adult fly head time points to high-throughput sequencing or Nascent-Seq (21, 22). Because there are no published sequencing data of head mRNA, we also sequenced head polyA+ RNA (RNA-Seq) in parallel. Using very stringent criteria for rhythmic gene expression, we identified more than 130 robust cycling transcriptional units in the Nascent-Seq data, of which more than one-third cycled in the parallel mRNA analysis. The reverse comparison indicated that only 19% of bona-fide cycling mRNAs were identified as cyclers in the Nascent-Seq data. The low percentage reflects the lower cycling amplitudes of most cycling mRNAs in the Nascent-Seq data, indicating a potent contribution of posttranscriptional regulation to the mRNA cycling amplitude. The same comparisons for core clock gene mRNAs revealed a qualitatively similar distinction between nascent RNA and mRNA cycling amplitudes. Taken together, the data indicate an important and general contribution of posttranscriptional regulation to mRNA cycling amplitude, complementing the transcriptional regulation foundation in the fly circadian system.
Results
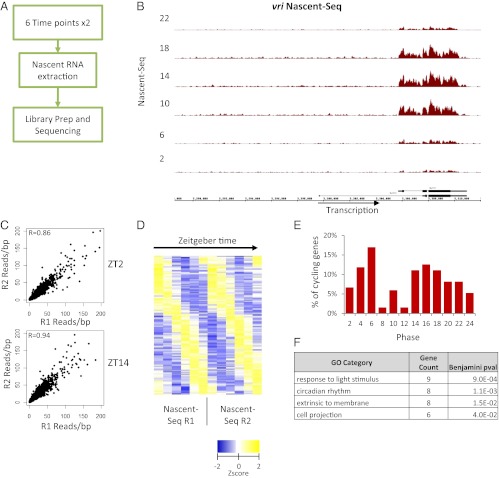
To address genome-wide circadian transcriptional regulation in the Drosophila system, we isolated nascent RNA from fly heads collected at six different times throughout a 12-h/12-h light/dark (LD) cycle. [Zeitgeber time (ZT), is the nomenclature for time in LD, with ZT0 referring to lights-on and ZT12 to lights-off.] We used well-established protocols for the purification of nascent RNA (21–24) and for the sequencing libraries and prepared 12 libraries from two independent replicas of six time points (Fig. 1A). The nascent RNA libraries were sequenced on the Illumina GA2 platform (Nascent-Seq). In brief, the subsequent analysis pipeline consisted of mapping the reads with TopHat (25) and then normalizing each sample by its total uniquely mapped reads. The transcription signal of individual genes was quantitated by calculating the average reads per base pair within exons normalized to sequencing depth. The exclusion of intron signal allowed a direct comparison with RNA-Seq and avoided complications caused by cotranscriptional splicing (21).
Fig. 1.
Nascent-Seq identifies 136 transcriptional cyclers in Drosophila heads. (A) Overview of the workflow used to prepare and sequence rhythmic nascent RNA. (B) Cycling Nascent-Seq signal over the vri locus for one set of replicates. Signal within the intron is enriched. (C) The reads per base pair coverage (red) was used to quantify the average coverage within exons. The correlation of this measure between the replicate time points is high, with coefficients of R = 0.86 and R = 0.94 for ZT2 and ZT14, respectively. (D) Fourier analysis of Nascent-Seq datasets identifies 136 nascent cyclers. Shown are the Z-scores of the reads per base pair coverage plotted as a heatmap. (E) Two main peaks of expression are observed, at ZT4–ZT6 and ZT12–ZT18. (F) A few GO categories were enriched, including circadian rhythms and response to stimulus.
Visualization of nascent RNA signal within core clock genes such as vri (Fig. 1B) indicates potent temporal oscillations, consistent with the known circadian regulation of vri transcription (26). Moreover, intron signal is apparent within the large vri first intron (Fig. 1B). There was more generally a large (ninefold) increase in the number of mapped reads within introns in the Nascent-Seq data as compared with the RNA-Seq data (Fig. S1 A and B). The duplicate time points also had a high degree of reproducibility. For example, the ZT2 and ZT14 duplicates had correlation coefficients of 0.85 and 0.94, respectively (Fig. 1C).
Because of this favorable initial characterization, the rhythmic regulation of the 12 time points was subjected to Fourier analysis (27), yielding scores (F24) used to determine cyclic fitness. Using stringent criteria (Materials and Methods), we identified 136 robust “nascent cyclers” (Fig. 1D). Their phase distribution was primarily bimodal, with a peak in the daytime between ZT4–ZT6 and another, less sharp peak in the nighttime between ZT12–ZT18 (Fig. 1E). They identified the expected enrichment Gene Ontology (GO) term, “circadian genes.” Based on nascent cycling amplitude, circadian genes are highly ranked: for example tim, 4; vri, 9; Clk, 12, Pdp1, 24, per, 28. A few additional GO terms, such as “light-responsive genes,” also are enriched in the nascent dataset (Fig. 1F). This category suggests an effect of the LD cycle on circadian transcriptional regulation during this LD paradigm (27). Other highly ranked cycling genes are well known from microarray studies, as discussed in more detail below (6, 7, 27–29).
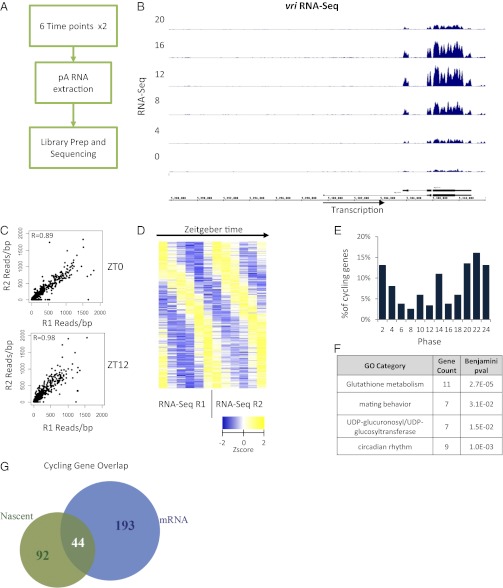
To compare the Nascent-Seq data with RNA-Seq data using the identical analytical tools (i.e., rather than comparing the Nascent-Seq data with previous microarray data), we prepared similar sequencing libraries from fly head mRNA samples, i.e., two replicates of each of six time points. We processed one replicate by paired-end sequencing and the other by single-end sequencing and used the analysis pipeline described above for Nascent-Seq (Fig. 2A).
Fig. 2.
RNA-Seq identifies more than 237 mRNA cyclers in Drosophila heads. (A) Overview of the workflow used to prepare and sequence rhythmic mRNA. (B) Cycling mRNA signal over the vri locus for one set of replicates. Signal within the intron is depleted. (C) The reads per base pair coverage (blue) was used to quantify the average coverage within exons. The correlation of this measure between the replicate time points is good, with coefficients of R = 0.89 and R = 0.98 for ZT0 and ZT12, respectively. (D) Fourier analysis of RNA-Seq datasets identifies 237 mRNA cyclers. Shown are the Z-scores of the reads per base pair coverage plotted as a heatmap. (E) The phase distribution of cycling is more spread out than the Nascent-Seq cyclers and increases gradually, peaking at ZT22. (F) A few GO categories were enriched, including circadian rhythms and glutathione metabolism. (G) A significant overlap between the Nascent and mRNA cycling genes is observed. Forty-four genes have robust Nascent and mRNA cycling which is indicative of the classic circadian transcription model.
As expected, vri showed robust RNA-Seq cycling (Fig. 2B) with a vanishingly small intron signal (compare with Fig. 1B). As in the Nascent-Seq samples, there was a high degree of reproducibility between duplicate time points, e.g., correlation coefficients of 0.84 and 0.98 for ZT0 and ZT12, respectively (Fig. 2C).
We obtained 237 robust mRNA cyclers with the Fourier cycling criteria used for the Nascent-Seq data (Fig. 2D). Interestingly, the phase distribution of the mRNA cyclers was slightly different from that of the nascent cyclers (compare Figs. 1E and 2E). For example, there is a monotonic decrease in the phase distribution of the RNA-Seq cyclers during the morning (ZT6 < ZT4 < ZT2; Fig. 2E), whereas there is a monotonic increase in the Nascent-Seq cyclers at the same times (ZT6 > ZT4 > ZT2; Fig. 1E).
Like the Nascent-Seq cyclers, clock gene mRNAs, as well as many cycling mRNAs previously identified with microarrays, were highly ranked in the cycling RNA-Seq data. Circadian and a few additional GO terms (such as “glutathione metabolism”) were also identified (Fig. 2F) (6, 7, 28, 29). The different noncircadian GO terms for Nascent-Seq and RNA-Seq cycling (Figs. 1F and 2F, respectively) indicate that, despite a highly significant overlap, most genes in the two datasets are distinct (P = 1e-5, χ2 test; P < 1e-9, Fisher’s exact test; Fig. 2G).
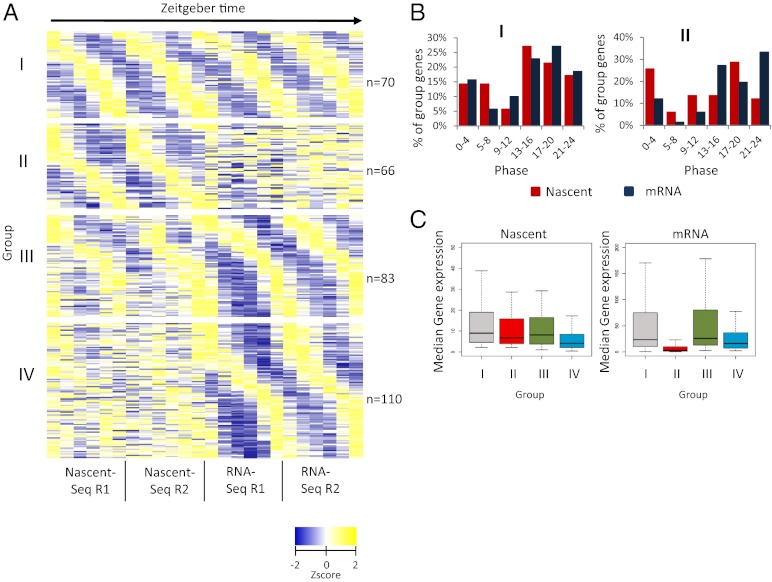
To compare the two cycling RNA populations in a more comprehensive fashion, we looked for reproducibility as well as phase correlations of individual genes between the nascent and mRNA datasets. The data could be organized into four simple groups (Methods) (Fig. 3A and Dataset S1). Group I contains genes with both robust nascent and robust mRNA cycling. Group II contains genes with robust nascent RNA cycling but little or no mRNA cycling, i.e., very poor F24 scores for putative mRNA cycling. Group III genes contain robust mRNA cyclers with weak but visually obvious nascent RNA oscillations. These genes failed to be identified in the nascent RNA analysis because they were below the stringent thresholds used to define cycling. Group IV genes also contain robust mRNA cyclers but even weaker, and perhaps no, nascent RNA cycling.
Fig. 3.
Four groups of cyclers are identified. (A) Four groups are identified based of cycling thresholds (Materials and Methods). Group I genes have robust nascent and mRNA cycling, whereas group II genes have robust nascent cycling but weak or flat temporal mRNA-expression profiles. Group III genes have robust cycling mRNA-expression profiles and weaker cycling nascent-expression profiles. Group IV genes have robust mRNA-expression profiles but very noisy or flat temporal nascent-expression profiles. Shown are the Z-scores of the reads per base pair coverage plotted as a heatmap. (B) Phase distribution of nascent RNA (red) and mRNA (blue) for the genes of groups I and II. Group II cycler mRNA phase distribution is not correlated with the nascent phase distribution, contrary to group I cyclers, which have very similar phase distributions. The phase was determined by assigning the time point with the maximum reads per base pair. (C) Group II genes have very weak mRNA expression compared with other group cyclers.
For group I genes, the phase distributions of the nascent and mRNA cycling are very similar in the two RNA populations, with peaks between ZT12 and ZT20 (Fig. 3B, group I). This distribution also is similar to the phases of the classical core circadian genes (10, 26, 30–34), which are under transcriptional regulation and all present within group I. The nascent and mRNA cycling phases of individual group I genes also are essentially identical (Fig. 3A and Fig. S2A), indicating that most, if not all, group I mRNA cycling occurs because of transcriptional regulation.
In contrast to group I, group II genes have poor mRNA F24 scores, making their phases difficult to assign and therefore difficult to compare with their nascent phases. To improve this comparison, group II mRNA phases were assigned the time point with the maximum reads per base pair. Not surprisingly, the two distributions are still quite different (Fig. 3B, group II), and the individual phases also are very different (Fig. S2B). Because the group II median mRNA expression level also is significantly lower than that of the other mRNA groups (Fig. 3C), the group II mRNA data are even more difficult to interpret.
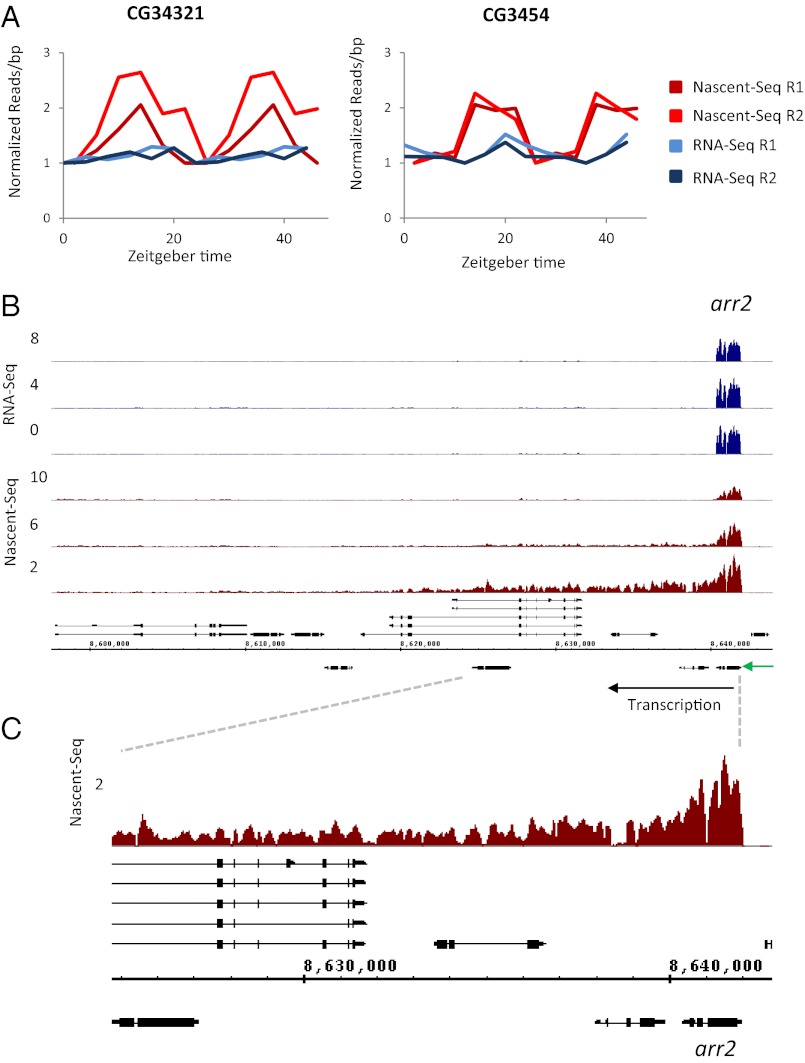
Only five group II genes had possible mRNA oscillations by visible inspection but with significantly lower mRNA cycling amplitude compared with their nascent oscillations. For example, CG34321 and CG3454 have significantly dampened RNA-Seq oscillations, and both peak later relative to the Nascent-Seq profiles (Fig. 4A). Interestingly, most group II genes (62%) had an unusual annotation feature, namely, 24 genes are closely positioned, in tandem on the same strand or as convergent genes on opposite strands. These genes include at least nine regions for which cycling transcription is caused by apparent Polymerase II read-through or a failure to terminate efficiently at the 3′ end of an upstream gene. Their transcription shows no indication of circadian regulation but only this lack of efficient termination at a specific time of the circadian cycle.
Fig. 4.
A subset of group II cyclers consists of read-through transcription at the beginning of the light phase. (A) In CG34321 and CG3454 the nascent cycling amplitude (red) is higher than the mRNA cycling amplitude (blue). Plotted are normalized reads per base pair averaged within exons. The data are double plotted. (B) A transcript appears to extend from the 3′ end of arr2 and transcribes across several downstream genes, which are located on both strands. No cycling signal is observed in the RNA-Seq data. Only the three day time points are shown for Nascent- and RNA- Seq profiles, in red and blue respectively. The expression profiles of the three night time points for the Nascent- and RNA-Seq datasets are indistinguishable from their ZT 10 and ZT 8 time points, respectively. (C) Zoomed-in view of ZT 2 Nascent-Seq read coverage (red) at the arr2 locus.
An example is ARR2, which encodes the major Drosophila visual arrestin. Inefficient termination at the ARR2 3′ end appears to read into downstream DNA for about 35 kb, transcribing through a group of several genes oriented in both sense and antisense directions (Fig. 4 B and C). Of the nine regions of group II genes, eight, like ARR2, are enriched for eye expression. Consistent with this tissue specificity, the read-through always occurs at the beginning of the light phase and declines thereafter (e.g., Fig. 4 B and C) (27). In addition, all these data were generated under LD conditions. We speculate that the low mRNA level of most of these genes was caused by a lack of proper transcription initiation resulting from the read-through transcription from the upstream ARR2 gene.
Group III genes are similar to group I genes despite somewhat weaker nascent RNA cycling. The group III nascent and mRNA phase distributions therefore are similar to each other and to the group I distributions, and the nascent phases of the individual genes also are similar to their mRNA phases (Fig. S2 C and D). The data therefore indicate that there may be no substantive distinction between group I and group III genes apart from the lower amplitudes of group III transcriptional oscillation.
Group IV genes have robust mRNA cycling but even weaker nascent cycling than group III genes. Not surprisingly, group IV genes also have much poorer agreement between their nascent and mRNA phases than group III and group I genes (Fig. S2 C and D). Although the median nascent expression of group IV genes is somewhat lower than that of the other groups (4.2 reads per base pair, compared with 9.0, 6.8, and 8.2 reads per base pair for groups I, II, III, respectively) (Fig. 3C), almost 80% of group IV genes have nascent expression levels above the threshold required for group I genes (Fig. S3A), which have robust nascent as well as mRNA cycling. This result suggests that sequencing depth is not the sole factor responsible for the less reliable nascent cycling of group IV. Furthermore, group IV has nascent RNA maximum/minimum (max/min) values considerably lower than their mRNA cycling amplitudes, as does group III (Fig. S3B), suggesting that posttranscriptional regulation makes a substantial contribution to the mRNA cycling amplitude of group III as well as group IV mRNA cyclers.
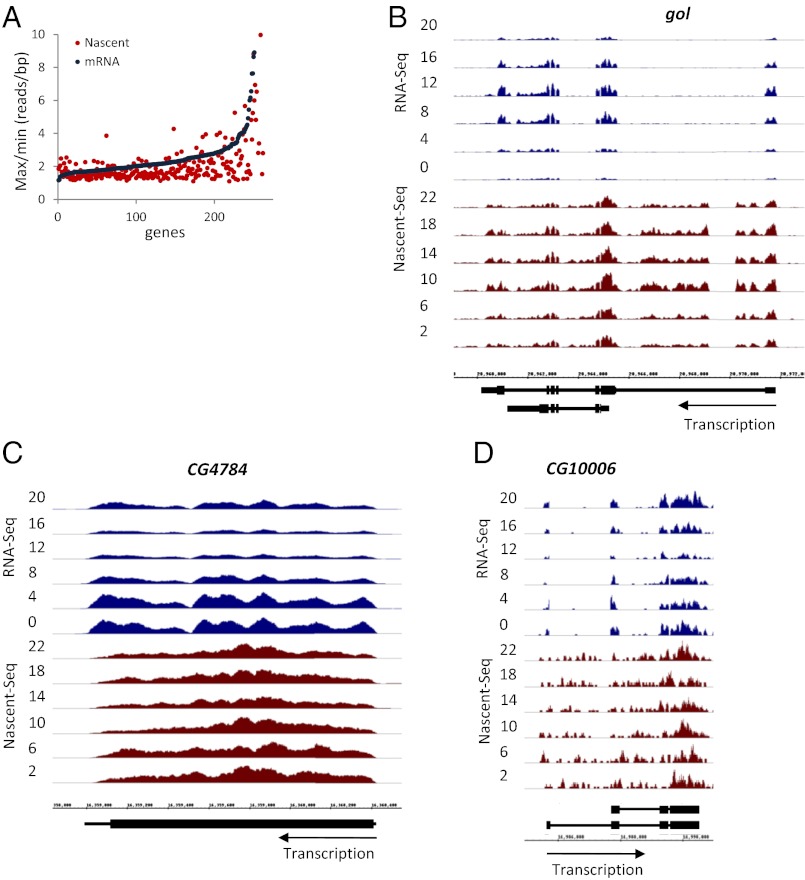
To examine the magnitude of this contribution, we considered all cycling mRNAs (groups I, III, and IV) and compared their nascent as well as their mRNA amplitudes (max/min). The plot is shifted to the right in favor of larger mRNA amplitudes, e.g., there are 24 genes with a twofold higher mRNA amplitude than nascent RNA amplitude but only two genes with a twofold higher nascent than mRNA amplitude (Fig. 5A and Fig. S3C). This result indicates that posttranscriptional regulation makes a significant contribution to the cycling amplitude of many mRNAs and turns out to be true even for some group I cyclers. Goliath (gol) is an example: Its mRNA cycling amplitude is threefold higher than its nascent RNA amplitude (Fig. 5B). gol also was of interest because it is a direct target of the CLK-CYC complex and is an eye-specific gene. As perhaps should be expected, CLK binding to gol occurs only in eyes (8), emphasizing the tissue heterogeneity that underlies these head datasets. This heterogeneity makes it difficult to compare the head datasets with recent RNA-Seq data from dissected fly brains (35).
Fig. 5.
Posttranscriptional contribution to mRNA cycling. (A) RNA-Seq cycling amplitudes for genes in groups I, III, and IV are higher than Nascent-Seq amplitudes, indicating a posttranscriptional contribution to the gene-expression profile. The ratio of max/min reads per base pair coverage for each dataset was calculated from the averaged time points and was plotted. (B) Visualization of Nascent-Seq (red) and RNA-Seq (blue) data at the gol locus. gol is a group I cycler and has a higher mRNA cycling amplitude than nascent RNA cycling amplitude. (C) Visualization of Nascent-Seq (red) and RNA-Seq (blue) data at the CG4784 locus. CG4784 is a group IV cycler and has a higher mRNA cycling amplitude than nascent RNA cycling amplitude. (D) Visualization of Nascent-Seq (red) and RNA-Seq (blue) data at the CG10006 locus. CG10006 is a group IV cycler and has a higher mRNA cycling amplitude than nascent RNA cycling amplitude.
CG4784 and CG10006 are group IV cyclers and also show a considerably higher mRNA than nascent mRNA cycling amplitude (Figs. 5 C and D and 6). CG10006 mRNA does not feature prominently in previous microarray studies, which defined mRNAs under circadian control. This data suggests that CG10006 may represent a class of cycling mRNAs that differ between RNA-Seq and microarray analyses. In contrast, CG4784 mRNA appears in many circadian studies and is one of the “top 50” cycling mRNAs based on these published microarray studies (Table S1 and Discussion). It therefore is a well-accepted cycling mRNA with little or no apparent transcriptional regulation.
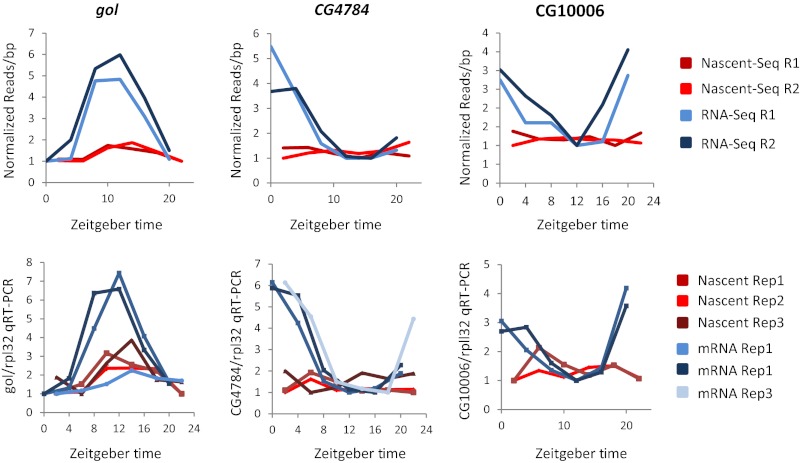
Fig. 6.
Validation of posttranscriptional candidates. Goliath, CG4784, and CG10006 cycling amplitudes are higher in the RNA-Seq data than in the Nascent-Seq data. All three candidates were validated by qRT-PCR using two independent samples each of Nascent and mRNA. An additional set of independent samples was assayed for gol and CG4784. Read per base pair quantitation of the sequencing data and of the qRT-PCR gene signal divided by RPL32 gene signal is plotted for each gene.
We verified the nascent and mRNA profiles of these three genes by quantitative RT-PCR (qRT-PCR) (Fig. 6). The curves were generated from four independent six-time-point RNA preparations (two for Nascent and two for mRNA), which were different from the four preparations used to generate the Seq data. qRT-PCR assays of gol and CG4784 were also done on an additional pair of six-time-point preparations (Fig. 6).
The qRT-PCR curves matched the sequencing curves well: All mRNA cycling amplitudes were well in excess of the nascent RNA amplitudes. The one exception was a single low-amplitude qRT-PCR mRNA curve for gol (Fig. 6). Although the two other qRT-PCR mRNA curves and both RNA-Seq curves were much more robust than the five low-amplitude nascent RNA curves, the unexpected low-amplitude qRT-PCR mRNA curve emphasizes that some aspects of the data are variable (Discussion).
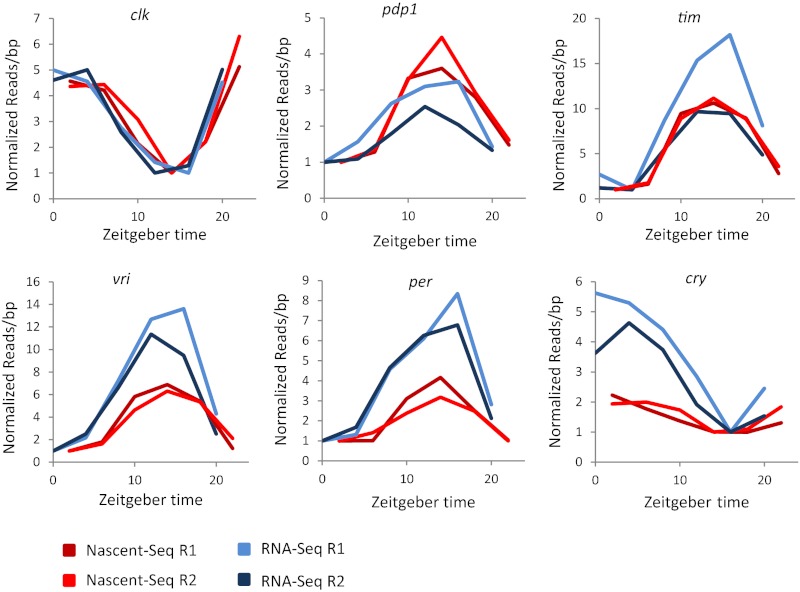
Last, we compared the Nascent-Seq data with the RNA-Seq data for the six best-studied core clock genes. We plotted the average read per base pair coverage by normalizing the data to the minimum of all time points (Fig. 7). By inspection, the mRNA and nascent RNA cycling curves of clk are very similar. This similarity can be explained by a short mRNA half-life with no role of circadian (cycling) posttranscriptional regulation. Although pdp1 mRNA appears to have a higher nascent than mRNA amplitude, this conclusion is compromised by the somewhat variable mRNA data. In contrast, the mRNA and nascent RNA curves of per, vri, and cry all have reproducibly higher mRNA than nascent amplitudes, indicating a substantive contribution of posttranscriptional regulation (Fig. S4). Although the data are more ambiguous for tim (one mRNA curve has a higher amplitude than the nascent curves and therefore resembles those of vri, per, and cry, whereas the other tim mRNA curve is similar to the nascent curves; Fig. 7), the clock-gene comparisons generally extend the conclusions from a previous study implicating posttranscriptional regulation in per and tim mRNA cycling (31).
Fig. 7.
Some core clock genes also are under posttranscriptional control. By inspection, per and cry have higher mRNA cycling amplitudes relative to their respective nascent cycling amplitudes. vri also has a smaller posttranscriptional contribution to its mRNA cycling amplitude. See text for more detail.
Discussion
Most studies characterizing circadian mRNA oscillations assume that they are caused by transcriptional regulation. This assumption is based on the robust and validated transcriptional regulation of core clock genes, which has been extrapolated to the hundreds and even thousands of oscillating mRNAs identified in different systems. However, little is known about the relative transcriptional and posttranscriptional contributions to these large numbers of cycling mRNAs. To address this issue, we sequenced nascent RNA from fly head circadian time points and, using very stringent criteria, identified more than 130 robustly cycling transcriptional units. Although circadian read-through transcription complicates the interpretation of this circadian Nascent-Seq dataset, its comparison with a parallel RNA-Seq dataset indicates substantial transcriptional regulation as well as a notable posttranscriptional contribution to the amplitude of rhythmic mRNA cycling. This is also the case for several core clock mRNAs, consistent with previous work (31). A similar Nascent-Seq vs. RNA-Seq strategy used in mammalian liver (36) and a recently published paper based on a different strategy for assessing liver transcriptional rhythms (37) come to a generally similar conclusion, namely, a widespread contribution of posttranscriptional regulation to circadian mRNA cycling.
Despite the presence of many identical genes in the two datasets, there also are distinct genes resulting in nonidentical GO lists. Many of these differences can be attributed to unique transcriptional features. For example, there are Nascent-Seq cyclers caused by cycling read-through transcription from a single adjacent gene. Almost all these genes (eight of nine) are enriched in the eye, and the rest of the data indicate that the read-through is light-regulated, perhaps like other cycling nascent RNAs and mRNAs that are light-regulated (27). This data probably accounts for the “response to light stimulus” GO term present only in the Nascent-Seq list. The read-through transcription also does not seem affect the downstream gene mRNA levels, probably reflecting the poor mRNA stability of improperly initiated RNAs and leaving open the function of this read-through transcription. We identified only five additional group II genes (8% of group II, 13 overall across groups I, III, and IV) with lower mRNA amplitude than nascent RNA amplitude. These mRNAs may have long half-lives, which dampen the amplitudes set by cycling transcription (for examples, see Fig. 4A) (31, 38).
This expected class of RNAs is based on the example of liver albumin mRNA, which has robust transcriptional oscillations but an exceptionally long cytoplasmic half-life that significantly dampens its transcriptional amplitude (39, 40). In contrast, a very short mRNA half-life, on the order of a couple of hours, is required for an mRNA cycling amplitude to be essentially indistinguishable from its transcription amplitude (31). This appears to be the case for Clk mRNA (Fig. 7). If, on the other hand, the cycling amplitude of an mRNA is greater than its transcriptional amplitude, some form of temporal posttranscriptional regulation is implicated, e.g., more potent mRNA turnover at one time of day than at another (38).
This difference in turnover appears to be the case for the large number of mRNA cyclers without sufficiently robust nascent cycling to pass the stringency thresholds (groups III and IV). However, the group III mRNAs are not solely dependent on posttranscriptional cycling, because they manifest some transcriptional cycling (Fig. 3A). In contrast, group IV mRNAs have very irreproducible or no nascent cycling, so their robust cycling should be more dependent on circadian posttranscriptional regulation. More generally, 46% of the robust pA cyclers (237 total) show only weak evidence of nascent RNA oscillations. Even for the 54% with strong transcriptional cycling, many of these mRNAs have considerably higher RNA-Seq than Nascent-Seq amplitudes. The data therefore indicate substantial posttranscriptional regulation, which at a minimum boosts the amplitude of the mRNA cycling.
We compared our cycling mRNAs with a list of the top 50 previously published fly head cycling mRNAs (Table S1), which is based simply on a correspondence between published microarray datasets. Despite identical conditions, e.g., LD entrainment, our RNA-Seq data failed to identify 22 of these 50 mRNAs. Although differences between RNA-Seq and microarray platforms might be responsible for this difference, it also could reflect the presence of unknown experimental variables, such as fly age, density, and food. Differences of this nature also might contribute to the poor correspondence between the different microarray results as well as to some irreproducibility within our own RNA-Seq data, e.g., the amplitude of gol mRNA (Fig. 6) and the amplitude of tim mRNA (Fig. 7). Nonetheless, many cycling mRNAs, including the core clock genes, continue to appear in different studies (8).
Several studies have implicated miRNAs in the posttranscription regulation of the circadian clock (41–44). For example, the nocturnin mRNA cycling profile is posttranscriptionally regulated by the microRNA mir122 in mammals (45). Although we did not observe any predicted enrichment for specific miRNA target sites in mRNAs with greater cycling amplitudes than nascent amplitudes, goliath associates with the AGO1 complex (41).
RNA-binding proteins also may dictate a circadian change in the rate of mRNA degradation. An effect of this nature is an integral part of the transcriptional feedback loop in Neurospora, where the key clock regulator FRQ not only regulates clock protein transcription in the nucleus but also associates with its own mRNA in the cytoplasm to affect frq mRNA stability (46). Moreover, FRQ interacts indirectly with RRP44, a component of the exosome (46). Mammalian hnRNP proteins also have been implicated in the degradation of clock gene mRNAs, namely per2 and cry1 mRNAs (47, 48). Although there is little comparable mechanistic information in the Drosophila system, Lark may function posttranscriptionally to regulate clock gene mRNAs (49, 50).
Importantly, some differences between mRNA and nascent amplitudes may not require the cyclical association of genes or RNAs with specific posttranscriptional regulators. The discrepancy also can be explained by a temporally constant RNA degradation threshold, below which there is a very little stable RNA accumulation (Fig. S5). Although analogous to translational regulation by miRNAs [which have been proposed to create mRNA level thresholds below which there is no protein expression (51)], a transcriptional threshold mechanism could apply broadly without implicating a large number of different regulatory molecules. Distinguishing between these possibilities will require additional experiments, but the genes identified in this study should help frame their direction.
Materials and Methods
Fly Strains.
Drosophila melanogaster yellow white (yw) and Canton-S (Cs) laboratory stock flies were used.
Nascent RNA Extraction.
The yw flies were entrained to 3 d of 12h/12h LD cycles at 25 °C. Flies were collected at 4-h intervals on the fourth day and frozen on dry ice. Fly heads were isolated with brass sieves. Nascent RNA was extracted as described (21, 22).
Total RNA Extraction.
Total RNA was extracted with TRIzol Reagent (Invitrogen) using the manufacturer’s protocol.
mRNA Extraction.
pA+ RNA was extracted from yw and Cs total RNA with Invitrogen Dynabeads oligo dT using the manufacturer’s protocol.
cDNA Synthesis.
Total RNA was DNased with Ambion TURBO DNase Free. The cDNA libraries for qRT-PCR then were made from 1 μg of Total RNA with SuperScript II (Invitrogen) and random primers according to the manufacturer’s protocol.
Primer Design.
PCR primers were designed from the exonic sequences. The sequences of the primers used are supplied in Table S2.
PCR and qRT–PCR.
PCR and qRT-PCR were performed with Platinum Taq (Invitrogen) and Syber Master Mix (Qiagen), respectively, using standard protocols as previously described (8).
Library Construction and Sequencing.
The sequencing library was constructed by using the standard Illumina protocol for mRNA using laboratory reagents. RNA libraries were made with 100 ng of mRNA-depleted nascent RNA (to remove potentially contaminating pA RNA) or 100 ng of polyA mRNA. Protocols can be provided on request (22). All libraries were sequenced on the Illumina Genome Analyzer II sequencing platform. All Nascent-Seq datasets were sequenced by the standard single-end protocol. RNA-Seq replicates 1 and 2 were sequenced by paired-end and single-end protocols, respectively. All raw sequencing data are available for download from the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE37232.
Top Microarray Cyclers.
Microarrays from refs. 27 and 41 were normalized using R and the GCRMA package. Fourier scores were calculated with R as described (52). Genes with max/min ratios above 1.75 and an F24 above 0.5 were classified as cycling with Microsoft Excel. Cycling genes were sorted by the number of microarray datasets containing a cycling probe for the gene and then by the least SD of phase across all microarray datasets. These criteria established the most robust microarray cyclers.
Sequencing Cyclers.
The average reads per base pair within exons was used as an approximation of expression for the nascent and pA datasets. For each replicate dataset, a gene was classified as a high-ranking cycler if at least one time point had an average of 2 reads per base pair and if the gene had an F24 score ≥0.6 with a max/min ratio of at least 1.75. Alternatively, the gene was classified as a low-ranking cycler if the maximum average reads per base pair was less than 2 or the max/min ratio was at least 1.5. If at least one replicate was identified as a high-ranking cycler and the other a low-ranking cycler, the transcript was identified as a cycler across the two replicate datasets if the correlation between the two replicate six-time-point data were ≥0.6, the F24 of the joint data points was >0.4, and the P value of the joint data points was <0.05. P values were calculated by performing 100,000 permutations of the 12 Nascent-Seq or RNA-Seq average reads per base pair within gene exons, calculating the F24 scores on the permutated data, and then determining the number of times the permutated F24 was greater than or equal to the actual F24. Last, the classification of cyclers into groups I–IV was done as follows: group I and II comprise only genes classified as high-ranking nascent cyclers. The average of the replicate time points within the Nascent-Seq or RNA-Seq datasets was calculated to determine the correlation between the datasets. Only correlations ≥0.8 were classified as group I. Group II comprises the remaining high-ranking nascent cyclers that did not satisfy the thresholds for group I. Group III comprises the high-ranking pA cyclers with Nascent-Seq and RNA-Seq correlations ≥0.8 as defined above. Group IV consists of the remaining high-ranking nascent cyclers.
Heatmaps.
Individual nascent and mRNA heatmaps were created using the R statistical software package. Heatmaps of groups I–IV were sorted by group and then by nascent phase for groups I and II and by mRNA phase for groups III and IV.
Venn Diagrams.
A Venn diagram was made using R and then PowerPoint. A χ2 test and Fisher’s exact test (two sided) was used to determine significance of overlap between groups.
Sequencing Alignment.
Nascent RNA libraries were aligned to the dm3 Drosophila melanogaster genome with TopHat (25) using the parameters “ -m 1 -F 0 –microexon-search –no-closure-search -G exon20110421.gtf –solexa1.3-quals -I 50000.” Cs paired-end samples were aligned with TopHat using the parameters “-r 50 -m 1 -F 0 -I 50000 -g 1 -G exon20110421.gtf,” and reads with overlapping ends were removed and remapped. Cs single-end samples were aligned with TopHat using the parameters “-m 1 -F 0 -g 1 –microexon-search –no-closuresearch -G exon20110421.gtf –solexa1.3-quals -I 50000.” We used annotation from the University of California, Santa Cruz Genes and Gene Prediction group, flybaseGene Track/table Apr. 2006 assembly, to aid TopHat in the alignment of splice junctions (exon20110421.gtf).
Sequencing Visualizations.
Sequencing visualizations were created with the Integrated Genome Browser using the Nascent-Seq Replicate 1 and RNA-Seq Replicate 2 sets. Visualization tracks were normalized by the number of uniquely mapped reads.
Supplementary Material
Acknowledgments
We thank Katharine Abruzzi for helpful comments. This research was supported by the Howard Hughes Medical Institute. J.R. was supported in part by Integrative Graduate Education and Research Traineeship Grant DGE 0549390 from the National Science Foundation and by Genetics Training Grant 5T32GM007122 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219969110/-/DCSupplemental.
References
- 1.Kula-Eversole E, et al. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci USA. 2010;107(30):13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 5.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 6.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107(5):567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 7.Claridge-Chang A, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32(4):657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 8.Abruzzi KC, et al. Drosophila CLOCK target gene characterization: Implications for circadian tissue-specific gene expression. Genes Dev. 2011;25(22):2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darlington TK, et al. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280(5 June 1998):1599–1603.
- 10.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343(6258):536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 11.Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: From on-DNA to off-DNA. Genes Dev. 2010;24(4):358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15(17):R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116(4):603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- 14.Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21(12):1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim EY, et al. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34(1):69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- 16.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22(13):1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145(3):357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307(5707):251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 19.Price JL, et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94(1):83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 20.Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6(5):e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khodor YL, et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25(23):2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez J, Menet JS, Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol Cell. 2012;47(1):27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14(11):7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40(4):571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99(6):661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 27.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2(3):e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda HR, et al. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277(16):14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 29.Ceriani MF, et al. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosc. 2002;22(21):9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89(24):11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16(23):7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sehgal A, et al. Rhythmic expression of timeless: A basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 33.Cyran SA, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112(3):329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 34.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93(5):791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 35.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22(7):1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-seq reveals novel features of mouse circadian transcriptional regulation. elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobshagen S, Kessler B, Rinehart CA. At least four distinct circadian regulatory mechanisms are required for all phases of rhythms in mRNA amount. J Biol Rhythms. 2008;23(6):511–524. doi: 10.1177/0748730408325753. [DOI] [PubMed] [Google Scholar]
- 39.Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63(6):1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- 40.Wuarin J, et al. The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci Suppl. 1992;16:123–127. doi: 10.1242/jcs.1992.supplement_16.15. [DOI] [PubMed] [Google Scholar]
- 41.Kadener S, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23(18):2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276(19):5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 43.Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltage-gated calcium channel alpha1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284(38):25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE. 2010;5(6):e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138(6):1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo KC, et al. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37(1):26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo KC, et al. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol. 2010;30(1):197–205. doi: 10.1128/MCB.01154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE. 2007;2(10):e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kojima S, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA. 2007;104(6):1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherji S, et al. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43(9):854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wijnen H, Naef F, Young MW. Molecular and statistical tools for circadian transcript profiling. Methods Enzymol. 2005;393:341–365. doi: 10.1016/S0076-6879(05)93015-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.