Abstract
Activation of STAT3 in cancers leads to gene expression promoting cell proliferation and resistance to apoptosis, as well as tumor angiogenesis, invasion, and migration. In the characterization of effects of ST3-H2A2, a selective inhibitor of the STAT3 N-terminal domain (ND), we observed that the compound induced apoptotic death in cancer cells associated with robust activation of proapoptotic genes. Using ChIP and tiling human promoter arrays, we found that activation of gene expression in response to ST3-H2A2 is accompanied by altered STAT3 chromatin binding. Using inhibitors of STAT3 phosphorylation and a dominant-negative STAT3 mutant, we found that the unphosphorylated form of STAT3 binds to regulatory regions of proapoptotic genes and prevents their expression in tumor cells but not normal cells. siRNA knockdown confirmed the effects of ST3-HA2A on gene expression and chromatin binding to be STAT3 dependent. The STAT3-binding region of the C/EBP-homologous protein (CHOP) promoter was found to be localized in DNaseI hypersensitive site of chromatin in cancer cells but not in nontransformed cells, suggesting that STAT3 binding and suppressive action can be chromatin structure dependent. These data demonstrate a suppressive role for the STAT3 ND in the regulation of proapoptotic gene expression in cancer cells, providing further support for targeting STAT3 ND for cancer therapy.
Keywords: H3K9me3, peptide inhibitor, prostate cancer, transcription factor
STAT3, a member of the STAT family, is a key signaling protein that transduces extracellular signals to the nucleus and regulates transcription of genes (1). Following ligand stimulation, STAT3 is phosphorylated on Y705 tyrosine residue, dimerizes, and translocates to the nucleus to bind its cognate DNA-response elements, activating gene transcription (1). Constitutively activated STAT3 mediates deregulated growth, survival, and angiogenesis (2, 3). STAT3 is widely recognized as a potential drug target for cancer therapy, and various approaches, including targeting of upstream tyrosine kinases and direct inhibitors of STAT3 dimerization, have been advanced to inhibit STAT3 signaling in cancers (4). However, unphosphorylated STAT3 (U-STAT3) has also been shown to influence gene transcription, both in response to cytokines and in cancer cells, albeit by mechanisms that are distinct from those activated by phosphorylated STAT3 (5). We have developed a highly selective inhibitor of STAT3 ND, ST3-Hel2A-2 (ST3-H2A2), that binds to the N-terminal domain (ND) and inhibits STAT3 signaling (6). STAT3 ND is involved in the interactions of two STAT dimers on neighboring sites to form a more stable tetramer and the interactions with histone-modifier proteins to induce changes in chromatin structure (reviewed in ref. 7). These complex interactions may greatly affect STAT3-dependent transcriptional activity, suggesting that the STAT3 ND mediates important regulatory functions of STAT3 in normal cells (8) and in cancer (9). ST3-H2A2 induces death in breast cancer cells MDA-MB-231 and MCF-7 but does not affect survival of normal mammary epithelial cells MCF-10A (6), suggesting that ND is an important mediator of STAT3-dependent malignant progression. However, the cellular mechanisms of the STAT3 ND inhibitor action remain to be resolved. This study focused on the cell death mechanisms triggered by targeting the STAT3 ND. These data demonstrate that inhibition of the STAT3 ND activates expression of proapoptotic genes and initiates apoptotic death in cancer cells, revealing a previously underappreciated role of the STAT3 ND in the suppression of proapoptotic gene expression.
Results
ST3-H2A2 Induces Apoptotic Death in Prostate Cancer Cells.
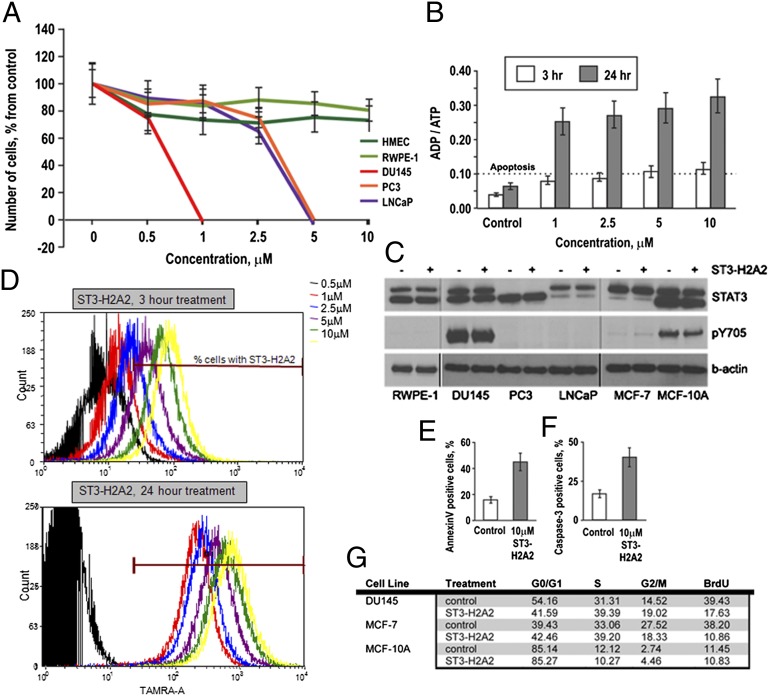
Treatment of androgen-dependent LNCaP and androgen-independent PC3 and DU145 prostate cancer cells with ST3-H2A2 results in dose-dependent growth inhibition and death of prostate cancer cells, whereas growth and viability of normal prostate epithelial cells RWPE-1 and human mammary epithelial cells (HMEC) are not altered (Fig. 1A). Scrambled peptides used as a negative control had no effect on cell growth or survival. ADP/ATP assay demonstrated that STAT3 ND inhibition triggers apoptotic death in prostate cancer cells (Fig. 1B). Although all studied cell lines express similar levels of STAT3 protein, only cancer cells are affected by STAT3 ND inhibition (Fig. 1C). The effect of ST3-H2A2 on survival of PC3 and LNCaP is consistent with Yang and Stark’s proposition that unphosphorylated STAT3 has an important role in oncogenesis (5).
Fig. 1.
STAT3 ND inhibitors induce apoptotic cell death in prostate cancer cells. (A) ST3-H2A2 toxicity in prostate cancer and normal epithelial cells. The cells were exposed to the compound for 48 h, and cell number was evaluated by MTT assay. (B) ADP/ATP ratios (>0.1) demonstrate that ST3-H2A2 triggers apoptotic death in DU145 cells. (C) Levels of tyrosine-phosphorylated (pY705) and total STAT3 in nontransformed RWPE-1, MCF-10A cells, and DU145, PC3, LNCaP, and MCF-7 cancer cells detected by Western blotting with antibodies against phospho-STAT3 Y705 and total STAT3 (C-20). (D) DU145 cells were treated with various doses of ST3-H2A2 labeled with tetramethyl rhodamine (TAMRA) for 3 or 24 h and analyzed by flow cytometry. When cells are treated with 10 μM ST3-H2A2 for 3 h; 93.6% cells contain the inhibitor. However, the intracellular concentration of ST3-H2A2 at this time point is about 2.5-fold lower than the concentration of the inhibitor when cells are treated with 1 μM ST3-H2A2 for 24 h. (E) AnexinV-PE binding is increased in DU145 cells after treatment with 10 μM ST3-H2A2 as detected by flow cytometry analysis. (F) 10 μM ST3-H2A2 induces caspase-3 cleavage in DU145 cells as detected by staining with rabbit anti-active caspase-3 antibody (BD Pharminger) and flow cytometry analysis. (G) Cell cycle analysis by flow cytometry and BrdU incorporation assays show 10 μM ST3-H2A2–triggered S-phase arrest in DU145 and MCF-7 cells after 3 h of exposure, but not in nontransformed MCF-10A.
Flow cytometry analysis of DU145 cells revealed accumulation of the tetramethylrhodamin (TAMRA)-labeled inhibitor in 93.6% cells after 3-h exposure to 10 μM ST3-H2A2-TAMRA (Fig. 1D). Therefore, cells were exposed to 10 μM ST3-H2A2 for 3 h for analysis in all cell-based and biochemical assays. AnnexinV binding and caspase-3 activation assays confirmed apoptotic cell death (Fig. 1 E and F). ST3-H2A2 induced cell cycle arrest in S- and G2/M-phases in cancer DU145 and MCF-7 cells but not in nontransformed MCF-10A cells as determined by cell cycle and BrdU incorporation assays (Fig. 1G).
To further confirm direct inhibitor binding to the STAT3 ND, we performed Förster resonance energy transfer (FRET) (6). HEK293 cells, stably expressing eGFP-tagged full-length STAT3 (eGFP-STAT3), the STAT3 ND (eGFP-STAT3-ND), or STAT3 ND-deletion mutant (eGFP-STAT3-ΔND) were treated with ST3-H2A2-TAMRA. The acceptor photobleaching demonstrated that, whereas ST3-H2A2 efficiently binds to eGFP-STAT3 and eGFP-STAT3-ND [normalized FRET efficiency (EFn) is 55.83 ± 2.00% and 54.91 ± 2.38%, respectively; t test, P = 0.0035 and P = 0.0039, respectively; Fig. S1], its binding to eGFP-STAT3-ΔND does not differ significantly from eGFP (t test, P = 0.103), suggesting preferential binding to the STAT3 ND. Microscale thermophoresis performed on lysates of STAT3-eGFP–expressing HEK293 cells provided additional proof of selective binding of the inhibitor (10). ST3-H2A2 binding resulted in significant changes in eGFP-STAT3 mobility in the temperature gradient and demonstrated an apparent dissociation constant of 7.95 ± 0.4 μM (Fig. S1). Because the inhibitor has to compete for the interaction with other protein partners of STAT3 present in the lysate, the apparent affinity can be lower than the actual affinity (11). No binding to GFP-STAT1 could be detected, further confirming inhibitor selectivity.
Inhibition of the STAT3 ND Induces Expression of Proapoptotic Genes.
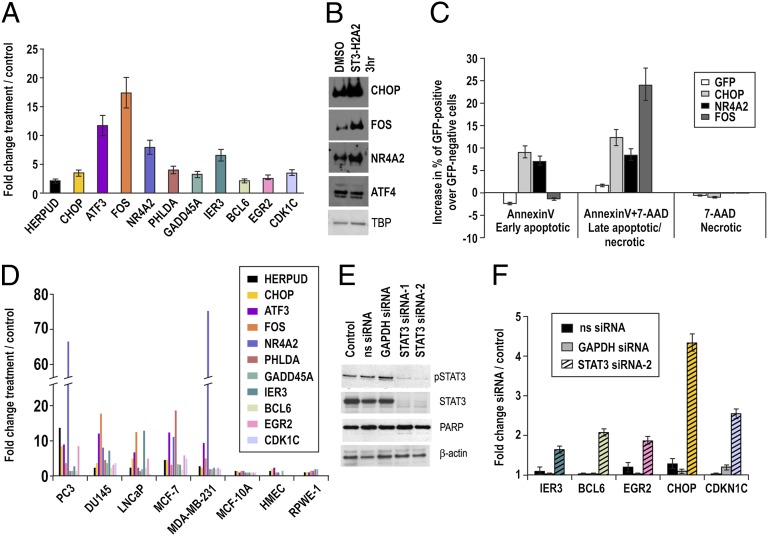
Exposure to ST3-H2A2 resulted in up-regulation of 147 genes and down-regulation of 11 genes compared with a control peptide (Table S1; GSE25866). qRT-PCR and Western blot analysis confirmed up-regulation of mRNA and protein levels (Fig. 2 A and B). Functional annotation clustering using DAVID software (12) showed that 17 of the up-regulated genes function as activators of apoptosis (Table S2). It has been shown that these genes induce growth arrest and apoptosis in cancer cells (13–15). Using AnnexinV binding assays and flow cytometry analysis of DU145 cells transfected with GFP-tagged GFP-tagged C/EBP-homologous protein (CHOP), FBJ murine osteosarcoma viral (v-fos) oncogene homolog (FOS) and nuclear receptor subfamily 4, group A, member 2 (NR4A2), we confirmed that overexpression of these proteins induced apoptosis in DU145 cells, whereas GFP alone did not affect cell survival (Fig. 2C; Fig. S2A). The loss-of-function experiments by knocking down of only one protein at a time (data for CHOP are shown in Fig. S2B) did not rescue ST3-H2A2–treated cells from death, suggesting that any single gene was not able to overcome redundancy of other genes in triggering apoptosis. Overall, our data suggest that induction of robust cell death is the result of the simultaneous up-regulation of multiple proapoptotic genes in DU145 prostate cancer cells. It is remarkable that the same genes were also induced by ST3-H2A2 in other cancer cells (PC3, LNCaP, MCF-7, MDA-MB-231) but not in the normal epithelial cells MCF-10A, HMEC, and RWPE-1 (Fig. 2D).
Fig. 2.
STAT3 ND inhibitor and STAT3 siRNA activate expression of proapoptotic genes. (A) qRT-PCR detected increased mRNA levels of selected genes in DU145 cells after 3-h exposure to ST3-H2A2. Expression levels were normalized using 18S rRNA levels. Expression levels in untreated cells were used as a baseline. Columns represent fold change; error bars indicate ±SD. (B) Protein levels of CHOP, NR4A2, and FOS were increased in response to ST3-H2A2 as demonstrated by Western blot analysis. In comparison, expression of ATF4 was not affected by ST3-H2A2 treatment. Levels of TBP (TATA binding protein) were used as loading controls. (C) Overexpression of GFP-CHOP, GFP-NR4A2, or GFP-FOS induces apoptosis in DU145 cells in 24 h as detected by AnnexinV-PE binding and 7-AAD assays using flow cytometry. Columns represent increase in percent of AnnexinV-PE and 7-AAD labeling in GFP-positive cells over GFP-negative cells; error bars indicate ±SD. (D) ST3-H2A2 increased expression levels of the selected genes in prostate and breast cancer cells, but not in normal epithelial cells as determined by qRT-PCR. (E) Both pSTAT3 and total STAT3 proteins levels were decreased after siRNA knockdown in DU145 prostate cancer cells as detected by Western blotting. Nonspecific scrambled siRNA (ns siRNA) was used as negative control; GAPDH siRNA was used as a positive control. Decreases in pSTAT3 levels were accompanied by the appearance of a weak band for cleaved PARP, which is an indicator of apoptosis. Membranes were reblotted with β-actin antibody for control of protein loading. (F) qRT-PCR validation of an increase in mRNA levels of selected genes after siRNA knockdown in DU145 cells.
Peptide inhibitors tend to be more selective than small molecules because they form multipoint interactions with extended surfaces (16). However, STAT3 ND is 72% homologous to its closest analog, STAT1 ND. To exclude the cross action, we performed a microarray analysis with the STAT1 ND inhibitor ST1-H2A2 (6). Gene expression changes induced by ST3-H2A2 and ST1-H2A2 differed profoundly (Table S3), further supporting high selectivity of the ST3-H2A2 inhibitor and underlining significant differences in the ND functions of the two members of STAT family. STAT3 siRNA knockdown (Fig. 2E) followed by RT-PCR confirmed that ST3-H2A2-up-regulated genes were also up-regulated by STAT3 siRNA (Fig. 2F).
Inhibition of the ND Decreases STAT3 Binding to Chromatin.
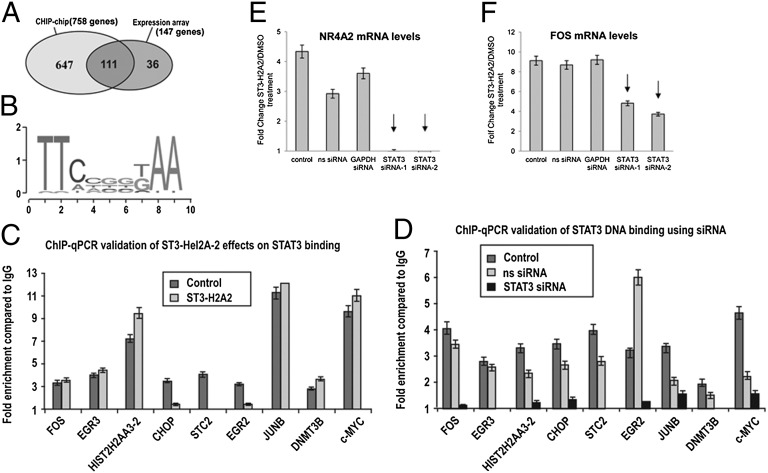
To determine whether up-regulated genes are direct STAT3 targets, we performed ChIP using anti-STAT3 antibody coupled with hybridizations to the tiling human promoter array (ChIP-chip) assays, followed by PARTEK model–based analysis (GSE25943). We found 758 genes to be bound by STAT3 in untreated DU145 cells and 286 STAT3-bound genes after a 3-h exposure to ST3-H2A2, with P < 0.001 and MAT score > 5. Comparison of STAT3 binding with gene expression changes shows that 111 of 147 up-regulated genes (75.5%) are bound by STAT3 (Fig. 3A). CisGenome Browser analysis identified the previously reported STAT3 consensus-binding motif (17) in the regulatory regions of STAT3-bound genes (Fig. 3B). STAT3 siRNA knockdown resulted in abrogation of STAT3 binding to identified DNA regions as determined by qPCR confirming specificity of the ChIP assays (Fig. 3C). These data indicate that genes up-regulated by ST3-H2A2 are direct STAT3 targets. ChIP-chip and ChIP-qPCR data revealed that ST3-H2A2 decreases STAT3 DNA binding to regulatory regions of CHOP, EGR2, and STC2 but does not affect binding to FOS, EGR3, JUNB, DNMT3B, or c-MYC (Fig. 3D), suggesting different STAT3 binding modes. Further support for this interpretation was provided by DNA affinity precipitation assays (DAPAs) demonstrating STAT3 binding to STAT consensus sites within identified genomic regions in vitro and by the observation that ST3-H2A2 abrogates STAT3 binding to some, but not all, DNA sequences (Fig. S3 A–C). These data correlate with the observed difference in the degree of effects of the STAT3 siRNA and ST3-H2A2 on gene expression. For example, CHOP induction levels are similar (4.4- and 5.6-fold change, respectively), whereas significant differences were observed for the genes such as FOS (1.7- and 21.4-fold change) and NR4A2 (1.5- and 9.2-fold change; Table S4). ChIP-chip data suggest that effects of ST3-H2A2 and siRNA on CHOP expression are similar because both treatments significantly decrease STAT3 DNA binding (Fig. 3 C and D; Fig. S3B). However, in case of FOS and NR4A2, STAT3 is depleted from the promoters by STAT3 siRNA only but remains bound after treatment with ST3-H2A2 (Fig. 3 C and D; Fig. S3B). To investigate whether activation of gene expression by ST3-H2A2 occurs because of effects on STAT3 rather than off-target effects, we knocked down the levels of STAT3 using siRNA, treated cells with ST3-H2A2, and measured mRNA levels by qRT-PCR (Fig. 3 E and F). Data presented for FOS and NR4A2 mRNAs demonstrate that knockdown of STAT3 levels prevented a ST3-H2A2–induced increase in gene expression, thus confirming that ST3-H2A2 effects on transcriptional activation are STAT3 dependent.
Fig. 3.
STAT3 binds to genes up-regulated in response to STAT3 ND inhibitor. (A) ChIP-chip assay defined 758 genes bound by STAT3 with P ≤ 0.001 and MAT score ≥ 5. Expression analysis showed 147 genes up-regulated on exposure to ST3-H2A2. The 111 up-regulated genes were bound by STAT3. (B) STAT3-bound sequences were enriched in the GAS consensus motifs. (C and D) qPCR-ChIP assays confirmed STAT3 binding to the genomic sequences identified by ChIP-chip assays. STAT3 binding was decreased following siRNA STAT3 knockdown (C) or treatment with ST3-H2A2 (D). (E and F) STAT3 knockdown diminishes the effects of ST3-H2A2 on transcriptional activation. DU145 cells were plated in six-well plates and transfected with nonspecific (ns siRNA), GAPDH siRNA, or STAT3 siRNA-1 and -2 using Trans-TKO reagent: 48 h after transfection, cells were treated either with ST3-H2A2 or DMSO for 3 h. Total RNA was extracted, and qRT-PCR using Taqman assays was performed to measure expression of FOS and NR4A2 mRNAs. Amplification of 18S rRNA was used as an endogenous control to standardize the amount of sample added to the reaction.
Unphosphorylated Form of STAT3 Binds to Regulatory DNA Regions of Proapoptotic Genes.
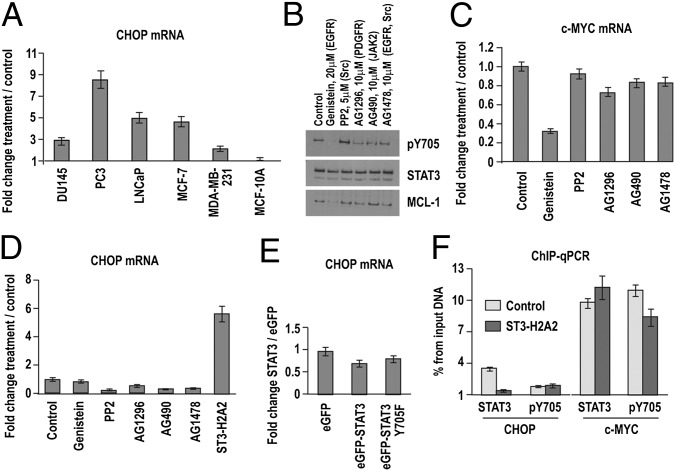
Expression of proapoptotic genes (e.g., CHOP) was induced in cancer cells with (DU145, MDA-MB-231) and without (LNCaP, PC3, and MCF-7) detectable levels of phosphorylated STAT3 (pSTAT3) (Fig. 4A). To evaluate the effects of inhibition of STAT3 phosphorylation on CHOP expression, we treated DU145 cells with the tyrosine kinase inhibitor Genistein. Decreased levels of pSTAT3 caused a drop in the expression of the known transcriptional targets MCL-1 and c-MYC (Fig. 4 B and C) but did not induce CHOP expression (Fig. 4D). Overexpression of the dominant-negative STAT3 Y705F mutant also did not change CHOP expression (Fig. 4E), suggesting that unphosphorylated STAT3 (U-STAT3) is responsible for proapoptotic gene suppression of this gene. ChIP assays demonstrated that only antibodies against total STAT3, but not pSTAT3, pulled down regulatory DNA regions of CHOP in the DU145 cells (Fig. 4F). For comparison, c-MYC promoter DNA was pulled down by antibodies against total STAT3 and pSTAT3 at similar levels (Fig. 4F). Because the antibody for total STAT3 recognizes pSTAT3 as well, the result suggests that, in the case of c-MYC, its promoter is preferentially bound by pSTAT3 (Fig. 4F).
Fig. 4.
STAT3 phosphorylation is not required for binding to the regulatory DNA regions. (A) ST3-H2A2 increased CHOP mRNA levels in cancer cells as detected by qRT-PCR. Genistein decreased levels of pSTAT3 and MCL-1 as demonstrated by Western blot (B) and c-MYC mRNA levels as demonstrated by qRT-PCR (C). qRT-PCR proved that Genistein (D) or dominant-negative mutant STAT3 pY705F (E) do not increase levels of CHOP mRNA. Expression levels were normalized using 18S rRNA levels. Expression levels in untreated cells were used as a baseline. Columns represent fold change; error bars indicate ±SD. (E) ChIP-qPCR with total (STAT3) and pSTAT3 (pY705) antibody shows that only antibody against total STAT3 precipitates CHOP DNA regions. However, both antibodies precipitate the c-MYC promoter. Columns represent fold enrichment over IgG antibody used as a negative control; error bars indicate ±SD.
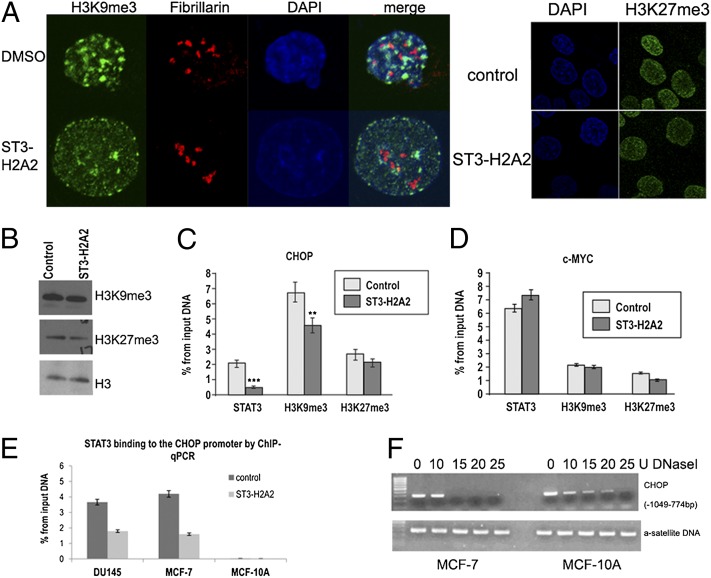
Recent reports by Shi et al. have demonstrated that the unphosphorylated STAT92E protein is important for heterochromatin maintenance through regulation of histone H3 Lys-9 trimethylation (H3K9me3) in Drosophila (18, 19). We observed that inhibition of STAT3 ND significantly reduced the number of loci with intense H3K9me3 staining in the nucleus (Fig. 5A) without a decrease in the total levels of H3K9me3 (Fig. 5B). ChIP-qPCR revealed that a decrease in STAT3 binding was accompanied by a modest but statistically significant decrease of H3K9me3 at the CHOP promoter region (Fig. 5C). Importantly, the levels of H3K9me3 on the c-MYC promoter were lower compared with that observed on the CHOP promoter in DU145 cells, and these levels were not further decreased following exposure to ST3-H2A2 (Fig. 5D). For comparison, localization and levels of H3K27me3, another chromatin modification associated with gene repression, were not affected by ST3-H2A2 (Fig. 5 A and C). ST3-H2A2 did affect H3K9me3 marks not only in cancerous cells but also in nontransformed MCF-10A cells (Fig. S4). No changes in heterochromatic marker H3K27me3, a nucleoli marker fibrillarin, or an active chromatin marker phospho-PolII have been detected, thus suggesting that effects on H3K9me3 are specific. Although ST3-H2A2 affected H3K9me3 in both normal and tumor cells, the ChIP-qPCR assay revealed that STAT3 did not bind to the CHOP promoter in nontransformed MCF-10A cells (Fig. 5E). We hypothesized that differences in STAT3′s effects on CHOP expression in normal and cancer cells were a result of the chromatin organization of this region. DNaseI treatment of the nuclei from DU145, MCF-7, and MCF-10A followed by PCR amplification of the CHOP genomic region containing the STAT3 binding site (Fig. 5F) has indeed demonstrated that the CHOP genomic region has DNaseI hypersensitive sites (DHS) in cancer cells but not in MCF10A cells. The data suggest an open, or at least more accessible, chromatin conformation in DU145 and MCF-7 cells allowing for STAT3 binding.
Fig. 5.
Effects of STAT3 ND inhibition on chromatin markers H3K9me3 and H3K27me3 in cancer and normal cells. (A) Laser scanning confocal microscopy reveals that ST3-H2A2 decreases the number of loci with high density of H3K9me3, but has no effect on H3K27me3 localization in the nuclei of DU145 cells. DNA was stained with DAPI. Nucleoli were visualized using antibodies against fibrillarin, a nucleoli marker. (B) Total levels of H3K9me3 and H3K27me3 were not changed by treatment with ST3-H2A2 as determined by Western blotting. The membranes were reblotted with histone H3 antibody for loading control. (C) Decreased STAT3 binding to the CHOP gene following the treatment with ST3-H2A2 was accompanied by partial demethylation of H3K9me3, but not H3K27me3, as demonstrated by ChIP-qPCR. IgG antibodies were used as a negative control; histone H3 antibodies were used as a positive control. (D) The levels of H3K9me3 were not affected by the c-MYC promoter. **P < 0.01, ***P < 0.005. (E) STAT3 binds CHOP promoter only in cancer DU145 and MCF-7 cells, but not in nontransformed MCF-10A cells as revealed by ChIP-qPCR. (F) Nuclei treatment with various doses of DNaseI followed by PCR with primers flanking STAT3 binding site within CHOP promoter (−1,049 to −774 bp) demonstrates that the CHOP promoter is localized in accessible chromatin in MCF-7 cells but in more condensed chromatin in MCF-10A cells. For comparison, a satellite DNA is found localized in condensed chromatin in both cell lines.
Discussion
STAT3 has been viewed as an activator of gene expression that drives tumorigenesis by increasing expression of prosurvival and proinflammatory genes (20, 21). However, several studies have described the suppressive effects of STAT3 on the expression of tumor suppressor genes, suggesting that additional mechanisms exist by which STAT3 promotes cancer cell survival and suppresses apoptosis (22–24). A search for genetic suppressive elements (GSEs) in breast cancer cells convincingly identified the NDs of STAT3 and STAT5 as major factors responsible for driving cancer cells proliferation and survival (9). Inhibition of the STAT3 ND resulted in breast cancer cell death (6). Activation of 17 proapoptotic genes, including CHOP, ERG2, NR4A2, PHLDA, and GADD45a, in response to the STAT3 ND inhibition explains the cytotoxic effects of ND inhibitor in cancer cells. Although the STAT NDs have been previously implicated in gene activation in response to cytokine signals, recent findings demonstrate a suppressive function for the STAT5 ND in leukemogenesis (25, 26). Similar to STAT3 ND, a role for the STAT5 ND has been proposed in repression of transcription, possibly through interactions with transcriptional repressors and via chromatin remodeling (25–27). Another study demonstrated that the repressive function of STAT5 correlates with the recruitment of Ezh2, leading to H3K27 trimethylation (28). Notably, the repressive function of STAT5 requires tetramer formation, and therefore its ND is necessary for Ezh2-mediated gene repression (28, 29). Our data suggest that STAT3 ND–dependent suppression occurs independently of tyrosine phosphorylation. Because tyrosine phosphorylation does not affect the ND structure, ST3-H2A2 is likely to bind to both phosphorylated and unphosphorylated forms of STAT3. The differential effects of the inhibitor on DNA binding can be caused by structural differences of dimers formed by P-STAT3 and U-STAT3. Structural data suggest that NDs do not interact within one P-STAT dimer (30, 31) but can form interactions either with another STAT dimer, leading to tetramer formation or with other transcription factors/regulators. Consequently, inhibition of the ND can affect interactions with other proteins but not phosphorylation-dependent STAT3 binding to DNA. A previous report confirmed that STAT3 ND is not essential for P-STAT3 functions in normal fibroblasts (32). Conversely, native gel electrophoresis, dual-focus fluorescence correlation spectroscopy (33), and FRET data suggest that dimerization of unphosphorylated STAT3 requires the ND (34). We recently showed that U-STAT3 is capable of binding to GAS DNA as a dimer (11). Therefore, the involvement of the ND in dimer stabilization and DNA binding of P-STAT3 and U-STAT3 differs dramatically.
The observed STAT3 ND–dependent suppression of proapoptotic genes correlates well with previously demonstrated involvement of the unphosphorylated STAT3 in regulation of gene expression and carcinogenic processes (5, 35). Reports by Yang et al. suggest an activating role for U-STAT3 but also include data consistent with a repressive function (35, 36). Interestingly, STAT3 ND inhibition had little, if any, effect on activation of proapoptotic gene expression in the nontransformed cells MCF-10A, HMEC, and RWPE-1 and therefore did not affect survival and proliferation of these cells. Chromatin accessibility analysis by DNaseI digestion revealed that the promoter of the CHOP gene resides in a more accessible chromatin in MCF-7 and DU145 cancer cells compared with MCF-10A cells. Recent studies from ENCODE-ChIP demonstrated that, on average, 98.5% of the occupancy sites of transcription factors are positioned within accessible chromatin defined by DNaseI hotspots (37). It is tempting to speculate that the more condensed conformation in nontransformed MCF-10A cells does not allow STAT3 binding and at the same time prevents transcription of CHOP and other genes through epigenetic mechanisms, whereas open conformation in cancer cells requires STAT3 to suppress CHOP expression. Our observations that STAT3 does not bind the more condensed CHOP promoter (in MCF-10A cells) are also in agreement with a recent report that STAT3 is recruited to already accessible chromatin sites in T-cell receptor-activated CD4+ T cells, but does not pioneer the access itself (38).
An open chromatin state of CHOP gene correlates with constitutively active endoplasmic reticulum stress response (ESR) in cancer cells (Fig. S5). Transcription factor ATF4, which regulates CHOP expression, is highly expressed in DU145 and MCF-7 cells but not in MCF-10A cells (Fig. S5). Because other proapoptotic genes detected in our study are also often activated during ESR (39), the data suggest that STAT3 ND suppression may be especially important for STAT3 functions under conditions of ESR in cancer cells. The identification of differences in epigenetic mechanisms that underlie differential activity of the STAT3 ND in normal and cancer cells can offer novel potential therapeutic targets for cancer treatment. The epigenetic mechanisms may also at least partially explain heterogeneity of gene expression profile changes observed in response to ST3-H2A2 in cancer cells. It also explains a heterogeneity in cellular responses previously observed in JAK-STAT signaling (40–42).
Although the CHOP promoter contains DNaseI hypersensitive sites in DU145 cells (Fig. S5), we found that it is enriched in the H3K9me3 histone mark. This chromatin mark has been associated with transcriptional repression but was also found in the promoters of transcriptionally active genes (43). The functional roles of H3K9me3 in the accessible chromatin have not been established, and it is not clear whether H3K9me3 is a normal counterpart in the transcription of all genes or whether it is aberrant readout of the histone code in cancer cells (43, 44). Changes in H3K9me3 may be involved in mediating activation of proapoptotic genes by STAT3 ND inhibition. However, the changes take place both in nontransformed and transformed cells. Consequently, other factors are likely to be contributing to selective activation in tumor cells. Taking into consideration the recently published report on STAT3 preferential binding to accessible chromatin (38), it appears likely that different groups of genes are targets for STAT3 binding in nontransformed and malignant cells.
In conclusion, our data demonstrate the STAT3 ND suppressive role in the regulation of gene expression in cancer cells. The study underscores the importance of cellular context, including activated signaling pathways and epigenetic mechanisms, for understanding and interpreting the functional roles of STAT3. Future profiling of chromatin accessibility and detailed characterization of histone code in the promoters of STAT3 target genes will add additional knowledge and understanding of the complex relationships between gene expression, chromatin organization, and STAT3 binding. The studies can offer opportunities for discovery of additional therapeutic targets for cancer treatment. Reprogramming, rather than completely inhibiting transcription factors with synthetic molecules exemplified by inhibitors of the STAT3 ND, presents a unique paradigm and a therapeutic approach that may provide a powerful therapeutic tool for the treatment of many pathological conditions.
Methods
Reagents.
RPMI 1640, DMEM/F12, FBS, and antibiotic-antimycotic solution were from Life Sciences. Antibodies against STAT3 and p-STAT3 were from Cell Signaling Technology. Antibodies against H3K9me3, NR4A2, and β-actin were from Abcam. Antibodies against c-FOS and CHOP were from BioLegend. Goat–anti-rabbit and anti-mouse HRP conjugates were from Jackson ImmunoResearch Laboratories. All chemicals were obtained from Sigma-Aldrich. The ST3-H2A2 inhibitor has been synthesized and purified as described (6). Cancer cell lines were obtained from American Type Culture Collection and were maintained as recommended.
Western Blotting.
Nuclear extracts were prepared using a kit from Millipore as recommended by the manufacturer. The histones were extracted using the EpiQuik Total Histone Extraction Kit (Epigentek). Western blotting was performed as described (6, 45).
Toxicity assays.
MTT (3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay were performed as described (6). ATP/ADP levels were measured using a kit from BioVision as recommended by the manufacturer.
Apoptosis Detection.
Cell cycle analysis and AnnexinV-PE and 7-AAD assays were performed as described (45). Activation of caspase-3 was detected using an antibody against the active caspase-3 isoform from BD Pharminger as recommended by the manufacturer.
Confocal Laser Scanning Microscopy.
DU145 cells were stained with antibodies against H3K9me3 (1:3,000; Abcam), fibrillarin (1:500; Abcam), p-PolII (1:200; Abcam), KAP-1 (1:500; Abcam), and H3K27me3 (1:250; Cell Signaling Technology) according to the manufacturers’ recommendations. DNA was stained with 2 μg/mL DAPI. Cells were observed using a Zeiss LSM 510 META confocal microscope (Carl Zeiss).
STAT3 Knockdown Using STAT3 siRNA.
DAPAs.
DAPA was performed as described (11).
Affymetrix Microarray Analysis and qRT-PCR.
Affymetrix expression microarray analysis was performed as described (46). Differentially expressed genes were identified based on a cutoff of false discovery rate (FDR) < 10% using Partek software 6.5 (www.partek.com). qRT-PCR was performed in triplicate as described (46).
ChIP-Chip and ChIP-qPCR.
ChIP-chip assays were performed using a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology). ChIP-enriched DNA replicates were amplified, labeled, and hybridized to Affymetrix GeneChip Human Tiling Promoter 1.0 arrays. Peaks of probe intensity (hits) were identified and associated with RefSeq genes (hg18) using Partek software. DNA sequences of the ChIP hits were extracted using the CisGenome browser (www.biostat.jhsph.edu/~hji/cisgenome) (47). Primers for quantitative PCR were designed using the IDT website (www.idtdna.com).
DNaseI Hypersensitive Sites Detection.
Nuclei were prepared using a kit from Millipore. Aliquots of 4 × 105 nuclei were digested with 0, 10, 15, 20, or 25 U of DNaseI for 10 min at 37 °C. The reactions were terminated by the addition of 5 M NaCl, RNase A, and Proteinase K. DNA was extracted by phenol-chloroform purification. PCR reactions were carried out with primers for the CHOP promoter or a-satellite DNA using the FastStart DNA polymerase kit from Roche.
Supplementary Material
Acknowledgments
We thank Drs. Alan Perantoni and Steve Byers for critical review of the manuscript and valuable suggestions; Drs. Richard Schlegel and Xuefeng Liu for providing HMEC cells; and LCCC Flow Cytometry, Microscopy and Imaging, and Tissue Culture Shared Resources for excellent assistance. This research was supported by American Cancer Society Grant IRG 97-152-17, the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and National Cancer Institute Award P30CA051008.
Footnotes
Conflict of interest statement: O.A.T. and N.I.T. are coinventors on a patent application No.12/601,711 “Peptide-based STAT inhibitors,” filed February 5, 2010 by the National Institutes of Health for ST3-H2A2.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE25866).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211805110/-/DCSupplemental.
References
- 1.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19(21):2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 4.Yue P, Turkson J. Targeting STAT3 in cancer: How successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18(4):443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 6.Timofeeva OA, et al. Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS Chem Biol. 2007;2(12):799–809. doi: 10.1021/cb700186x. [DOI] [PubMed] [Google Scholar]
- 7.Timofeeva OA, Tarasova NI. Alternative ways of modulating JAK-STAT pathway: Looking beyond phosphorylation. JAK-STAT. 2012;1(4):274–284. doi: 10.4161/jkst.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19(21):2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 9.Primiano T, et al. Identification of potential anticancer drug targets through the selection of growth-inhibitory genetic suppressor elements. Cancer Cell. 2003;4(1):41–53. doi: 10.1016/s1535-6108(03)00169-7. [DOI] [PubMed] [Google Scholar]
- 10. Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1:100, 10.1038/ncomms1093. [DOI] [PubMed]
- 11.Timofeeva OA, et al. Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J Biol Chem. 2012;287(17):14192–14200. doi: 10.1074/jbc.M111.323899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Unoki M, Nakamura Y. EGR2 induces apoptosis in various cancer cell lines by direct transactivation of BNIP3L and BAK. Oncogene. 2003;22(14):2172–2185. doi: 10.1038/sj.onc.1206222. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez AM, et al. Induction of the endoplasmic reticulum stress protein GADD153/CHOP by capsaicin in prostate PC-3 cells: A microarray study. Biochem Biophys Res Commun. 2008;372(4):785–791. doi: 10.1016/j.bbrc.2008.05.138. [DOI] [PubMed] [Google Scholar]
- 15.Nagai MA, Fregnani JH, Netto MM, Brentani MM, Soares FA. Down-regulation of PHLDA1 gene expression is associated with breast cancer progression. Breast Cancer Res Treat. 2007;106(1):49–56. doi: 10.1007/s10549-006-9475-6. [DOI] [PubMed] [Google Scholar]
- 16.Cirillo D, Pentimalli F, Giordano A. Peptides or small molecules? Different approaches to develop more effective CDK inhibitors. Curr Med Chem. 2011;18(19):2854–2866. doi: 10.2174/092986711796150496. [DOI] [PubMed] [Google Scholar]
- 17.Ehret GB, et al. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276(9):6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 18.Shi S, et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38(9):1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi S, et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10(4):489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darnell JE. Validating Stat3 in cancer therapy. Nat Med. 2005;11(6):595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- 21.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu G, et al. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25(17):7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu G, et al. Overexpression of a dominant-negative signal transducer and activator of transcription 3 variant in tumor cells leads to production of soluble factors that induce apoptosis and cell cycle arrest. Cancer Res. 2001;61(8):3276–3280. [PubMed] [Google Scholar]
- 24.Zhang Q, et al. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci USA. 2005;102(19):6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, et al. STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output. Exp Hematol. 2007;35(11):1684–1694. doi: 10.1016/j.exphem.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriggl R, et al. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7(1):87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Li G, et al. STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood. 2010;115(7):1416–1424. doi: 10.1182/blood-2009-07-234963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal M, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol. 2011;12(12):1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrar MA, Harris LMH. Turning transcription on or off with STAT5: when more is less. Nat Immunol. 2011;12(12):1139–1140. doi: 10.1038/ni.2163. [DOI] [PubMed] [Google Scholar]
- 30.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93(5):827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, et al. IL-6 signaling via the STAT3/SOCS3 pathway: Functional analysis of the conserved STAT3 N-domain. Mol Cell Biochem. 2006;288(1-2):179–189. doi: 10.1007/s11010-006-9137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogt M, et al. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci. 2011;124(Pt 6):900–909. doi: 10.1242/jcs.072520. [DOI] [PubMed] [Google Scholar]
- 34.Cimica V, Chen HC, Iyer JK, Reich NC. Dynamics of the STAT3 transcription factor: Nuclear import dependent on Ran and importin-β1. PLoS ONE. 2011;6(5):e20188. doi: 10.1371/journal.pone.0020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65(3):939–947. [PubMed] [Google Scholar]
- 36.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21(11):1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joo JH, Liao G, Collins JB, Grissom SF, Jetten AM. Farnesol-induced apoptosis in human lung carcinoma cells is coupled to the endoplasmic reticulum stress response. Cancer Res. 2007;67(16):7929–7936. doi: 10.1158/0008-5472.CAN-07-0931. [DOI] [PubMed] [Google Scholar]
- 40.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ecker A, et al. The dark and the bright side of Stat3: Proto-oncogene and tumor-suppressor. Front Biosci. 2009;14:2944–2958. doi: 10.2741/3425. [DOI] [PubMed] [Google Scholar]
- 42.de la Iglesia N, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22(4):449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiencke JK, Zheng S, Morrison Z, Yeh RF. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2008;27(17):2412–2421. doi: 10.1038/sj.onc.1210895. [DOI] [PubMed] [Google Scholar]
- 44.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timofeeva OA, et al. Serine-phosphorylated STAT1 is a prosurvival factor in Wilms’ tumor pathogenesis. Oncogene. 2006;25(58):7555–7564. doi: 10.1038/sj.onc.1209742. [DOI] [PubMed] [Google Scholar]
- 46.Timofeeva OA, et al. Enhanced expression of SOS1 is detected in prostate cancer epithelial cells from African-American men. Int J Oncol. 2009;35(4):751–760. [PMC free article] [PubMed] [Google Scholar]
- 47.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.