Abstract
The Piwi protein subfamily is essential for Piwi-interacting RNA (piRNA) biogenesis, transposon silencing, and germ-line development, all of which have been proposed to require Piwi endonuclease activity, as validated for two cytoplasmic Piwi proteins in mice. However, recent evidence has led to questioning of the generality of this mechanism for the Piwi members that reside in the nucleus. Drosophila offers a distinct opportunity to study the function of nuclear Piwi proteins because, among three Drosophila Piwi proteins—called Piwi, Aubergine, and Argonaute 3—Piwi is the only member of this subfamily that is localized in the nucleus and expressed in both germ-line and somatic cells in the gonad, where it is responsible for piRNA biogenesis and regulatory functions essential for fertility. In this study, we demonstrate beyond doubt that the slicer activity of Piwi is not required for any known functions in vivo. We show that, in transgenic flies with the DDX catalytic triad of PIWI mutated, neither primary nor secondary piRNA biogenesis is detectably affected, transposons remain repressed, and fertility is normal. Our observations demonstrate that the mechanism of Piwi is independent of its in vitro endonuclease activity. Instead, it is consistent with the alternative mode of Piwi function as a molecule involved in the piRNA-directed guidance of epigenetic factors to chromatin.
Keywords: small RNA, RNase H fold, transposable element, sterility
The Piwi protein family plays a fundamental role in sexual reproduction. Not only are Piwi proteins highly enriched in the gonads of all animals studied to date, but their reproductive phenotypes have been revealed in diverse model organisms, such as Caenorhabditis elegans, Drosophila, zebrafish, and mice (1). Piwi proteins are paralogs of Argonaute proteins, sharing the same domain architecture and the property of binding small noncoding RNAs (2–6). Argonaute proteins bind to small interfering RNAs (siRNAs) or microRNAs (miRNAs) and are effectors of RNA interference (RNAi) and miRNA pathways; conversely, Piwi proteins bind to Piwi-interacting RNAs (piRNAs) and are involved in the piRNA pathway in both the germ line and soma during gametogenesis (1, 7, 8).
In Drosophila, there are three Piwi proteins—Piwi, Aubergine (Aub), and Argonaute 3 (Ago3) (9)—all of which have been shown to possess small RNA-dependent endonuclease (slicer) activity by in vitro assays (10, 11), catalyzed by an aspartate catalytic triad (DDX) that is positioned within the core of the RNase H-fold (12). This activity has been proposed to be essential for the amplification of piRNAs through a mechanism called the ping-pong cycle that occurs in the cytoplasm and uses cRNA targets, such as transposon transcripts, as precursors (13, 14). In addition, the slicer activity has been proposed play a role in silencing target transposons through cleaving their transcripts (13).
Among the three Piwi proteins in Drosophila, only Piwi is expressed in both germ-line and somatic cells in the gonad and is involved in both germ-line and somatic piRNA pathways (15, 16). Piwi is also distinct from the other members of the subfamily in terms of subcellular localization. We showed previously that Piwi is a nuclear protein that associates with an epigenetic factor, Heterochromatin Protein 1a, and binds to piRNA-complementary sequences in the genome and that Piwi deficiencies lead to classical epigenetic phenotypes in Drosophila (15, 17, 18). These observations led us to propose that piRNA molecules can guide Piwi to specific genomic loci by sequence complementarity, where Piwi recruits epigenetic modifiers (17–19). In this epigenetic model, the slicer activity of Piwi seems superfluous. Indeed, the involvement of the slicer activity of Piwi has been questioned recently based on experiments performed in an ovarian somatic cell line (20). However, this analysis was not definitive. It was based on a single cultured cell type, relied on RNAi that only incompletely reduced endogenous levels of Piwi and tested only one transposon element. Thus, these results could not predict the consequence of abolishing the slicer function in an organism. Therefore, we sought to test the role of the endonuclease activity of Piwi in all known biological processes that involve Piwi function, such as piRNA biogenesis, transposon silencing, germ-line development, and gametogenesis.
Results
Slicer Mutations Do Not Affect the Stability or Localization of Piwi in Vivo.
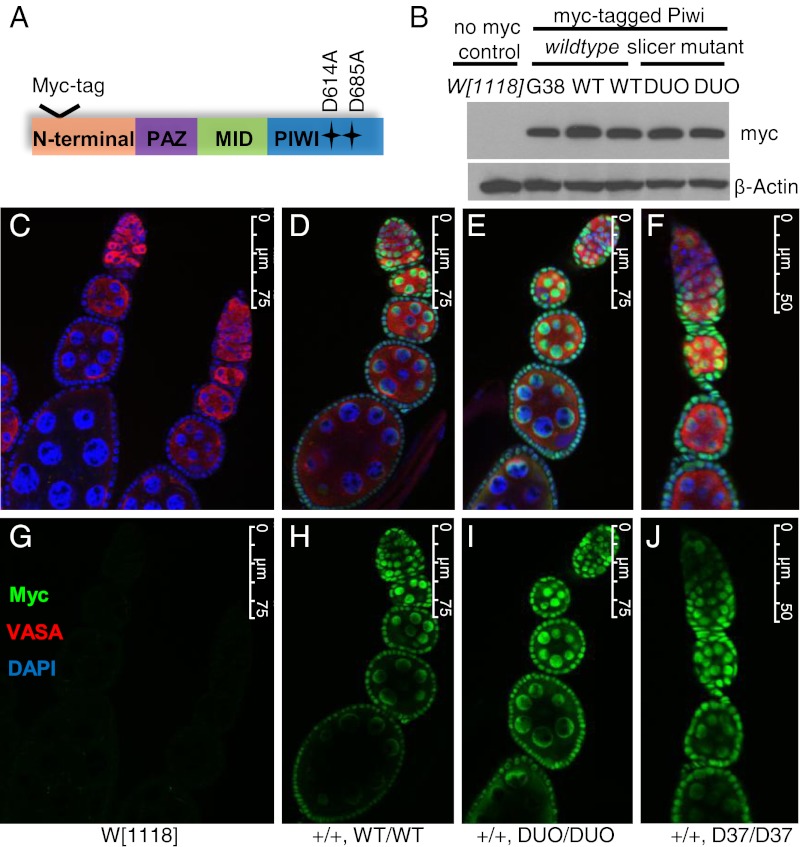
We replaced the two critical aspartate residues, D614 and D685, that form the catalytic triad with alanine, as reported in vitro (16) (Fig. 1A). We used FlyC31 site-specific recombination technology to introduce the slicer-deficient Myc–Piwi transgene (“DUO,” which means “two” in Latin, herein refers to two-point mutations) or a wild-type Myc–Piwi control transgene (WT) into a well-characterized genomic site called attP2 to generate transgenic Drosophila strains (21). The WT Myc–Piwi gene has been shown to be fully functional compared with the endogenous piwi gene (22). The insertion of the DUO and WT transgenes precisely at the same site in the genome eliminates potentially different influences of the surrounding genomic context toward the expression of the inserted transgenes. Additionally, we generated another DUO Myc–Piwi transgenic line by conventional P-element transformation, with a WT Myc–Piwi transgenic line (G38) previously generated the same way as controls (15). The expression levels of WT and DUO Myc–Piwi proteins in ovaries are very similar among all transgenic lines (Fig. 1B). Likewise, the localization pattern of slicer-deficient Myc–Piwi (green) both in the germ-line (red for Vasa, a germ-line marker) and in somatic cells was indistinguishable from that of WT Myc–Piwi (Fig. 1 C–J). Together, these results confirm that these two mutations have no effect on the stability or localization of Piwi in vivo.
Fig. 1.
Slicer-deficient Piwi shows a normal level of expression and pattern of localization. (A) Schematic representation of the domain architecture of PIWI, highlighting the two critical aspartate residues essential for endonuclease activity, which were replaced by alanine in the slicer-deficient transgene. (B) Western blot showing similar levels of WT and DUO Piwi proteins in ovaries. The wild-type nontransgenic line (W1118) is a negative control. Myc–Piwi transgenic line G38 was described (15). Protein levels of two independent site-specific transgenics are shown for wildtype Myc–Piwi at the attP2 site (WT) and Myc–Piwi slicer mutant (DUO). All transgenes are in the W1118;piwi+ background. (C–J) Immunofluorescence micrographs of ovarioles from transgenic flies. Myc (green) was costained with Vasa (red) and DAPI (blue). C and G show no Myc staining in Myc-negative W1118 ovaries. D and H show the normal localization of WT Myc–Piwi, whereas E and I show the normal localization of DUO Myc–PIWI (D614A, D685A). F and J show the localization of D37 Myc–PIWI (D614A, D685A).
None of the Essential Functions of Piwi in Somatic and Germ-Line Cells Requires its Slicer Activity.
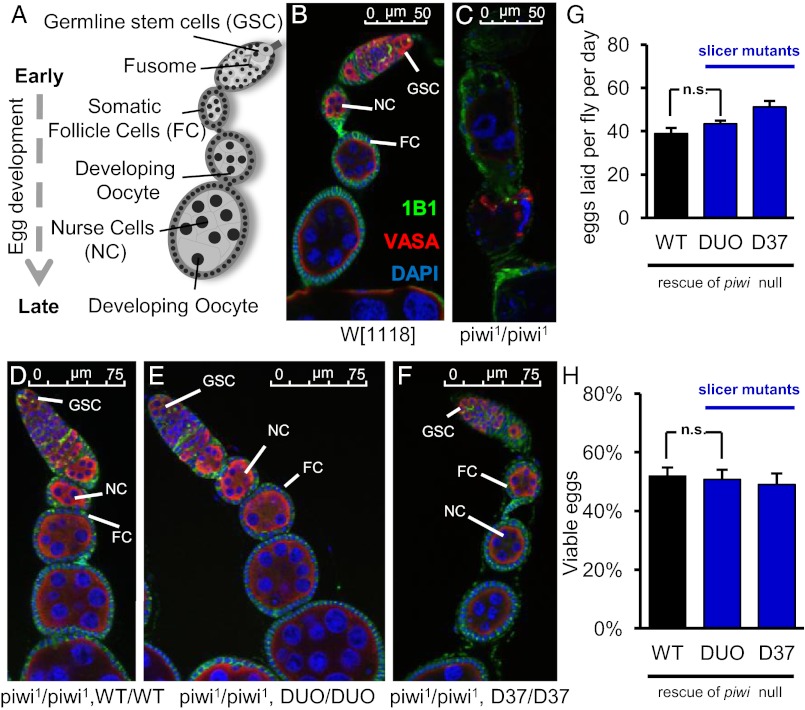
To assess the biological role of the slicer activity of Piwi, we crossed the DUO and WT transgenic lines into the homozygous piwi1 mutant background as outlined in Fig. S1. To our surprise, ovaries from the resulting piwi1/piwi1;DUO/DUO genotype are morphologically indistinguishable from piwi1/piwi1;WT/WT and wild-type flies (Fig. 2 A–F), as indicated by normal 1B1 staining (green) that outlines somatic cells and highlights spectrosomes and fusomes in the germ line as well as Vasa staining (red) that labels germ-line cells. We then quantified the fertility of the slicer-deficient flies by counting the number of eggs laid per fly per day. These flies laid as many eggs as the wild-type control (Fig. 2G). Furthermore, the progeny of the slicer-deficient flies were as viable as those of the wild-type control (Fig. 2H). In fact, we have been keeping piwi1/piwi1;DUO/DUO flies as a healthy stock since November 2009. Thus, the piwi1/piwi1;DUO/DUO flies without slicer activity have no detectable defect or reproductive disadvantage. These results indicate that all essential functions of Piwi in both somatic and germ-line cells are independent of its slicer activity.
Fig. 2.
Slicer-deficient piwi mutants display normal germ-line stem cell division, oogenesis, and viability. (A) Schematic diagram of a normal fly ovariole. (B–F) Immunofluorescence images of ovarioles dissected from W[1118] strain (B), piwi1/piwi1 mutant (C), piwi1/piwi1;WT/WT (D), piwi1/piwi1;DUO/DUO (E), and piwi1/piwi1;D37/D37 (F). Vasa staining (red) highlights germ-line cells. 1B1 staining (green) outlines somatic cells, spectrosomes, and fusomes. DAPI reveals nuclear morphology. (G and H) Average fertility (G) and viability (H) of transgenic rescue fly strains are graphed with SE indicated. piwi1 mutant flies fail to lay any egg due to ovarian defects as shown in C.
Slicer Activity of Piwi Is Not Required for piRNA Biogenesis.
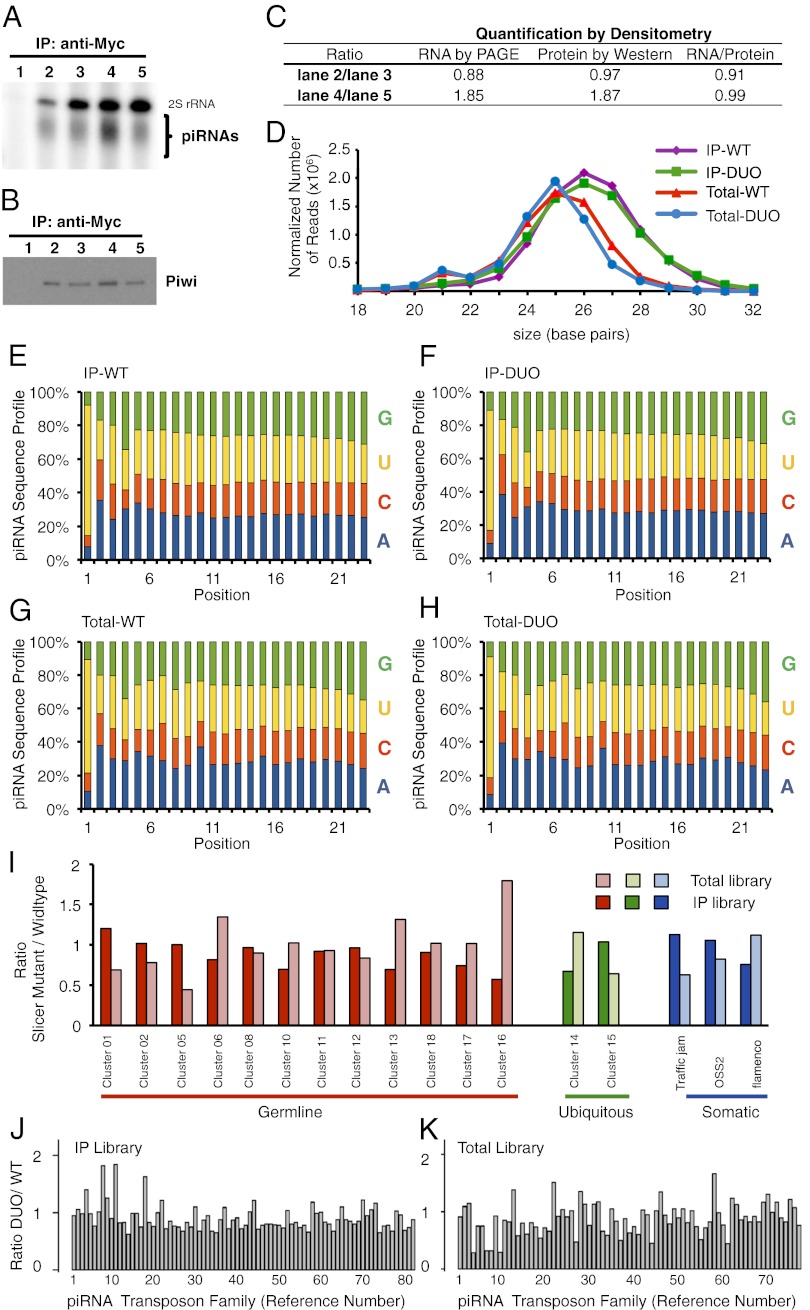
To directly test the involvement of the slicer activity of Piwi in piRNA biogenesis, we purified Piwi–piRNA complexes from piwi1/piwi1;DUO/DUO and piwi1/piwi1;WT/WT ovaries by immunoprecipitation using a high-affinity and high-specificity Myc antibody. Piwi-associated piRNAs were visualized by RNA-PAGE (Fig. 3A) and normalized to protein loading controls as quantified by Western blotting (Fig. 3 B and C). By comparing the pull-down efficiency of DUO and WT Myc–Piwi in a wild-type background, we confirmed that the slicer point mutations do not impair piRNA binding in vivo. Furthermore, the amount of piRNAs relative to Piwi protein does not differ between slicer-deficient and wild-type strains in the piwi1 background.
Fig. 3.
Duo mutant has no detectable defects in piRNA biogenesis. (A–C) Total levels of Piwi-associated piRNAs from +/+;WT/WT (lane 2), +/+;DUO/DUO (lane 3), piwi1/piwi1;WT/WT (lane 4), and piwi1/piwi1;DUO/DUO (lane 5) ovaries, resolved by denaturing PAGE (A). (B) Immunoprecipitation efficiency by Western blotting. (A and B) Lane 1 corresponds to W[1118], no-myc negative control. (C) Quantification by densitometry and normalization of piRNA/Piwi protein. (D) Size profile of ovarian piRNAs as revealed by deep sequencing. (E–H) Sequence profiles of piRNAs (∼23–28 nt) from IP libraries of piwi1/piwi1;WT/WT (E) and piwi1/piwi1;DUO/DUO (F) flies and from Total small RNA libraries of piwi1/piwi1;WT/WT (G) and piwi1/piwi1;DUO/DUO (H) flies. (I) Relative abundance of piRNAs derived from 17 known piRNA clusters from fly ovarian Piwi co-IP and total small RNA libraries, calculated as fold change of slicer mutant (DUO) over wildtype (WT). Only unique piRNAs are counted in this calculation, and the abundance is scaled to sequencing depth. The 17 clusters are grouped into three categories: germ-line–enriched (red), somatic-enriched (blue), and a ubiquitous class, comprising piRNAs similarly abundant in germ line and soma (green). Dark colors represent the Piwi co-IP libraries, and light colors represent the Total small RNA libraries. (J and K) Relative abundance of unique piRNAs derived from each transposon class in IP (J) and Total (K) small RNA libraries, shown as fold change of slicer mutant over wildtype. The abundance is scaled to the sequencing depth.
To further examine whether the slicer mutant Piwi is defective in piRNA biogenesis, we deep-sequenced four piRNA libraries prepared from either total small RNAs or Piwi coimmunoprecipitated samples from piwi1/piwi1;WT/WT and piwi1/piwi1;DUO/DUO genotypes, which we refer to as Total-WT, Total-DUO, IP–WT and IP–DUO, respectively. DUO and WT ovaries show similar distribution of both primary and ping-pong–derived secondary piRNA sequences in both Total and IP libraries (Fig. 3D and Fig. S2). Approximately 13% of sequences from Total RNA libraries are miRNAs. In contrast, both IP-WT and -DUO libraries only retained ∼1% of miRNA sequences, which could be Piwi-associated miRNAs (23). Regardless of whether the ∼1% of miRNAs reflect nonspecific background, this finding shows the similar effectiveness of both WT and DUO Piwi proteins in binding to piRNAs. In both IP–WT and IP–DUO libraries, ∼40% of piRNAs are comprised of transposon-derived piRNAs. After bioinformatic subtraction of spurious cellular RNA contaminants and miRNAs, we observed in both total small RNA libraries two peaks typical of piRNA profiles at 21 and 25 nt, corresponding to the sizes of endo-siRNAs and piRNAs, respectively (Fig. 3D). Furthermore, the IP–WT and IP–DUO piRNAs showed almost identical peaks at 26 nt—the expected peak size of Piwi-associated piRNAs. These results indicate that the slicer mutation has no visible effect on the biogenesis of either the PIWI-associated or total piRNA populations.
Slicer Activity of Piwi Is Not Required for piRNA Amplification via the Ping-Pong Mechanism.
To further resolve whether the slicer activity of Piwi contribute to the ping-pong mechanism, we analyzed the nucleotide sequence composition of the piRNA pools from the four libraries. The WT and DUO libraries show a striking resemblance of nucleotide composition at all corresponding positions. In particular, the first nucleotide exhibits strong uracil bias (Fig. 3 E–H). In addition, both total small-RNA libraries displayed a similar and weak adenine bias in the 10th nucleotide (Fig. 3 G and H), as expected for ping-pong–generated piRNAs. Expectedly, such a bias does not exist in either IP–WT or IP–DUO libraries (Fig. 3 E and F), because Piwi-associated piRNAs are primary piRNAs. Importantly, in either total or IP libraries, the WT and DUO ovaries showed indistinguishable sequence composition. Therefore, the slicer activity of Piwi is not required for the ping-pong mechanism.
The above conclusion is based on the fact that DUO mutation showed no effect on piRNAs at the population level, which does not rule out the possibility that DUO mutations may affect piRNA biogenesis at specific loci. To test this possibility, we examined whether the slicer activity of Piwi is involved in the biogenesis of 17 known piRNA clusters, which comprise 12 somatic-enriched, two germ-line–enriched, and three ubiquitous clusters (16, 24, 25). As shown in Fig. 3I, no significant change was observed for any of these clusters in the slicer mutant. Finally, we compared the level of piRNAs corresponding to specific transposon families and found that no piRNA from any specific transposon was affected by the slicer mutation (Fig. 3 J and K), except that total piRNA corresponding transposons HOBO, LOOPER1, POGON1, and POGO (Fig. 3K, at positions 4, 7, 8, and 10 of the horizontal axis) are decreased to ∼50% of the original levels. However, this decrease is not seen in PIWI-associated piRNAs corresponding to these transposons (Fig. 3J), and its impact on fertility and viability is undetectable and therefore inconsequential. To further verify our conclusion, we compared the abundance of piRNAs associated with 140 transposons in DUO and WT flies to those in Yb and papi mutants by the same bioinformatics analysis (Fig. S3). The Yb mutation caused significant decrease of piwi-bound piRNAs, whereas the papi mutation did not show any detectable effect. We observed that the piRNA abundance in the DUO mutant is very similar to WT and less variable than that of the papi mutant, let alone the Yb mutant. Together, our results indicate that the slicer activity of Piwi is not involved in the biogenesis of primary or secondary piRNAs in either somatic or germ-line cells.
Slicer Activity of Piwi Is Not Required for Transposon Silencing.
Lastly, we assessed whether the slicer activity of Piwi is involved in repression of transposons. To this end, we compared the levels of several Drosophila transposon transcripts (invader1, idefix, mdg1, blood, gtwin, HetA, diver, and 1360) between our slicer-deficient flies and wild-type flies by quantitative RT-PCR (qRT-PCR) assay. None of the transposons tested were significantly up-regulated in piwi1/piwi1,DUO/DUO or piwi1/piwi1,D37/D37 ovaries (Fig. 4 A–C), consistent with the normal development and fertility of these transgenic flies. These results demonstrate that the function of Piwi in repressing transposons is not mediated by its endonuclease activity.
Fig. 4.
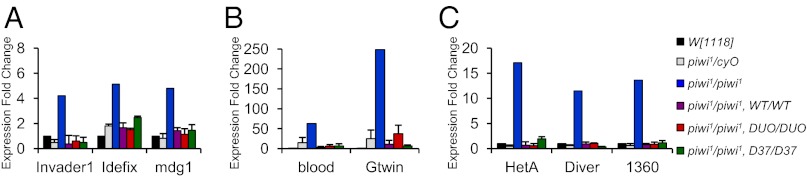
Piwi’s catalytic triad is not necessary to suppress transposons. The expression level of various transposons (invader1, idefix, mdg1, blood, gtwin, HetA, diver, and 1360) in ovaries was measured by qRT-PCR. After normalization of transposon levels per actin, the fold increase for each strain was calculated as a ratio over the wildtype W[1118] strain. The transposons are presented in three panels (A, B, and C) according to the magnitude of Piwi suppression towards their expression.
Discussion
We have shown that a catalytically null mutant of Piwi (D614A, D685A) fully rescues all of the known defects of piwi1 mutant—the most severe mutant allele of Piwi. These slicer-deficient flies have normal ovary morphology, fertility, and viability, and they retain the ability of generating piRNA molecules and repressing transposon elements. These results indicate that Piwi does not require slicer activity in vivo, despite the conservation of the catalytic triad and in vitro observations (10). Our results are consistent with the recent observation that Miwi2, a mouse Piwi protein that is enriched in the nucleus (26), does not require the slicer catalytic triad for its essential functions in spermatogenesis, piRNA biogenesis, and transposon suppression in the mouse testis (27). These congruent findings across species underscore the distinct and conserved nuclear function of Piwi proteins that is independent of their slicer activity. There are multiple lines of evidence supporting the notion that transposon silencing is regulated by the Piwi–piRNA complex in the nucleus (8, 28). The association of Piwi with chromatin and with epigenetic factors offers an attractive model for transposon repression via an epigenetic mechanism guided by piRNA sequence-specific recognition of the target transposon sequences (17, 18). There is increasing evidence suggesting that small RNAs hold answers to many unsolved questions in genome regulation, and this study suggests that careful in vivo validation of the proposed or in vitro-derived functions of the main players may hold the key to unraveling these mechanisms.
Materials and Methods
DNA Constructs and Site-Directed Mutagenesis.
Site-directed mutagenesis was performed by using the Stratagene Quick Change II kit on the pPMB1-6 P-element transformation construct that contains myc-tagged piwi genomic sequences (22), using the following primers:
Forward D614A: GATGACAATTGGCTTTGCCATTGCGAAGAGCACAC;
Reverse D614A: GTGTGCTCTTCGCAATGGCAAAGCCAATTGTCATC;
Forward D685A: CGAATCGTATTTTATCGAGCCGGTGTGAGCTCCGGC;
Reverse D685A: GCCGGAGCTCACACCGGCTCGATAAAATACGATTCG.
Mutagenesis was achieved by subcloning a 5.6-kb HindIII–HindIII fragment of the piwi gene into pBluescript, followed by site-directed mutagenesis and subcloning a 2.3-kb NheI–NheI subfragment containing the relevant point mutations back into pPMB1-6. For site-directed transgenesis, the wildtype and mutant (D614A,D685A) Myc–Piwi genomic rescue fragments were cloned by high-fidelity PCR amplification (Phusion; NEB) from pPMB1-6 into the XbaI and SpeI sites on pCa4B2G (GenBank accession no. EU420017.1) (21). All constructs were confirmed by sequencing in full.
For convenience we provide the FASTA sequence and positional maps for pPMB1-6, pCa4B2G–Myc–Piwi in Datasets S1 and S2.
Drosophila Transgenic Strains and Husbandry.
All Drosophila melanogaster strains were reared between 18 and 25 °C on yeast/molasses agar medium. The Piwi mutant strain used, piwi1, has been described (22, 29).
Transgenic flies were generated by the following two independent methods.
P-element transformation of the slicer mutant.
PMB1-6 mutant (D614A, D685A) construct was coinjected with transposase into white-eyed W[1118] stocks. Positive red-eyed transformants were balanced with a double balancer strain (Sco/CyO; Sb/TM6b) and made homozygous. We mapped the integration site of transformant fly line D37 to an intergenic region on chromosome 3 (3R:13) and confirmed the point mutations by sequencing a PCR product from transgenic fly genomic DNA.
Site-specific transgenesis by FlyC31 technology.
WT (wildtype Myc–Piwi) and DUO (catalytically deficient Myc–Piwi) transgenic fly lines were generated by injection of pCa4B2G–Myc–Piwi wild-type or point mutant (D614A, D685A) constructs into a transgenic line containing white−, X-linked integrase, and an attP2 site at 68A4 by Duke University Model System Genomics. Red-eyed transformant males were crossed with w[1118] virgin females before balancing with Sco/CyO; Sb/TM6b to remove the integrase transgene from the background. Site-specific integration of the transgenic constructs at the attP2 site was confirmed by PCR of junctional sequences. The D614A, D685A point mutations were also confirmed by sequencing genomic PCR from DUO transgenic line, which were found absent in the WT transgenic line.
All transgenic strains used in this study contain a single copy transgene.
Western Blotting, Immunofluorescence, and Antibodies.
Western blotting was performed by using the standard protocol with the following primary antibodies: mouse anti-Myc (Cell Signaling; 9B11) at a dilution of 1:1,000, mouse anti-Piwi (P3G11 and P4D2 mixed; Haru Siomi laboratory, Keio University, Tokyo, Japan.) at 1:5,000 dilution, and mouse anti-β-actin (Sigma Aldrich; A1978) at 1:2,000.
Ovarian immunofluorescence protocol has been described (30). Antibodies used for ovary immunofluorescence were mouse anti-Myc (Cell Signaling; 9B11, 1:1,000 dilution); rabbit anti-Vasa (Paul Lasko laboratory, McGill University, Montreal, Canada) at 1:10,000 dilution; mouse anti-1B1 (Developmental Studies Hybridoma Bank; 1:100 dilution); anti-mouse Alexa 488 (Invitrogen), 1:500 dilution; Anti-Rabbit Alexa 456 (Invitrogen) 1:500 dilution. Images were acquired with a Leica TCS SP5 Spectral Confocal microscope using sequential scanning mode.
Fertility and Viability Tests.
To assess fertility, virgin female flies were collected and paired with two or three males from the same genotype in individual vials. Flies were transferred to clean vials every 24 h, and the number of eggs laid was counted. Egg laying per fly per day was averaged across days 3–9 for several flies of each genotype. Sample size, N, corresponds to the number of vials analyzed. To reduce variability, we assayed fertility for each genotype in parallel using the same batch of yeast–molasses medium. Both fertility and viability assays were carried out at 25 °C. Viability was scored by counting the number of adult flies enclosed per vial normalized per egg counted in that vial.
Transposon Expression Analysis.
RNA was isolated from pooled ovaries dissected from each genotype by using the RNeasy kit (Qiagen) with on-column DNase digestion. cDNA was prepared by using the High-capacity cDNA Reverse Transcription Kit from Applied Biosystems. Real-time PCR was performed with 2× ready SYBR green Mastermix (BioRad) by using the primer pairs (Table 1). For all genotypes except piwi1/piwi1, biological triplicates of at least 10 ovaries per sample were averaged. For piwi1/piwi1, many more ovaries were pooled to produce one datapoint given their small size and rudimentary nature.
Table 1.
Primer pairs for real-time PCR analysis of transposon expression
| Transposon | Primer |
| Blood | |
| Forward | TGCCACAGTACCTGATTTCG |
| Reverse | GATTCGCCTTTTACGTTTGC |
| Gtwin | |
| Forward | TTCGCACAAGCGATGATAAG |
| Reverse | GATTGTTGTACGGCGACCTT |
| HetA | |
| Forward | CGCGCGGAACCCATCTTCAGA |
| Reverse | CGCCGCAGTCGTTTGGTGAGT |
| Diver | |
| Forward | GGCACCACATAGACACATCG |
| Reverse | GTGGTTTGCATAGCCAGGAT |
| 1360 | |
| Forward | GGAGCTCTGCGTATAGCCAACTT |
| Reverse | ACCTAAACCGCCGAGTCCTG |
| Invader1 | |
| Forward | GTACCGTTTTTGAGCCCGTA |
| Reverse | AACTACGTTGCCCATTCTGG |
| Idefix | |
| Forward | TGAAGAAAAGAAGGGCGAGA |
| Reverse | TTCTGCTGTTGATGCTTTGG |
| mdg1 | |
| Forward | AACAGAAACGCCAGCAACAGC |
| Reverse | CGTTCCCATGTCCGTTGTGAT |
PIWI–piRNA Co-IP.
A total of 100 pairs of ovaries were dissected in ice-cold PBS and homogenized with a pestle in Ovary Lysis Buffer [20 mM Hepes, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.1% SDS, 0.1% Deoxycholate.Na2, 1% (vol/vol) Triton X-100, 5% (vol/vol) glycerol supplemented with 1 mM fresh DTT, 1× complete mini EDTA-free Proteinase inhibitor mixture (Roche), and 0.5 U/μL RNase OUT (Invitrogen)]. Approximately 200 μL of soluble fraction was recovered by centrifugation, adjusted to 400 μL, and precleared with 100 μL of 50% bead slurry (GE Protein A beads washed and resuspended in ovary lysis buffer). Precleared lysate was recovered by centrifugation and incubated with mouse anti-Myc antibody (3 μL per sample) for 12 h at 4 °C after which 50 μL of 50% Protein A bead slurry was incubated for 1 h. Beads were washed twice with ovary lysis buffer for 5 min and twice with radioimmunoprecipitation assay buffer with inhibitors. To control for protein immunoprecipitation efficiency, 10% of beads were saved by boiling on SDS-loading buffer and Western blotting. The rest was used to recover PIWI-associated RNAs by TRIzol LS extraction and small RNA precipitation. To visualize total piRNA levels on a gel, 10% of the total isolated small RNAs were subjected to phosphonucleotide kinase reaction with P-32 γ-ATP and electrophoresed on a 15% polyacrylamide–6 M urea denaturing gel. The gel was covered in plastic-wrap and exposed to a phosphoimager screen for quantification.
Small RNA Profiling.
Ovaries from piwi1/piwi1,WT/WT and piwi1/piwi1,DUO/DUO genotypes were isolated to prepare four piRNA libraries from either total small RNAs (Total-WT and Total-DUO) or from Piwi co-IP samples (IP–WT and IP–DUO). Small RNAs were gel purified, and libraries were prepared according to the Illumina prep kit. After cloning, 16- to 29-nt small RNAs were isolated for Solexa sequencing.
Bioinformatic Analysis.
Clipping.
In total, >75 million small RNAs were obtained from Solexa sequencing. Total-WT library yielded ∼10 million small RNAs, whereas each of the other three libraries yielded >20 million small RNAs. Linkers were clipped off by using our homemade program, and 18- to 32-nt sequences were kept. In detail, a linker sequence is searched against the small RNA sequence pools. If the linker sequence is detected to match to a sequence, the portion that is beyond the matching region is removed. Otherwise, the linker is trimmed 1 nt off and subjected to a similar search. During the search process, mismatch is not allowed for any match that is <9 nt. Otherwise, one mismatch is allowed for a search. The average clipping rate is 0.81.
Mapping.
For downstream analysis, only sequences perfectly mapped to the D. melanogaster Release 5 genome (excluding Uextra) were retained. Libraries were normalized to sequencing depth to allow for cross-analysis.
Annotation.
To implement the origin annotation of piRNAs represented in our libraries, we used the miRBase Release 16 for miRNA-derived small RNAs, repeat masker annotation for transposon-derived RNAs, and ensembl gene annotation for gene-derived RNAs. Up to 2 mismatches were allowed with the insertion/deletion included to account for incomplete annotation of noncoding RNAs.
Cluster analysis.
We selected 17 piRNA clusters from different scientific groups, which covers clusters that were expressed preferentially in germ-line, somatic cells, together with the clusters that showed no preference: 15 piRNA clusters reported by Hannon and coworkers (24) to produce the greatest number of piRNAs; 1 piRNA cluster (Traffic Jam) from Siomi and coworkers (16); and 1 piRNA cluster (OSS2) located in Chr2R: 936,722–943,706 from Lau, Lai, and coworkers (25). For this analysis, we only considered uniquely mapped small RNAs, defined as mapping perfectly to only one position in the D. melanogaster Release 5 genome.
Transposon analysis.
We selected ∼80 transposons to show their piRNA ratio between DUO and WT for both IP and Total libraries. The criteria for choosing transposons were set to include only transposons for which at least 20 piRNA sequences were present in WT.
The sequencing data analyzed in this paper have been deposited to the Gene Expression Omnibus database (accession no. GSE34032).
Supplementary Material
Acknowledgments
We thank members of the H.L. laboratory—in particular Drs. John Saxe, Vamsi Gangaraju, Travis Thomson, and Hongyin Qi—for input toward this project. pCa4B2G was provided by Dr. Norbert Perrimon, and the P3G11 and P4D2 antibodies were provided by Dr. Mikiko Siomi. All transgenic injections were performed by Jamie Roebuck (Duke University Model System Genomics). This work was supported by National Institutes of Health Grant DP1 CA174418 (to H.L.). N.D. was supported by a Fonds de Recherche en Santé Québec Master’s Training Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34032).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213283110/-/DCSupplemental.
References
- 1.Juliano C, Wang J, Lin H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 5.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 6.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 7.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 8.Klenov MS, et al. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108(46):18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senti KA, Brennecke J. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet. 2010;26(12):499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20(16):2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 12.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23(24):4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell. 2010;19(5):687–697. doi: 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 16.Saito K, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461(7268):1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 17.Brower-Toland B, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21(18):2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, Yin H. A novel epigenetic mechanism in Drosophila somatic cells mediated by Piwi and piRNAs. Cold Spring Harb Symp Quant Biol. 2008;73:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito K, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24(22):2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox DN, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16(19):1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 24.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137(3):522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau NC, et al. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19(10):1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoji M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 27.De Fazio S, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480(7376):259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 28.Ishizu H, Nagao A, Siomi H. Gatekeepers for Piwi-piRNA complexes to enter the nucleus. Curr Opin Genet Dev. 2011;21(4):484–490. doi: 10.1016/j.gde.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124(12):2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 30.Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120(4):947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.