Abstract
Daily cyclical expression of thousands of genes in tissues such as the liver is orchestrated by the molecular circadian clock, the disruption of which is implicated in metabolic disorders and cancer. Although we understand much about the circadian transcription factors that can switch gene expression on and off, it is still unclear how global changes in rhythmic transcription are controlled at the genomic level. Here, we demonstrate circadian modification of an activating histone mark at a significant proportion of gene loci that undergo daily transcription, implicating widespread epigenetic modification as a key node regulated by the clockwork. Furthermore, we identify the histone-remodelling enzyme mixed lineage leukemia (MLL)3 as a clock-controlled factor that is able to directly and indirectly modulate over a hundred epigenetically targeted circadian “output” genes in the liver. Importantly, catalytic inactivation of the histone methyltransferase activity of MLL3 also severely compromises the oscillation of “core” clock gene promoters, including Bmal1, mCry1, mPer2, and Rev-erbα, suggesting that rhythmic histone methylation is vital for robust transcriptional oscillator function. This highlights a pathway by which the clockwork exerts genome-wide control over transcription, which is critical for sustaining temporal programming of tissue physiology.
Keywords: epigenomics, systems biology
The molecular clockwork consists of well-characterized transcriptional components [including Clock, brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like (Bmal) 1, Period, Cryptochrome, and nuclear receptor subfamily 1, group D, member 1/2 (Rev-erbα/β)], as well as more recently discovered posttranscriptional processes (1–4). Despite progress in understanding the make up of circadian transcriptomes and proteomes, which are thought to encompass more than 10% of known genes and proteins, the systems-level mechanisms driving their rhythmic abundance have remained unclear (5–7).
Based on observations that a few specific clock-regulated genomic loci undergo changes in chromatin state over the circadian cycle (8–10), we hypothesized that this might be more generally applicable at other clock-controlled gene (CCG) loci. We investigated this by performing chromatin immunoprecipitation (ChiP) and high-throughput sequencing (ChIP-seq) on mouse liver tissue collected over the circadian cycle to delineate global changes in the epigenome over a 24-h time frame.
Results and Discussion
We focused mostly on the activation mark, H3K4me3 (histone H3 trimethylated at lysine 4) (11), which exhibited a clear circadian profile at thousands of genomic loci (Fig. 1 A and B). This was in stark contrast to H3K9me3 (histone H3 trimethylated at lysine 9), which is associated with transcriptional inhibition (12), and unmodified histone H3, the binding of which changed only at relatively few loci (Fig. 1A). Therefore, genomic scale changes in the evolutionarily conserved activation mark H3K4me3 are regulated in a circadian manner at thousands of gene loci.
Fig. 1.
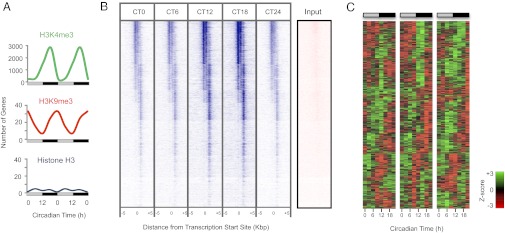
Circadian cycling of chromatin marks at promoter regions in mouse liver. (A) Whole-genome cycling of activating (H3K4me3) and inhibitory (H3K9me3) chromatin marks but not unmodified histone H3, over the circadian day and night. Data are double-plotted for clarity. Light gray bars indicate circadian daytime; black bars signify circadian night. (B) Heat map of H3K4me3 binding signal at CT0 to CT18 (with CT0 replotted), from –5 kb to +5 kb surrounding the center of all coding gene loci. ChIP-seq with anti-H3K4me3 antiserum was performed, and data were analyzed as described in SI Appendix. Each line represents a single H3K4me3-binding region. CT, circadian time (animals maintained in constant darkness, with subjective dawn represented by CT0, and dusk represented by CT12). The input (nonenriched) DNA signal at the same locus is shown for comparison. (C) Heat map of genes exhibiting circadian expression that also undergo rhythmic change in H3K4me3 binding at their promoter regions. Normalized expression is shown for three independent biological replicate sets of liver, sampled every 3 h in constant conditions.
We next examined the relevance of these pervasive changes for rhythmic gene expression. Initially, we cross-correlated rhythmic H3K4me3-binding sites with genes that had been shown previously to be rhythmic by high-resolution gene expression profiling (7). This revealed that 23% (887/3821) of rhythmically methylated loci were associated with circadian transcription patterns (SI Appendix, Table S1 and Fig. S1A). To confirm these findings, in the same liver sample set used for the ChIP-seq experiments, we performed expression profiling with a different microarray platform and found that these transcripts also oscillated in three biological replicates per time point (Fig. 1C and SI Appendix, Table S2). At the genomic scale, loci that did not undergo rhythmic histone modification were 3.16-fold (95% confidence interval, 2.94–3.40) less likely to exhibit circadian transcription (χ2 = 997.2; P < 0.0001; SI Appendix, Fig. S2). Thus, rhythmic histone modification over the circadian day and night is highly associated with rhythmic gene expression. Interestingly, however, there was no obvious relationship between the H3K4me3 mark being more prevalent at circadian time (CT)18 and the number of transcripts peaking at that particular time point compared with others (SI Appendix, Fig. S1B).
To determine the mechanisms by which circadian changes in histone H3 methylation might be brought about, we assayed the expression of components of the activating signal cointegrator-2 like complex, which facilitates trimethylation of H3K4 (13). Only the histone methyltransferase mixed lineage leukemia 3 (MLL3) showed rhythmic abundance over the circadian cycle, with peak expression at the end of the rodents’ inactive phase (Fig. 2A). This was mirrored by changes in MLL3 protein abundance, which was clearly disrupted in mCryptochrome (mCry) 1/2 double-knockout livers (14), illustrating that MLL3 rhythmicity is dependent on a functional transcriptional clock (Fig. 2B). Thus, MLL3 is a clock-controlled factor that could potentially regulate the circadian epigenomic and transcriptional profiles we observed.
Fig. 2.
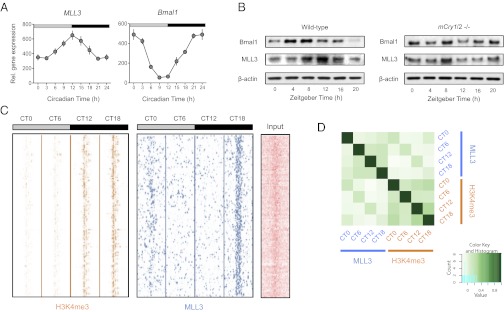
MLL3 is a circadian factor that binds to the genome rhythmically. (A) Expression of MLL3 (and the control gene, Bmal1) in mouse liver over the circadian cycle. Data are means ± SEM for three biological replicate liver samples per time point. (B) Abundance of MLL3 (and Bmal1) protein in wild-type and cryptochrome-deficient (mCry1/2−/−) mouse livers sampled over the day and night. Immunoblots representative of at least three independent time courses are shown. β-Actin blots are shown as gel loading controls. (C) Heat maps showing H3K4me3- and MLL3-binding signal at hundreds of promoter regions, from –5 kb to +5 kb surrounding the center of each binding site. Each line represents an H3K4me3 site and MLL3-binding region. CT, circadian time (animals maintained in constant darkness, with subjective dawn represented by CT0, and dusk represented by CT12). The input (nonenriched) DNA signal at the same locus is shown for comparison. (D) Heat map representing MLL3-binding events compared with those of H3K4me3. For each of the differentially bound sites, the reads per kilobase per million mapped reads (RPKM)-fold (RPKM of ChIP divided by RPKM of control) was calculated for each sample, and a Pearson correlation value for each sample pair computed. The heat map shows the correlation scores.
To test this, we determined MLL3-binding sites across the genome, using the same samples that we used for our histone H3 analysis. At rhythmic H3K4me3 loci that generated circadian transcripts, MLL3 was bound at 12% (460/3821) of transcription start sites (Fig. 2C and SI Appendix, Table S3 and Fig. S3). Furthermore, comparison of MLL3 binding and H3K4 methylation at the genomic scale, using DiffBind (15), revealed substantial cross-correlation (Fig. 2D). Interestingly, although the genomic distribution of H3K4me3 sites remained similar over the circadian cycle, there was a sizeable shift of MLL3-binding sites to intergenic regions at the end of the circadian night, which may be relevant for the regulation of noncoding RNA expression (SI Appendix, Fig. S4). These data, thus, imply that MLL3 is rhythmically bound at epigenetically regulated circadian loci in the liver.
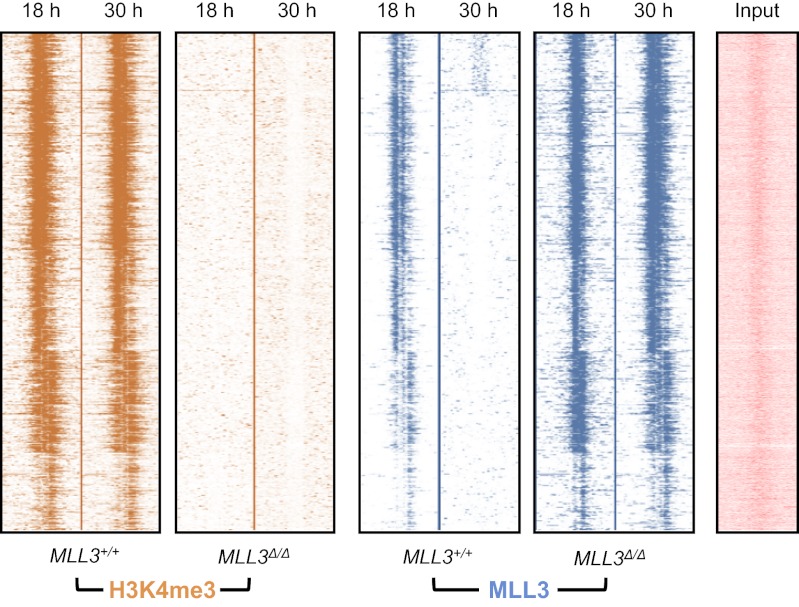
To dissect mechanism further, and specifically show that methyltransferase activity of MLL3 is important for generating the transcriptional changes that we observe, we performed further ChIP-seq experiments with transgenic mouse embryonic fibroblasts (MEFs) from mice harboring a mutant (catalytically inactive) form of MLL3. At the genomic scale, catalytic inactivation of MLL3 in the MLL3Δ/Δ cells had a major impact, resulting in loss of trimethylation of H3K4 at thousands of loci (Fig. 3A). Moreover, in these cells, there was arrhythmic, but augmented, MLL3 binding at loci that were rhythmically bound in wild-type (MLL3+/+) cells sampled at the same time point after cell synchronization (Fig. 3A). One intriguing possibility is that this rather widespread loss of trimethylation results from a dominant-negative effect of the MLL3Δ/Δ allele upon other cognate methyltransferases. With this in mind we, filtered for MLL3-specific methylation by only focusing on loci that, having been methylated in wild types, became unmethylated in MLL3Δ/Δ cells.
Fig. 3.
Histone methyltransferase activity of MLL3 is important for circadian gene expression at the genomic scale. Heat maps showing H3K4me3- and MLL3-binding signal at promoter regions in wild-type (MLL3+/+) or methyltransferase-deficient (MLL3Δ/Δ) MEFs. Regions from –5 kb to +5 kb surrounding the center of each binding site are shown. Each line represents an H3K4me3- and MLL3-binding region. Time after dexamethasone (DEX) synchronization is shown. The input (nonenriched) DNA signal at the same locus is shown for comparison.
This filtered set of regions was then mapped back on to our liver cistromic datasets to reciprocally determine expression profiles for these loci, assuming that these would be similarly regulated in liver tissue. We, thus, found that 13% (119/887) of rhythmically methylated genes are likely to be directly regulated by MLL3 (SI Appendix, Table S4). Functional analysis of these MLL3-regulated genes revealed some interesting targets including S-adenosyl homocysteine hydrolase and peroxiredoxin 6 (Prdx6), both of which are characterized clock-controlled genes in the liver (5, 16) (SI Appendix, Table S5). In particular, S-adenosyl homocysteine hydrolase is essential for adequate removal of S-adenosyl homocysteine (SAH), which is a product of methyltransferases (including MLL3) that use S-adenosyl methionine as a methyl donor for their reactions (17). Accumulation of SAH inhibits enzyme activity (18). Thus, hydrolase activity removes product inhibition by SAH, which is essential for maintenance of methyltransferase activity. In addition, functionally related components in the one-carbon pathway are also MLL3-regulated circadian genes (SI Appendix, Fig. S5), suggesting that MLL3 regulates the availability of methyl groups, which is essential for its own function, as well as for that of other methyltransferase enzymes. Importantly, this regulation is dependent on the liver clock, rather than on systemic (19) or feeding-related (20) cues (SI Appendix, Tables S6 and S7).
Given that MLL3 activity had an impact on the expression of a large number of clock-controlled genes, we investigated whether it might also influence the rhythmicity of components of the “core” transcriptional machinery, namely Reverb-α (Nr1d1), mPeriod (mPer) 1, mPer2, mCry1, mCry2, and Bmal1. Chromatin state maps of their genomic loci displayed circadian variation in H3K4me3 levels, suggesting further that MLL3 could potentially affect their expression, either directly or indirectly (Fig. 4A). To probe this further, we made stable bioluminescent MEF reporter cell lines that gave real-time readouts of transcriptional activation at each clock gene’s promoter region. Following synchronization with dexamethasone, we sampled wild-type (MLL3+/+) and catalytically inactive (MLL3Δ/Δ) cells, monitoring promoter activity for at least 5 d in the absence of external time cues (that is, under “constant conditions”) (Fig. 4B). In all reporter lines, there was a stark difference between the genotypes, with robust reporter expression in wild types, but severely compromised transcriptional output in the MLL3Δ/Δ MEFs. These findings are echoed by data from a recent RNA inference screen for circadian modifiers in human cells (SI Appendix, Fig. S6) (21). Correspondingly, temporal patterning of precursor mRNA (pre-mRNA) and mature mRNA in MLL3Δ/Δ cells was clearly different to that seen in MLL3+/+ MEFs (Fig. 4C). Lack of MLL3 methyltransferase activity, thus, significantly disrupts “clock gene” transcriptional activity (Fig. 4 B and C), as well as its own expression (SI Appendix, Fig. S7), implying that MLL3 plays an important role in controlling both core and “output” genes associated with the transcriptional clockwork.
Fig. 4.
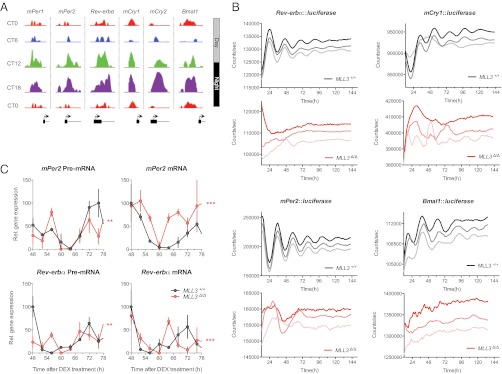
MLL3 activity is integral to the maintenance of the core transcriptional clockwork. (A) Chromatin-state maps showing H3K4me3-binding peaks for the core clock genes mPer1-2, mCry1-2, Rev-erbα, and Bmal1 over the circadian cycle in mouse liver. Binding peaks are all plotted on the same scale. Genomic loci are shown schematically below each map, showing the first exon of each gene and its transcription start site (indicated by an arrow). CT, circadian time (animals maintained in constant darkness, with subjective dawn represented by CT0, and dusk represented by CT12). (B) Real-time bioluminescent reporter assays in wild-type (MLL3+/+) or methyltransferase-deficient (MLL3Δ/Δ) MEFs stably transfected with Reverb-α::luciferase, mCry1::luciferase, mPer2::luciferase, or Bmal1::luciferase constructs. Assays were performed under constant conditions, at 37 °C, after synchronization with DEX. Data shown were baseline-subtracted using biological rhythms analysis software system (Materials and Methods). Representative traces from three biological replicates for each cell line are shown. (C) Expression of mPer2 and Reverb-α precursor mRNA (pre-mRNA) and mature mRNA in MEFs. Following synchronization with DEX, wild-type (MLL3+/+) or methyltransferase-deficient (MLL3Δ/Δ) MEFs were harvested and assayed by quantitative real-time PCR using primers specific for each gene’s pre-mRNA or mature mRNA. Data are means ± SEM for three biological replicates per time point. Two-way ANOVA revealed a significant (genotype × time) interaction (**P < 0.01; ***P < 0.001).
Together, our findings strongly support the idea that daily changes in chromatin architecture at the genomic scale take place in mammalian tissue and that almost a fifth of circadian gene expression in the liver is associated with variation of a single activating chromatin mark (H3K4me3). A significant proportion of these changes, resulting in rhythmic transcription at the systems level, can be attributed to MLL3 activity. Whereas 13% of these transcriptional changes might be direct effects of rhythmic MLL3-dependent histone methylation at the loci in question, the remainder might possibly be an indirect effect of the action of MLL3 upon transcription of core clock genes and other methyltransferases. In contrast to the recently described role played by another trithorax-group methyltransferase, MLL1, in modulating specific Clock-bound genomic loci (9), our data suggest that recruitment of MLL3 does not require Bmal1 (SI Appendix, Fig. S8) (22) and, therefore, nor its heterodimerization partner Clock, supporting a complementary and independent mechanism of action. This is consistent with the fact that MLL1 and MLL3 form mutually exclusive complexes to modulate chromatin state, with differing constituents (23). It is increasingly apparent that the temporal phasing between chromatin modifications and transcription will depend on multiple influences and that a combinatorial assessment of these phenomena is required to dissect these factors more fully (24), as well as disentangling the importance of posttranscriptional and posttranslational processes in sculpting circadian mRNA and protein profiles (1, 5, 25). Because misregulation of MLL3 activity is associated with multiple cancers (26–28), and our data link its function to the competency of the core transcriptional clockwork and its downstream effectors, this provides a framework to understand the role of MLL3, histone methylation, and the circadian clock in maintaining normal tissue physiology, and in pathologies such as cancer.
Materials and Methods
ChIP.
All animal experimentation was licensed by the Home Office under the Animals (Scientific Procedures) Act, 1987, and was reviewed by the University of Cambridge Local Ethical Review committee. Liver tissue was harvested from n = 4 adult male C57BL/6 mice once every 3 h on the second cycle after transfer from 12L:12DR to 12DR:12DR (L, light [220 µW⋅cm−2] and DR, dim red light [<5 µW⋅cm−2]) and immediately frozen and then stored at −80 °C before use. Before sampling, animals were stably entrained to a 12-h L:12-h DR cycle for 3–4 wk. Liver tissue (30 mg per final ChIP) was rapidly chopped into small (approximately 5 × 5 × 2.5 mm3) while defrosting using a sterile scalpel and immediately submerged in 4% (wt/vol) formaldehyde (Sigma) and incubated at room temperature with gentle shaking for 10 min. Glycine (1.25 M) was added to a final concentration of 125 mM and incubated for a further 10 min to quench the formaldehyde. Tissue was then dounce-homogenized briefly to break up the tissue and then passed through a 100-µm cell strainer (BD Biosciences), and a cell suspension was harvested. The cells were washed in ice-cold PBS and then incubated with 2 mL of Farnham lysis buffer at 4 °C for 15 min to release nuclei. After spinning down, pelleted nuclei were then lysed with 1 mL radioimmunoprecipitation assay (RIPA) buffer and incubated for a further 15 min at 4 °C. Chromatin was sheared using a Diagenode Bioruptor at 4 °C. Sheared chromatin was transferred to 1.5-mL microcentrifuge tubes and spun at 14,000 rpm (Eppendorf 5415R) for 15 min at 4 °C to pellet debris. For each liver from n = 4 animals per time point, duplicate ChIPs were performed with each antibody. ChIP-grade antibodies were purchased from Abcam: anti-H3K4me3 (ab8580), anti-H3K9me3 (ab8898), anti-histone H3 (ab1791), and anti-MLL3 (ab71200). Fifty microliters of Protein G magnetic beads (Dynal/Invitrogen) were washed three times and then incubated with 5 µg of antibody in 300-µL volumes for 4 h at 4 °C on a rotator. Excess, or nonspecifically bound, antibody was removed by washing a further three times in PBS/BSA. The beads were then added to 900 µL of sheared chromatin in 1.5-mL DNA Lo-bind tubes (Eppendorf) and incubated at 4 °C overnight with gentle rotation. Beads were washed six times in 1 mL of LiCl Wash Buffer for 3 min each, followed by a brief wash in Tris-EDTA (TE) buffer (pH 7.4). The beads were then suspended in TE and transferred to a clean microcentrifuge tube, and TE was removed. IP elution buffer (200 μL) was added at room temperature, and then the beads were incubated for 45 min at 65 °C with continuous shaking on a thermomixer (at 800 rpm). The eluate was then removed from the beads and incubated overnight at 65 °C to reverse cross-linking. DNA was extracted using a Qiagen Minelute PCR purification kit (using 12 μL of Buffer EB to elute DNA) and then quantified using Picogreen assays (Invitrogen) in a microplate as per the manufacturer’s instructions. Input chromatin was reverse–cross-linked in parallel and assayed with a Nanodrop spectrophotometer (because of the much higher DNA concentration), and a sample was run on a 2% agarose gel to check fragment size distributions were correct. See SI Appendix for further details.
Multiplex Library Preparation and Sequencing.
For H3K4me3, H3K9me3, histone H3, and input DNA samples, these were prepared for sequencing on an Illumina Genome Analyzer II following the manufacturer’s recommended library preparation protocol using 50 ng of chromatin-immunoprecipitated DNA. All libraries were multiplexed, and four indexed pooled samples (corresponding to four time points: CT0, -6, -12, -18) sequenced per lane. See SI Appendix for further details.
Bioinformatics: Alignment and Peak Detection.
Raw paired-end 50-bp reads (in FASTQ format) were aligned to the University of California, Santa Cruz (UCSC) mm9 genome build with Bowtie using prebuilt indexes downloaded from the host website (29). Alignment files were then converted from the standard Bowtie output format to ALN files (for use with CisGenome) using a custom Perl script. Alignments were then analyzed using a 2-sample comparison (chromatin-immunoprecipitated sample vs. input control for each time point) with CisGenome Version 1, using a cutoff false discovery rate of 10% and a sliding window size of 100 (30). Output files were converted into various formats for browsing datasets and to display data. The CisGenome and UCSC browsers were used for data visualization, using custom tracks (31). Raw sequence data (FASTQ format), alignments (Bowtie format), and peak data (as CisGenome/Affymetrix BAR format) have been deposited in the GEO database (accessions nos. GSE23550 and GSE37396). See SI Appendix for further details.
Microarrays.
Total RNA was isolated from liver samples of C57/Bl6 mice over a circadian time course as described previously (16). Per time point, n = 3 biological replicate samples were collected and processed individually. RNA from each individual biological replicate sample was hybridized on an Affymetrix mouse Gene ST1.0 microarray (GEO accession no. GSE37396), using the manufacturer’s protocol. Microarray analysis was performed using GeneSpring GX 11, with intensity values being normalized using the robust multiarray average algorithm. See SI Appendix for further details.
Real-Time PCR for Gene Expression Analysis.
RNA from individual livers (see ChIP methods above) was extracted with TRIzol reagent (Invitrogen) and purified with RNAeasy Mini Kits (Qiagen). Total RNA was then used for reverse transcription via a High Capacity cDNA Archive Kit (Applied Biosystems). The resulting cDNA was then diluted 1:5 and used in duplicate 10-μL PCR reactions according to the manufacturer’s protocol (TaqMan Gene Expression Master Mix; Applied Biosystems) with validated TaqMan Gene Expression Assays (Applied Biosystems), for Nr1d1, assay mm00520708_m1 and for mPer2, assay mm00478113_m1. For control reactions, mouse β-actin mRNA was amplified from the same samples. Real-time PCR was performed with an ABI 7900HT (Applied Biosystems) system. See SI Appendix for further details.
Gel Electrophoresis and Immunoblotting.
Wild-type and mCry1−/− mCry2−/− animals (C57BL/6 background, 12 wk old) were individually housed under 12-h light, 12-h dark cycles and constant temperature (21 ± 2 °C). Livers from animals were harvested every 4 h over the 24-h cycle (ZT0, -4, -8, -12, -16, -20), flash-frozen and stored at −80 °C. Lysates were diluted with denaturing lithium dodecyl sulfate sample buffer (Invitrogen) with 1:10 β-mercaptoethanol to a final protein concentration 2 μg⋅μL−1 and heated to 70 °C for 10 min before loading on gels. Ten micrograms of protein per lane were loaded for immunoblotting. We used NuPAGE Novex 4–12% Bis-Tris gradient gels for Bmal1 and β-actin analyses, and 3–8% Tris-acetate gels for MLL3 (Life Technologies). Gels were run according to the manufacturer’s protocol with a nonreducing Mes SDS buffer system (for Bis-Tris gels), or Tris-acetate SDS buffer (for Tris-acetate gels). Protein transfer to nitrocellulose for blotting was performed and the nitrocellulose was then washed briefly and then blocked for 30 min in 0.5% wt/wt BSA/nonfat dried milk (Marvel) in Tris-buffered saline/0.05% Tween-20 (TBST). After three brief washes in TBST, membranes were incubated in antibody diluted in blocking buffer (0.5% milk/BSA) overnight at 4 °C. The following day, membranes were washed for 5 min three times (in TBST) and then incubated with 1:10,000 HRP-conjugated secondary antibody (Sigma-Aldrich) for 30 min. Four more 10-min washes were then performed before performing chemiluminescence detection. See SI Appendix for further details.
Bioluminescence Reporter Vectors.
Promoter regions from mouse Bmal1 and Per2 (30, 32) were amplified by PCR or digested directly from the parent vector for Cry1 (31, 33) and subcloned into the pGL4.20 vector (Promega) between the KpnI and HindIII restriction enzyme sites. For Rev-erbα (Nr1d1), a 1,003-bp fragment (mouse chromosome 11:98636454–98637456, mm9 annotation) was amplified by PCR from BAC clone RP23-395E10 and then cloned into pGL4.20 using restriction-free cloning (16, 34). Clones were sequence-verified to confirm correct insert sequences. See SI Appendix for further details.
Cell Culture and Bioluminescence Assays.
Cells were cultured at 37 °C, 5% CO2 in a standard humidified incubator. To create stable luciferase reporter lines, 1 μg of each vector was transfected using GeneJuice Transfection Reagent (catalog no. 70967-3; Novagen), and then cells selected for approximately 2 wk with puromycin-supplemented medium (final concentration of 2 μg⋅mL−1) in 35-mm-diameter cell culture dishes. Bioluminescence assays were performed at 37 °C using 96-well plate reader (CentroLIA LB 960; Berthold Technologies), with an integration time of 56 s per well, every 90 min, using a standard protocol (7, 35). See SI Appendix for further details.
Statistical Analysis.
Parametric statistics (one- and two-way ANOVA) and χ2 tests were performed using Graphpad Prism Version 5 software.
Supplementary Material
Acknowledgments
We thank J. Lee for sharing MEF cell lines. This work was supported by the Wellcome Trust (083643/Z/07/Z), the European Molecular Biology Organization Young Investigators Programme, the Medical Research Council Centre for Obesity and Related metabolic Disorders, and the National Institute for Health Research Cambridge Biomedical Research Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.S. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE23550 and GSE37396).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214168110/-/DCSupplemental.
References
- 1.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469(7331):554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogenesch JB, Ueda HR. Understanding systems-level properties: Timely stories from the study of clocks. Nat Rev Genet. 2011;12(6):407–416. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 4.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 6.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 9.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17(12):1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333(6051):1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 14.van der Horst GTJ, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 15.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy AB, et al. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007;45(6):1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- 17.Campagna-Slater V, et al. Structural chemistry of the histone methyltransferases cofactor binding site. J Chem Inf Model. 2011;51(3):612–623. doi: 10.1021/ci100479z. [DOI] [PubMed] [Google Scholar]
- 18.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8(9):724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 19.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang EE, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139(1):199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho Y-W, Hong S, Ge K. Affinity purification of MLL3/MLL4 histone H3K4 methyltransferase complex. Methods Mol Biol. 2012;809:465–472. doi: 10.1007/978-1-61779-376-9_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe Y, et al. Frequent alteration of MLL3 frameshift mutations in microsatellite deficient colorectal cancer. PLoS ONE. 2011;6(8):e23320. doi: 10.1371/journal.pone.0023320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X-X, et al. Somatic mutations of the mixed-lineage leukemia 3 (MLL3) gene in primary breast cancers. Pathol Oncol Res. 2011;17(2):429–433. doi: 10.1007/s12253-010-9316-0. [DOI] [PubMed] [Google Scholar]
- 28.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H, Wang F, Dyer NP, Wong WH. CisGenome Browser: A flexible tool for genomic data visualization. Bioinformatics. 2012;26(14):1781–1782. doi: 10.1093/bioinformatics/btq286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 33.Fustin JM, O’Neill JS, Hastings MH, Hazlerigg DG, Dardente H. Cry1 circadian phase in vitro: Wrapped up with an E-box. J Biol Rhythms. 2009;24(1):16–24. doi: 10.1177/0748730408329267. [DOI] [PubMed] [Google Scholar]
- 34.van den Ent F, Löwe J. RF cloning: A restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67(1):67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Hastings MH, Reddy AB, McMahon DG, Maywood ES. Analysis of circadian mechanisms in the suprachiasmatic nucleus by transgenesis and biolistic transfection. Methods Enzymol. 2005;393:579–592. doi: 10.1016/S0076-6879(05)93030-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.