Abstract
Interactions between swimming cells and surfaces are essential to many microbiological processes, from bacterial biofilm formation to human fertilization. However, despite their fundamental importance, relatively little is known about the physical mechanisms that govern the scattering of flagellated or ciliated cells from solid surfaces. A more detailed understanding of these interactions promises not only new biological insights into structure and dynamics of flagella and cilia but may also lead to new microfluidic techniques for controlling cell motility and microbial locomotion, with potential applications ranging from diagnostic tools to therapeutic protein synthesis and photosynthetic biofuel production. Due to fundamental differences in physiology and swimming strategies, it is an open question of whether microfluidic transport and rectification schemes that have recently been demonstrated for pusher-type microswimmers such as bacteria and sperm cells, can be transferred to puller-type algae and other motile eukaryotes, because it is not known whether long-range hydrodynamic or short-range mechanical forces dominate the surface interactions of these microorganisms. Here, using high-speed microscopic imaging, we present direct experimental evidence that the surface scattering of both mammalian sperm cells and unicellular green algae is primarily governed by direct ciliary contact interactions. Building on this insight, we predict and experimentally verify the existence of optimal microfluidic ratchets that maximize rectification of initially uniform Chlamydomonas reinhardtii suspensions. Because mechano-elastic properties of cilia are conserved across eukaryotic species, we expect that our results apply to a wide range of swimming microorganisms.
Keywords: algal surface accumulation, swimming rectification
Surface interactions of motile cells play crucial roles in a wide range of microbiological phenomena, perhaps most prominently in the formation of biofilms (1) and during the fertilization of mammalian ova (2). However, despite their widely recognized importance, the basic physical mechanisms that govern the response of swimming bacteria, algae, or spermatozoa to solid surfaces have remained unclear. This predicament is exemplified by the current debate (3–6) about the relevance of hydrodynamic long-range forces and steric short-range interactions for the accumulation of flagellated cells at liquid–solid interfaces. From a general perspective, improving our understanding of cell surface scattering processes promises not only new insights into structure, dynamics, and biological functions of flagella and cilia, it will also help to advance microfluidic techniques for controlling microbial locomotion (7, 8), with potential applications in diagnostics (9), therapeutic protein synthesis (10), and photosynthetic biofuel production (11–14). That microfluidic circuits provide an excellent test bed for developing and assessing new strategies for the control of cell motility was recently demonstrated by the rectification of random bacterial swimming through microscopic wedge-shaped barriers (7, 8). However, because eukaryotic and prokaryotic swimming strategies differ substantially from each other (6, 15–19), it is unclear whether design principles that exploit surface collisions to achieve control of bacterial locomotion are transferrable to motile eukaryotes.
Aiming to elucidate the role of eukaryotic cilia in cell–surface interactions, we report here a detailed experimental investigation of surface scattering for bull spermatozoa and Chlamydomonas reinhardtii algae (simply referred to as Chlamydomonas herein). Bull sperm and other mammalian spermatozoa are “pusher” swimmers that generate propulsion by undulating a single posterior cilium (Fig. 1A). By contrast, a WT Chlamydomonas cell is a “puller” that achieves locomotion by the breaststroke-like beating (15–17) of a pair of anterior flagella (Fig. 2A). Chlamydomonas algae have long been appreciated as premier model organisms in biology (20–22), in particular for studying photosynthesis (11, 23) and ciliary (15, 20, 24) functions in eukaryotes. More recently, they have also attracted considerable interest as possible sources of therapeutic proteins (10) and renewable biofuels (11–14, 25–28). Against this backdrop, our second goal is to demonstrate the feasibility of microfluidic rectification schemes for these organisms.
Fig. 1.

Surface scattering of bull spermatozoa is governed by ciliary contact interactions, as evident from the scattering sequences of individual cells at two temperature values: (A) T = 10 °C and (B) T = 29 °C. The background has been subtracted from the micrographs to enhance the visibility of the cilia. The cyan-colored line indicates the corner-shaped boundary of the microfluidic channels (see Movies S1 and S2 for raw imaging data). The horizontal dotted line in the last image in B defines θ = 0. (Scale bars: 20 μm.) (C) The probability distributions of scattering angles θ from the corner peak at negative angles, due to the fact that the beat amplitude of the cilia exceeds the size of the cell body (sample size: n = 116 for T = 10 °C and n = 115 for T = 29 °C). At higher temperatures, the cilia exhibit a larger oscillation amplitude and beat frequency (29), resulting in a larger swimming speed and shifting the typical scattering angles to larger absolute values.
Fig. 2.
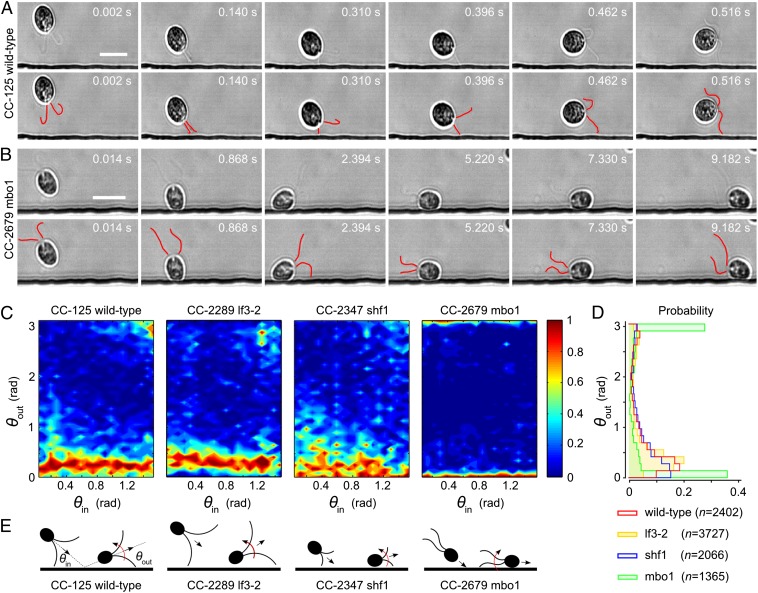
Surface scattering of Chlamydomonas is governed by ciliary contact interactions. (A) Scattering sequence for WT Chlamydomonas CC-125 (Movie S3). (Upper) Original micrographs. (Lower) Cilia manually marked red. Results for the long-flagella mutant lf3-2 and the short-flagella mutant shf1 look qualitatively similar (Movies S4 and S5). (Scale bar: 20 μm.) (B) The mutant pusher mbo1 remains trapped for several seconds (Movie S6). (Scale bar: 20 μm.) (C) The conditional probability distributions P(θout|θin) indicate that, for all four strains, memory of the incidence angle is lost during the collision process, due to multiple flagellar contact with the surface. (D) The cumulative scattering distribution P(θout) shows how cilia length and swimming mechanisms determine the effective surface-scattering law. (E) Schematic illustration of the flagella-induced scattering and trapping mechanisms.
Rectification of bacterial run-and-tumble motion in microfluidic ratchets (7, 8) is believed to result from the swimmers’ tendency to align their motion along the ratchet barriers, either by steric (4–6) or by hydrodynamic (3) surface interactions, although the exact mechanism is not well understood. Whereas steric alignment with surfaces seems intuitively plausible for rod-shaped bacteria such as Escherichia coli or Bacillus subtilis, additional hydrodynamic alignment is thought to arise from the fact that the posterior flagellar bundle of a bacterium creates a pusher-like dipolar flow during locomotion (19). This flow points outward along the body axis and inward along the lateral directions (6). The presence of a wall couples the swimmer’s translation and rotation and causes it to align parallel to the surface (3). By contrast, the anterior flagella of WT Chlamydomonas pull the organism through the fluid, thereby generating a far-field flow topology (16, 17) that looks roughly opposite to that of a bacterium. Hence, far-field hydrodynamics suggests that Chlamydomonas should either turn away from or collide head-on with a nearby no-slip surface, but the complex time-dependent flow structure (16, 17) close to the cell body makes it difficult to predict the scattering dynamics in the vicinity of the surface. It is therefore not possible to infer from general hydrodynamic arguments whether it is at all feasible to design microfluidic structures that are capable of rectifying algal swimming. Moreover, purely hydrodynamic considerations completely neglect direct contact interactions between cilia or flagella and solid surfaces. Unfortunately, this potentially important scattering mechanism (4) is not included in currently prevailing theoretical models of microbial swimming near solid boundaries (3).
Here, we present direct experimental evidence that the scattering of bull spermatozoa and Chlamydomonas algae off a solid boundary is, in fact, mainly determined by the contact interactions between their flagella and the surface, whereas hydrodynamic effects only play a secondary role. Building on these insights, we derive a simple criterion to predict an efficient ratchet design for Chlamydomonas and confirm its validity experimentally, thereby demonstrating that robust rectification of algal locomotion is possible. More generally, our results show that the interactions between swimming microorganisms and surfaces are more complex than previously recognized, suggesting the need for a thorough revision of currently accepted paradigms. Because mechano-elastic properties of eukaryotic cilia are conserved across eukaryotic species, we expect flagella–surface interactions to play a similarly important role for a wide range of natural microswimmers, thus promising new diagnostic tools and microfluidic sorting devices for sperm (9) and other motile cells.
Results
To identify the dynamical details of eukaryotic cell–surface interactions, we studied the surface scattering of bull spermatozoa and four different Chlamydomonas algae strains in quasi-2D microfluidic channels (height ∼ 25 μm), using high-speed microscopic imaging (Materials and Methods).
Scattering of Individual Sperm Cells from Solid Boundaries.
We analyzed the scattering of individual sperm cells from corner-shaped channel boundaries at high and low temperatures (Fig. 1 A and B; Movies S1 and S2). Direct observation reveals that the short-range interaction of the cilium with the boundary determines how a spermatozoon swims along a solid surface. In a typical scattering event, a sperm cell closely follows the boundary until it reaches a corner and departs at an angle θ, defined here relative to the initial swimming direction such that θ = 0 corresponds to the surface tangent (Fig. 1B). We determined the temperature-dependent distributions of θ from more than 200 scattering events by tracking the position of the cell body up to a distance of 70 μm from the corner (Fig. 1C). The histograms show that sperm–surface interactions are typically characterized by negative scattering angles θ < 0 due to the fact that the beat amplitude of the ciliary motion is much larger than the size of the cell body. Hence, ciliary contact with the surface tends to turn the spermatozoa toward the boundary, thereby preventing their escape from flat and weakly curved surfaces.
To illustrate how differences in the ciliary beating patterns affect the surface interaction of spermatozoa, we exploit the fact (29) that amplitude and frequency of the ciliary motion increase monotonically with temperature T in the range 5 °C < T < 38 °C. This variability is caused by a change in motor activity (30), approximately described by an Arrhenius law ∝ exp(−Δ/kT) (29), where Δ denotes the activation energy and k the Boltzmann constant. If ciliary contact governs the surface interactions of sperm, then one should expect that the absolute mean scattering angle  increases with temperature. By comparing the scattering distributions at low and high temperatures, we find that this is indeed the case (Fig. 1C). At low temperature T = (10 ± 1) °C, the experimental data yield a mean scattering angle
increases with temperature. By comparing the scattering distributions at low and high temperatures, we find that this is indeed the case (Fig. 1C). At low temperature T = (10 ± 1) °C, the experimental data yield a mean scattering angle 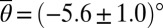 , whereas
, whereas 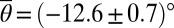 at a high temperature T = (29 ± 1) °C. In particular, these results suggest that, at higher temperatures, sperm cells can become more easily trapped at strongly curved surfaces. With regard to future biotechnological applications, this self-trapping by ciliary beating can provide a useful mechanistic basis for sorting and rectifying spermatozoa (9).
at a high temperature T = (29 ± 1) °C. In particular, these results suggest that, at higher temperatures, sperm cells can become more easily trapped at strongly curved surfaces. With regard to future biotechnological applications, this self-trapping by ciliary beating can provide a useful mechanistic basis for sorting and rectifying spermatozoa (9).
Flagella-Induced Scattering of Individual Algae from Solid Boundaries.
To further test the idea that ciliary contact dominates eukaryotic cell–surface interactions, we studied the surface scattering of four different Chlamydomonas strains (The Chlamydomonas Resource Center, www.chlamy.org): the WT CC-125, the long-flagella mutant CC-2289 lf3-2, the short-flagella mutant CC-2347 shf1, and the moving-backward-only mutant CC-2679 mbo1 (Fig. 2). All four Chlamydomonas strains share an essentially identical geometrical structure, but whereas the WT and the mutants lf3-2 and shf1 are puller-type swimmers that differ only in flagella length [6−8 μm for shf1 (31) compared with 11−13 μm for WT and 12−22 μm for lf3-2 (32)], the backward-swimmer mbo1 has a persistent undulatory swimming gait and can be considered a pusher similar to bacteria and spermatozoa (Movies S3, S4, S5, and S6). For each strain, we recorded more than 1,300 boundary-scattering events in quasi-2D microfluidic channels (height ∼ 25 μm).
High-speed imaging of individual cell trajectories for both WT and mutants reveals that the interaction of Chlamydomonas with the channel wall is also strongly affected by the short-range contact forces between the flagella and the surface (Fig. 2 A and B; Movies S3, S4, S5, and S6). In the case of the three puller swimmers (WT, lf3-2, and shf1), the flagella prevent the cell body from touching the surface while simultaneously creating an effective torque that turns the organism away from it (Fig. 2 A and E). By contrast, for the backward-swimming mutant mbo1, posterior thrust by the flagella pushes the cell body onto the surface (Fig. 2 B and E). Subsequently, the mechanical contact of the cilia with the boundary leads to a net torque that keeps rotating the alga toward the surface (Fig. 2E). As a result, mbo1 cells remain trapped at the channel wall for several seconds compared with ≲ 0.5 s for WT.
Fig. 3.
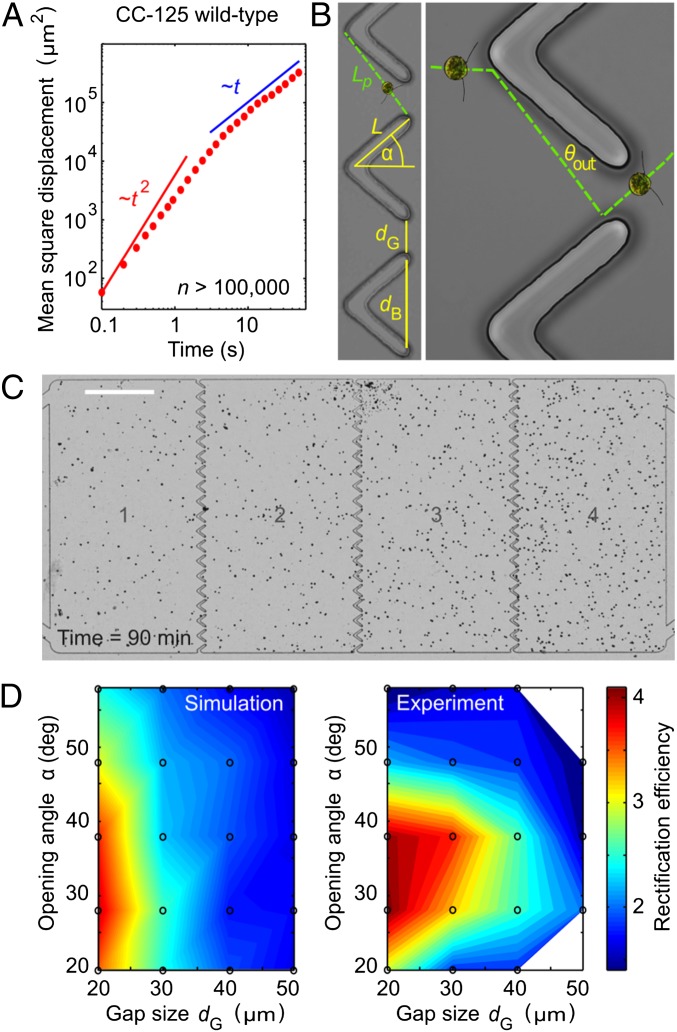
Rectification of algal locomotion in microfluidic ratchets. (A) When confined in a quasi-2D chamber (height, 25 μm), WT CC125 perform run-and-turn motions, moving ballistically (average speed 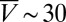 μm/s) on short time scales (<2 s) and diffusively on larger time scales. (B) Ratchet geometry and schematic representation of secondary algal scattering. (C) Rectified steady state for a chamber with four compartments (Movie S7). (Scale bar: 0.5 mm.) (D) In both simulations of the minimal model (Materials and Methods) and experiments, the rectification efficiency
μm/s) on short time scales (<2 s) and diffusively on larger time scales. (B) Ratchet geometry and schematic representation of secondary algal scattering. (C) Rectified steady state for a chamber with four compartments (Movie S7). (Scale bar: 0.5 mm.) (D) In both simulations of the minimal model (Materials and Methods) and experiments, the rectification efficiency 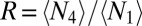 exhibits a maximum near to the theoretically predicted optimal ratchet parameters. Each data point (circles) represents an average over three to five different experiments. Rectification efficiencies are linearly interpolated. The SD for different experiments is less than 30% of the mean value.
exhibits a maximum near to the theoretically predicted optimal ratchet parameters. Each data point (circles) represents an average over three to five different experiments. Rectification efficiencies are linearly interpolated. The SD for different experiments is less than 30% of the mean value.
The contact force that is exerted by a flagellum onto the surface per stroke can be estimated from the flagellar beat frequency (∼50 Hz) and the experimentally observed angular displacement amplitude per beat (∼0.2 rad), which gives a typical angular speed ω ∼ 10 rad/s. Assuming a spherical cell body (radius a ∼ 5 μm), the torque T can be obtained from T ∼ ξω, where the rotational drag coefficient is given by ξ = 8πηa3. Using η = 10−3 Pa⋅s for water, we find T ∼ 30 pN⋅μm and, furthermore, by assuming a flagella length L ∼ 10 μm, the typical force F ∼ T/L ∼ 3 pN. These estimates, which are based on the observed rotation of the cell body near the surface, are consistent with the values obtained from recent measurements of freely swimming Chlamydomonas algae (17, 33), suggesting that reorientation at the wall is primarily determined by flagellar contact.
To quantify the different surface scattering laws for each of the four Chlamydomonas strains in detail, we measured the incidence and scattering angles θin and θout (Materials and Methods) and determined the conditional scattering distributions P(θout|θin), defined as the probability of being scattered into the interval [θout, θout + dθout] for a given incidence angle θin (Fig. 2C). For WT Chlamydomonas, P(θout|θin) is independent of the incidence angle θin and exhibits a narrow peak at 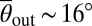 . The typical scattering angle
. The typical scattering angle 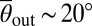 is larger for the long-flagella mutant lf3-2, whereas for the short-flagella mutant shf1, the maximum of P(θout|θin) is shifted to smaller angles
is larger for the long-flagella mutant lf3-2, whereas for the short-flagella mutant shf1, the maximum of P(θout|θin) is shifted to smaller angles 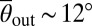 . The systematic increase of the typical scattering angle with flagella length, in conjunction with the above force estimates, implies that, in all three cases, the characteristic escape angle
. The systematic increase of the typical scattering angle with flagella length, in conjunction with the above force estimates, implies that, in all three cases, the characteristic escape angle 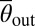 is set geometrically through the length of the cilia, the diameter of the cell body, and the typical distance of the latter from the surface at the moment of departure (Movies S3, S4, and S5). For shf1, the cell body can come closer to the wall than for the WT, and the resulting lubrication forces may also affect the escape dynamics. The relatively larger spread in the distribution of scattering angles for shf1 is compatible with previous observations of stronger intrinsic fluctuations in shorter flagella (34). By contrast, the pusher-mutant mbo1 generally remains close to the surface, θout ∼ 0 or θout ∼ π (Movie S6). The differences between the scattering laws of the four different strains are also clearly evident from the mean scattering distributions P(θout), obtained by averaging P(θout|θin) over all incoming angles (Fig. 2D). The symmetric bimodal shape of P(θout) for mbo1 signals a complete loss information about the incidence angle, whereas in the case of the three puller strains, the swimmers still remember their incoming directions but not the exact values of θin. Quantitatively similar probability distributions characterize scattering laws in thicker chambers (80 and 300 μm).
is set geometrically through the length of the cilia, the diameter of the cell body, and the typical distance of the latter from the surface at the moment of departure (Movies S3, S4, and S5). For shf1, the cell body can come closer to the wall than for the WT, and the resulting lubrication forces may also affect the escape dynamics. The relatively larger spread in the distribution of scattering angles for shf1 is compatible with previous observations of stronger intrinsic fluctuations in shorter flagella (34). By contrast, the pusher-mutant mbo1 generally remains close to the surface, θout ∼ 0 or θout ∼ π (Movie S6). The differences between the scattering laws of the four different strains are also clearly evident from the mean scattering distributions P(θout), obtained by averaging P(θout|θin) over all incoming angles (Fig. 2D). The symmetric bimodal shape of P(θout) for mbo1 signals a complete loss information about the incidence angle, whereas in the case of the three puller strains, the swimmers still remember their incoming directions but not the exact values of θin. Quantitatively similar probability distributions characterize scattering laws in thicker chambers (80 and 300 μm).
Optimal Rectification of Algal Locomotion in Microfluidic Ratchets.
Knowledge of flagella-induced scattering can be used to design microscopic obstacles that will passively guide the random swimming of microorganisms in a desired direction (Fig. 3). The design principles for mbo1 mutants and spermatozoa (9) are similar to those for E. coli (7, 8), as these species glide along surfaces after collision. The challenge is to find the optimal rectification geometry for puller organisms, which scatter at a finite angle θout > 0 off solid surfaces. To demonstrate the optimization procedure, we focus on the WT Chlamydomonas strain, which has the peak angle 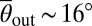 , and consider wedge-shaped obstacles, like those successfully used in bacterial rectification (7, 8) (Fig. 3B). Optimal rectification requires maximizing the ratio jF/jB between the algae currents in forward and backward directions. The diffusive backward flux jB is driven by gradients in the algal concentrations across the obstacle rows (Fig. 3C) and can be minimized by decreasing the gap distance dG between neighboring obstacles relative to the effective width dB of a single barrier, dG ≪ dB (Fig. 3B). At the same time, the forward current jF can be maximized by adjusting the wedge angle α of the barriers to exploit secondary scattering (Fig. 3B). Assuming a fixed angle θout, basic geometric considerations yield the criterion 2α + θout ≤ π/2 for secondary scattering in the forward direction. Thus, for ideal deterministic scatterers with
, and consider wedge-shaped obstacles, like those successfully used in bacterial rectification (7, 8) (Fig. 3B). Optimal rectification requires maximizing the ratio jF/jB between the algae currents in forward and backward directions. The diffusive backward flux jB is driven by gradients in the algal concentrations across the obstacle rows (Fig. 3C) and can be minimized by decreasing the gap distance dG between neighboring obstacles relative to the effective width dB of a single barrier, dG ≪ dB (Fig. 3B). At the same time, the forward current jF can be maximized by adjusting the wedge angle α of the barriers to exploit secondary scattering (Fig. 3B). Assuming a fixed angle θout, basic geometric considerations yield the criterion 2α + θout ≤ π/2 for secondary scattering in the forward direction. Thus, for ideal deterministic scatterers with 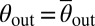 , one would expect maximal rectification for
, one would expect maximal rectification for 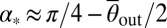 , yielding α* ∼ 37° for WT Chlamydomonas. This estimate should, however, be viewed as an upper bound for the optimal opening angle, because the measured scattering distributions P(θout) have large-angle tails (Fig. 2D) that shift the optimal α-value to smaller angles.
, yielding α* ∼ 37° for WT Chlamydomonas. This estimate should, however, be viewed as an upper bound for the optimal opening angle, because the measured scattering distributions P(θout) have large-angle tails (Fig. 2D) that shift the optimal α-value to smaller angles.
We tested this prediction in numerical simulations of a minimal 2D model and by performing quasi-2D microfluidic experiments (Fig. 3). In our simulations (see Materials and Methods for details), microswimmers are represented by noninteracting point particles whose motion captures the main observed features of Chlamydomonas trajectories. Each particle moves ballistically at a constant speed V, performs random turns after exponentially distributed run-periods (15) with persistence time τ ∼ 1.5 s, and scatters from boundaries at a random angle θout. Both V and θout are drawn from distributions that mimic the experimentally measured speed distributions and scattering laws (Materials and Methods). The value of τ is consistent with estimates from the mean square displacement of WT Chlamydomonas, as measured in the quasi-2D microfluidic chambers (height, 25 μm) used for the rectification experiments (Fig. 3A). Compared with 3D chambers, the typical swimming speed of the algae (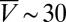 μm/s in quasi-2D chambers) is reduced by approximately a factor of 2 due to the presence of nearby no-slip surfaces, which also tend to suppress hydrodynamic interactions (35).
μm/s in quasi-2D chambers) is reduced by approximately a factor of 2 due to the presence of nearby no-slip surfaces, which also tend to suppress hydrodynamic interactions (35).
In both experiments and simulations, we considered chambers with four compartments separated by rows of wedge-shaped barriers with gaps of length dG (Fig. 3 B and C). For each wedge angle α, the arm length L of the barriers was chosen such that the length of the longest possible deterministic trajectory leading through the gap after scattering is equal to the persistence path length Lp of the algae (Fig. 3B). We then systematically quantified the rectification efficiency by scanning the (dG, α) parameter space (Fig. 3D). Here, the rectification efficiency is defined by the ratio R = 〈N4〉/〈N1〉, where 〈Ni〉 denotes the steady-state time average of the number of swimmers in compartment i. Both simulations and experiments show maximal rectification for parameter values (dG = 20 μm, α ≃ 30°) that are close to the theoretically predicted optimum (Fig. 3D). Thus, although the minimal model neglects variability in swimming behavior and hydrodynamic effects, it captures the main features of the experiments.
Generally, our numerical and experimental results support the hypothesis that rectification of microorganisms in environments with broken reflection symmetry is a universal phenomenon (36–43). The ratchets mimic Maxwell’s demon, but there is no conflict with thermodynamics due to the nonequilibrium nature of living systems.
Discussion
Our analysis of individual sperm–surface and alga–surface interactions shows that, in contrast to prevailing theoretical views, multiple flagellar contact determines the surface scattering of these eukaryotic cells. The puller-swimmer C. reinhardtii exhibits a complex ciliary contact dynamics over the course of the scattering process, which spans a large number of successive swimming strokes and leads to an almost complete erasure of memory about the incidence angle. Direct contact of the flagella with nearby walls may also be the main factor in the surface interactions of other motile algae, as the general structure of cilia is highly conserved across many eukaryotic species. The flagella-induced self-trapping of bull spermatozoa and the Chlamydomonas mutant mbo1 suggests that similar steric mechanisms could also be responsible for the long surface-residence times of other pusher-type microorganisms (4, 6), possibly even in the case of bacteria, where the role of hydrodynamic interactions as the main determinant of surface scattering has recently been questioned (4–6). With regard to future theoretical studies, our experimental results anticipate that a detailed qualitative and quantitative understanding of microbial surface interactions will require models that account for the elastic properties of eukaryotic cilia and bacterial flagella.
Finally, as an illustration of how empirically measured surface-scattering laws can be exploited to control locomotion of unicellular green algae in biotechnological applications, we demonstrated robust rectification of random swimming for WT Chlamydomonas algae. In contrast to microfluidic rectification devices for rod-like prokaryotic (pusher) swimmers (7, 8), the optimal ratchet geometry for algae (pullers) exploits secondary scattering. The proposed design can be serialized (Materials and Methods) and parallelized to facilitate large-scale microfluidic implementation. Combining the results of the scattering analysis and the subsequent rectification study suggests the possibility of integrating different ratchet geometries to create selection devices that sort microorganisms according to structure and dynamics of their propulsive appendages. More generally, the present investigation implies that suitably designed microstructured surfaces can yield new diagnostic tools to quantify function and response of eukaryotic cilia (mechano-sensing, desynchronization after contact, etc.), which may help to improve our understanding of transport processes in the respiratory or reproductive systems of higher organisms.
Materials and Methods
Sperm Sample Preparation.
Cryogenically frozen bull spermatozoa were purchased from Genus Breeding. For each experiment, a sample of 250 μL was thawed in a water bath (37 °C) for 15 s. The sample was washed three times by centrifuging at 500 × g for 5 min and resuspending the pellet in a basic medium containing 72 mM KCl, 160 mM sucrose, 2 mM Na-pyruvate, and 2 mM Na-phosphate buffer at pH 7.4 (29).
Algal Growth.
C. reinhardtii strains CC-125 WT, CC-2347 shf1-277, CC-2289 lf3-2, and CC-2679 mbo1 (The Chlamydomonas Resource Center, www.chlamy.org) were grown axenically in Tris-acetate-phosphate (TAP) medium (22) and constantly mixed on an orbital shaker in a diurnal growth chamber (KBW400; Binder). The daily cycle was as follows: 14 h at 100 μE⋅m−2⋅s−1 Photosynthetically Active Radiation (Fluora; OSRAM) and 10 h in dark at 24 °C. Cells were harvested during the exponential growth phase. To achieve large concentrations of highly motile algae (≥106 cells/cm3), cultures were centrifuged in Eppendorf tubes at 100 × g for 10 min, and subsequently, fast-swimmming algae were selected according to their ability to swim upward against gravity. Thereafter, fresh TAP was added to the cell pellet, and the tubes were placed in the diurnal chamber for at least 1 h to allow cells to recover.
Microfluidics.
Quasi-2D channels were manufactured using standard soft lithography techniques (44). The master mold was produced from SU8 2015 (MicroChem), spun to a 25-μm-thick layer, and exposed to UV light through a high-resolution mask to obtain the desired structures. The microfluidic chip containing the channels was cast from polydimethylsiloxane (PDMS) (Sylgard 184; Dow Corning) and bonded to a PDMS-covered glass slide after oxygen plasma treatment of the surfaces. Microchannels for the sperm experiments were designed such that the sperm cells could be injected in a region that was spatially separated from the observation area. In the observation area, microchannels (100 μm wide) were arranged in a zig-zag pattern with 90° corners, where the scattering events were imaged (Fig. 1 A and B). The temperature was measured by a calibrated thermistor, which was inserted into the PDMS chip 2 mm away from the observation region. To prevent adhesion of the spermatozoa to the walls of the channel, BSA at 5 mg/mL was added prior to the injection into the chamber. After injection of the sperm sample into the microchannel, motile spermatozoa could escape from the injection site into the observation region. The concentration of motile spermatozoa in this region did not exceed 1% volume fraction. In both spermatozoa and Chlamydomonas experiments, the microbial solutions were introduced through inlets that were plugged with unpolymerized PDMS afterward. This procedure prevents fluid flows through the chambers and ensures conservation of the total number of cells over the course of the experiment. In the Chlamydomonas experiments, the concentration of the algae was kept below a 2% volume fraction. For the rectification studies, each channel was subdivided into four chambers of size 2 × 1 × 0.025 mm, separated by wedge-shaped barriers (Fig. 3). We treated PDMS surfaces of the channels prior the experiments with 10% (wt/vol) polyethylene glycol (molecular weight, 8,000; Sigma) solution in water for 30 min to prevent adhesion of the algae to the walls and then flushed them gently with fresh TAP. For the given parameters and uniform initial concentration profile across each chamber, rectified steady states were achieved typically after 90 min (in the dark).
Microscopy.
To identify the swimming characteristics of individual spermatozoa and Chlamydomonas and their scattering distributions (Figs. 1 and 2), cell trajectories were reconstructed by applying a custom-made particle tracking-velocimetry algorithm to image data taken with a Nikon TE2000-U inverted microscope (10× objective, 10 fps). The flagella dynamics close to the boundary (Figs. 1 A and B and 2 A and B; Movies S1, S2, S3, S4, S5, and S6) was captured with a Fastcam SA-3 Photron camera (500–2,000 fps, 40×/NA 1.3 oil immersion and 60×/NA 1.0 water immersion objectives). For sperm, white light was used, and for Chlamydomonas, bright field illumination under red light (λ > 620 nm) was used to minimize phototaxis. Sperm-surface scattering angles θ were determined by tracking the cell body up to a distance of 70 μm from the corner. For Chlamydomonas, incidence and scattering angles θin and θout were obtained by measuring the slope of the trajectory at a distance of 20 μm in either direction from the scattering point (defined as the location at which the distance from the wall becomes minimal). In the rectification experiments, algae concentrations in the microfluidic chambers were measured by averaging intensity profiles of image data obtained with a confocal scanning microscope (Zeiss LSM 700; 5× objective). Here, the transmitted light photomultiplier tube (PMT) mode was used while exposing the chambers to laser light (639 nm) at the lowest intensity to minimize phototactic response of the algae.
Numerical Simulations.
We simulated the dynamics of n = 750 self-propelled, noninteracting point particles in a 2D box using MATLAB. Box size and ratchet geometries were chosen to match the experimental setup (Fig. 3). To account for the finite radius of Chlamydomonas, a virtual layer of thickness a = 5 μm was added to boundaries and obstacles. Particles move ballistically at a constant speed V until undergoing random turns or colliding with boundaries or obstacles. Initial particle speeds V were sampled from a gamma-distribution Γ(x; k, ν) with parameters k = 4.2 and ν = 7.3 μm/s, obtained from a best fit to the experimentally measured speed distribution. Run times between successive random turns are sampled from independent exponential distributions with mean τ. New directions after turn events are sampled uniformly from the unit circle. We simulated two types of boundary collision scenarios as simplified approximations to the experimentally observed WT scattering behavior: deterministic collisions with an outgoing angle θout = 16° and randomized forward scattering. In the latter case, the outgoing angles θout were sampled from a truncated superposition of three Gaussians distributions 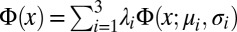 while rejecting angles θout outside of the interval (0, π). The distributions parameters were chosen as (λ1, λ2) = (0.48, 0.26), with normalization requiring λ3 = 1 − λ1 − λ2, and (μ1, μ2, μ3) = (0.43, 0.26, 3.06) and (σ1, σ2, σ3) = (0.58, 0.14, 0.55), obtained from a best fit to the WT scattering distribution. In the deterministic case, our simulations confirmed maximal rectification for α ∼ 37°. Simulation results in Fig. 3D show averages from three runs with τ = 1.5 s for randomized scattering. Generally, numerical results were found to be qualitatively robust under moderate variations of the model parameters, but the optimal wedge angle is sensitive to changes in the scattering distribution.
while rejecting angles θout outside of the interval (0, π). The distributions parameters were chosen as (λ1, λ2) = (0.48, 0.26), with normalization requiring λ3 = 1 − λ1 − λ2, and (μ1, μ2, μ3) = (0.43, 0.26, 3.06) and (σ1, σ2, σ3) = (0.58, 0.14, 0.55), obtained from a best fit to the WT scattering distribution. In the deterministic case, our simulations confirmed maximal rectification for α ∼ 37°. Simulation results in Fig. 3D show averages from three runs with τ = 1.5 s for randomized scattering. Generally, numerical results were found to be qualitatively robust under moderate variations of the model parameters, but the optimal wedge angle is sensitive to changes in the scattering distribution.
Data Resources and SI Movie Information.
A MATLAB script of the source code, simulation data and experimental raw data, and additional experimental movies can be downloaded from http://damtp.cam.ac.uk/user/gold/datarequests.html. Scale bars are 20 μm in Movies S1 and S2; 10 μm in Movies S3, S4, S5, and S6; and 0.5 mm in Movie S7.
Serialization (Markov Model).
At low-to-intermediate algal volume fractions, as realized in our experiments, the dynamics of the Chlamydomonas population on the microfluidic chip can be described by a Markov model (7). This approach allows estimation of the total rectification with an increasing number of compartments. Assuming that the chip consists of i = 1,…, N identical compartments (Fig. 2D), the time evolution of the relative concentration pi(t) of algae in the ith compartment is governed by
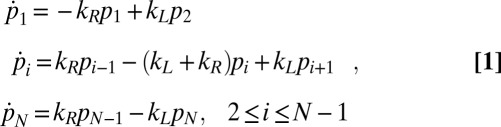 |
where kL/R denote the rates for transitions to the left/right neighboring compartment (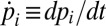 and
and 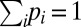 ). The stationary distribution
). The stationary distribution 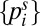 , corresponding to the eigenvector of the eigenvalue λ = 0 of the transition matrix Kij, defined by
, corresponding to the eigenvector of the eigenvalue λ = 0 of the transition matrix Kij, defined by 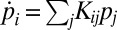 , is obtained as
, is obtained as
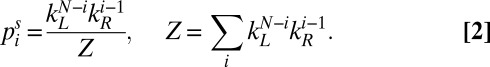 |
The effective rates kL/R can be estimated by fitting 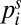 to the experimentally measured stationary distribution. Assuming kR > kL, Eq. 2 implies that rectification increases with the number of chambers N as
to the experimentally measured stationary distribution. Assuming kR > kL, Eq. 2 implies that rectification increases with the number of chambers N as 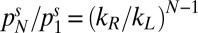 . Assuming detailed balance and diffusive backward flux suggests that kR/kL = pR(dG + dB)/dG, where pR is probability that an alga is guided through the gap after having entered the barrier region. From our experimental data, we estimate pR ∼ 0.3.
. Assuming detailed balance and diffusive backward flux suggests that kR/kL = pR(dG + dB)/dG, where pR is probability that an alga is guided through the gap after having entered the barrier region. From our experimental data, we estimate pR ∼ 0.3.
Supplementary Material
Acknowledgments
We thank Howard Berg and Paul Chaikin for invaluable discussions and Elgin Columbo for providing the spermatozoa. This work was supported in part by the Biotechnology and Biological Sciences Research Council (BBSRC), an Engineering and Physical Sciences Research Council (EPSRC) postdoctoral fellowship (to M.P.), and European Research Council (ERC) Advanced Investigator Grant 247333 (to R.E.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210548110/-/DCSupplemental.
References
- 1.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30(2):285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 2.Quill TA, et al. Hyperactivated sperm motility driven by CatSper2 is required for fertilization. Proc Natl Acad Sci USA. 2003;100(25):14869–14874. doi: 10.1073/pnas.2136654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berke AP, Turner L, Berg HC, Lauga E. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys Rev Lett. 2008;101(3):038102. doi: 10.1103/PhysRevLett.101.038102. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Tang JX. Accumulation of microswimmers near a surface mediated by collision and rotational Brownian motion. Phys Rev Lett. 2009;103(7):078101. doi: 10.1103/PhysRevLett.103.078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, et al. Accumulation of swimming bacteria near a solid surface. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;84(4 Pt 1):041932. doi: 10.1103/PhysRevE.84.041932. [DOI] [PubMed] [Google Scholar]
- 6.Drescher K, Dunkel J, Cisneros LH, Ganguly S, Goldstein RE. Fluid dynamics and noise in bacterial cell-cell and cell-surface scattering. Proc Natl Acad Sci USA. 2011;108(27):10940–10945. doi: 10.1073/pnas.1019079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galajda P, Keymer J, Chaikin P, Austin RH. A wall of funnels concentrates swimming bacteria. J Bacteriol. 2007;189(23):8704–8707. doi: 10.1128/JB.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert G, Liao D, Austin RH. Collective escape of chemotactic swimmers through microscopic ratchets. Phys Rev Lett. 2010;104(16):168102. doi: 10.1103/PhysRevLett.104.168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denissenko P, Kantsler V, Smith DJ, Kirkman-Brown J. Human spermatozoa migration in microchannels reveals boundary-following navigation. Proc Natl Acad Sci USA. 2012;109(21):8007–8010. doi: 10.1073/pnas.1202934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasala BA, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J. 2010;8(6):719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godman J, Balk J. Genome analysis of Chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics. 2008;179(1):59–68. doi: 10.1534/genetics.107.086033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stripp ST, et al. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc Natl Acad Sci USA. 2009;106(41):17331–17336. doi: 10.1073/pnas.0905343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eroglu E, Melis A. Photobiological hydrogen production: Recent advances and state of the art. Bioresour Technol. 2011;102(18):8403–8413. doi: 10.1016/j.biortech.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Mayfield S, Wong PK. Forum. Chemical engineering: Fuel for debate. Nature. 2011;476(7361):402–403. doi: 10.1038/476402a. [DOI] [PubMed] [Google Scholar]
- 15.Polin M, Tuval I, Drescher K, Gollub JP, Goldstein RE. Chlamydomonas swims with two “gears” in a eukaryotic version of run-and-tumble locomotion. Science. 2009;325(5939):487–490. doi: 10.1126/science.1172667. [DOI] [PubMed] [Google Scholar]
- 16.Drescher K, Goldstein RE, Michel N, Polin M, Tuval I. Direct measurement of the flow field around swimming microorganisms. Phys Rev Lett. 2010;105(16):168101. doi: 10.1103/PhysRevLett.105.168101. [DOI] [PubMed] [Google Scholar]
- 17.Guasto JS, Johnson KA, Gollub JP. Oscillatory flows induced by microorganisms swimming in two dimensions. Phys Rev Lett. 2010;105(16):168102. doi: 10.1103/PhysRevLett.105.168102. [DOI] [PubMed] [Google Scholar]
- 18.DiLuzio WR, et al. Escherichia coli swim on the right-hand side. Nature. 2005;435(7046):1271–1274. doi: 10.1038/nature03660. [DOI] [PubMed] [Google Scholar]
- 19.Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245(5425):380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 20.Rüffer U, Nultsch W. Flagellar photoresponses of Chlamydomonas cells held by micropipettes: II. Change in flagellar beat pattern. Cell Motil Cytoskeleton. 1991;18:269–278. [Google Scholar]
- 21.Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 22.Harris EH. The Chlamydomonas Sourcebook. Oxford, UK: Academic Press; 2009. [Google Scholar]
- 23.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiu K, Nakayama Y, Iida H, Sokabe M, Yoshimura K. Mechanoreception in motile flagella of Chlamydomonas. Nat Cell Biol. 2011;13(5):630–632. doi: 10.1038/ncb2214. [DOI] [PubMed] [Google Scholar]
- 25.Melis A, Happe T. Hydrogen production. Green algae as a source of energy. Plant Physiol. 2001;127(3):740–748. [PMC free article] [PubMed] [Google Scholar]
- 26.Esper B, Badura A, Rögner M. Photosynthesis as a power supply for (bio-)hydrogen production. Trends Plant Sci. 2006;11(11):543–549. doi: 10.1016/j.tplants.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell. 2010;9(4):486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das D, Veziroglu TN. Advances in biological hydrogen production processes. Int J Hydrogen Energy. 2008;33:6046–6057. [Google Scholar]
- 29.Rikmenspoel R. Movements and active moments of bull sperm flagella as a function of temperature and viscosity. J Exp Biol. 1984;108:205–230. doi: 10.1242/jeb.108.1.205. [DOI] [PubMed] [Google Scholar]
- 30.Riedel-Kruse IH, Hilfinger A, Howard J, Jülicher F. How molecular motors shape the flagellar beat. HFSP J. 2007;1(3):192–208. doi: 10.2976/1.2773861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuchka MR, Jarvik JW. Short-flagella mutants of Chlamydomonas reinhardtii. Genetics. 1987;115(4):685–691. doi: 10.1093/genetics/115.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barsel S-E, Wexler DE, Lefebvre PA. Genetic analysis of long-flagella mutants of Chlamydomonas reinhardtii. Genetics. 1988;118(4):637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayly PV, et al. Propulsive forces on the flagellum during locomotion of Chlamydomonas reinhardtii. Biophys J. 2011;100(11):2716–2725. doi: 10.1016/j.bpj.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldstein RE, Polin M, Tuval I. Emergence of synchronized beating during the regrowth of eukaryotic flagella. Phys Rev Lett. 2011;107(14):148103. doi: 10.1103/PhysRevLett.107.148103. [DOI] [PubMed] [Google Scholar]
- 35.Liron N, Mochon S. Stokes flow for stokeslet between to parallel flat plates. J Eng Math. 1976;10:287–303. [Google Scholar]
- 36.Wan MB, Olson Reichhardt CJ, Nussinov Z, Reichhardt C. Rectification of swimming bacteria and self-driven particle systems by arrays of asymmetric barriers. Phys Rev Lett. 2008;101(1):018102. doi: 10.1103/PhysRevLett.101.018102. [DOI] [PubMed] [Google Scholar]
- 37.Sokolov A, Apodaca MM, Grzybowski BA, Aranson IS. Swimming bacteria power microscopic gears. Proc Natl Acad Sci USA. 2010;107(3):969–974. doi: 10.1073/pnas.0913015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Leonardo R, et al. Bacterial ratchet motors. Proc Natl Acad Sci USA. 2010;107(21):9541–9545. doi: 10.1073/pnas.0910426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Astumian RD, Hänggi P. Brownian motors. Phys Today. 2002;55(11):33–39. [Google Scholar]
- 40.Hänggi P, Marchesoni F. Artificial Brownian motors: Controlling transport on the nanoscale. Rev Mod Phys. 2009;81:387–442. [Google Scholar]
- 41.Grimm A, Stark H, van der Maarel JRC. Model for a Brownian ratchet with improved characteristics for particle separation. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79(6 Pt 1):061102. doi: 10.1103/PhysRevE.79.061102. [DOI] [PubMed] [Google Scholar]
- 42.Grimm A, Stark H. Hydrodynamic interactions enhance the performance of Brownian ratchets. Soft Matter. 2011;7:3219–3227. [Google Scholar]
- 43.Reimann P. Brownian motors: Noisy transport far from equilibrium. Phys Rep. 2002;361:57–265. [Google Scholar]
- 44.Xia YN, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.