Abstract
Conversion of CO2 for the synthesis of chemicals by photosynthetic organisms is an attractive target for establishing independence from fossil reserves. However, synthetic pathway construction in cyanobacteria is still in its infancy compared with model fermentative organisms. Here we systematically developed the 2,3-butanediol (23BD) biosynthetic pathway in Synechococcus elongatus PCC7942 as a model system to establish design methods for efficient exogenous chemical production in cyanobacteria. We identified 23BD as a target chemical with low host toxicity, and designed an oxygen-insensitive, cofactor-matched biosynthetic pathway coupled with irreversible enzymatic steps to create a driving force toward the target. Production of 23BD from CO2 reached 2.38 g/L, which is a significant increase for chemical production from exogenous pathways in cyanobacteria. This work demonstrates that developing strong design methods can continue to increase chemical production in cyanobacteria.
Keywords: metabolic engineering, synthetic biology, biofuel, renewable energy
Amid rising global energy demands and pressing environmental issues, interest is growing in the production of fuels and chemicals from renewable resources. Petroleum consumption reached 37.1 quadrillion BTU in the United States in 2008, of which a large majority (71%) was liquid fuel in the transportation sector. Petroleum and natural gas account for 99% of the feedstocks for chemicals, such as plastics, fertilizers, and pharmaceuticals in the chemical industry (1). Considering rapidly increasing world population and exhaustion of fossil fuels, the development of sustainable processes for energy and carbon capture to produce fuels and chemicals is crucial for human society.
Energy and carbon capture by cyanobacteria is also directed toward mitigating increasing atmospheric CO2 concentrations. According to the US Energy Information Administration (2), world energy-related CO2 emissions in 2006 were 29 billion metric tons, which is an increase of 35% from 1990. Accelerating accumulation of atmospheric CO2 is not only a result of increased emissions from world growth and intensifying carbon use, but also from a possible attenuation in the efficiency of the world’s natural carbon sinks (3). As a result, atmospheric levels of CO2 have increased by ∼25% over the past 150 y and it has become increasingly important to develop new technologies to reduce CO2 emissions. Many creative solutions have been proposed and argued for carbon capture, each with varied environmental side-effects and costs (4). Sequestration by photosynthetic microorganisms in which CO2 is biologically converted to valuable chemicals is an important addition to the toolbox for overall capture of CO2 (5–7).
Photosynthetic microorganisms, including cyanobacteria, are currently being engineered for platforms to convert solar energy to biochemicals renewably (5–7). These microorganisms possess many advantages over traditional terrestrial plants with regard to biochemical production. For example, the photosynthetic efficiency of photosynthetic microorganisms is higher than plants, and photosynthetic microorganisms can be cultivated in locations that do not compete with traditional agricultural crops (8). Cyanobacteria are collectively responsible for almost 50% of global photosynthesis and are found in a wide range of environments (9). Although cyanobacteria have many similar features with algae in this context, many cyanobacterial species feature more simple genetic structures and faster growth rates (5). As a result, genetic engineering methods for cyanobacteria are more advanced than those for algae (10–12).
Cyanobacteria have the biochemical machinery required to fix CO2, but lack the critical components to generate fuels and chemicals efficiently. An increased understanding of cellular systems enables us to construct novel systems using synthetic biology, assembling the components and control systems into new combinations (6, 7, 13). This approach can be applied to produce valuable chemicals, that the cyanobacteria host strains do not produce naturally, by constructing new biosynthetic pathways (6). Recently, cyanobacteria have been engineered to produce various chemicals, including isobutyraldehyde (14), isobutanol (14), 1-butanol (15), ethylene (16), isoprene (17), acetone (18), fatty acids (19), and fatty alcohols (20), through exogenous biosynthetic pathways. This approach in cyanobacteria is significantly less developed compared with a model organism such as Escherichia coli. However, results in E. coli cannot be directly translated into cyanobacteria. For example, an engineered E. coli strain containing the 1-butanol pathway produced more than 30 g/L 1-butanol (21), but the cyanobacterial strain with the same pathway produced only trace amounts of 1-butanol (22). We thus seek to establish an efficient method for biosynthetic pathway construction in cyanobacteria.
In this work, we aim to establish that matching the choice of target chemicals and pathways to the chosen photosynthetic production host is beneficial for cyanobacterial chemical production. First, low toxicity toward cells is important. Isobutyraldehyde production in Synechococcus elongatus PCC7942 has achieved the highest production rate in the literature; however, because of high toxicity of isobutyraldehyde this system requires constant isolation of the product to achieve high productivity (14). Second, because photosynthesis produces oxygen, activities of oxygen-sensitive enzymes are usually problematic in cyanobacteria, with the result that pathways such as 1-butanol and acetone from anaerobic organisms are inefficient in cyanobacteria (18, 22). Third, chemicals that are secreted from cells without overexpression of transporters or additional genetic modifications to cells are ideal to make the system simple and inexpensive. Finally, providing irreversible reactions, which create a driving force to direct flux toward the target chemicals, is important for construction of efficient pathways (15, 21, 23).
In this work we used exogenous 2,3-butanediol (23BD) production (Fig. 1) in S. elongatus as a model system and followed a systematic approach in which various enzymes were compared both in E. coli and S. elongatus to optimize chemical production. Microbial fermentation of 23BD has been under investigation for many years as an alternate route for chemical production (24, 25). 23BD can be converted by dehydrogenation to methyl ethyl ketone, which is a liquid fuel additive and useful industrial solvent (26). Furthermore, the catalytic conversion of 23BD to 1,3-butadiene, which is a precursor for a diversity of polymer and copolymer materials, has been well established (27). 23BD has also been used in the manufacturing of plasticizers, inks, fumigants, and explosives (28). Concurrently, methods for improving product recovery efficiency, which is an important determinant of the total production cost and a major limitation of biochemical production from photosynthetic organisms (29, 30), have been well-established (24, 31). This current work will provide a stronger understanding of the potential of cyanobacteria as a platform for production of valuable biochemicals, such as 23BD, and also provide important fundamental insight to facilitate future progress along such lines.
Fig. 1.
The pathway for acetoin and 23BD production in S. elongatus PCC7942. The acetoin/23BD production pathway contains three enzymatic steps from pyruvate.
Results
Exogenous 23BD Exhibits Low Toxicity in S. elongatus.
To increase the titer and duration of chemical production, low toxicity or constant removal of the product is necessary. Because constant removal and purification of small concentrations during production is not cost-effective on an industrial scale, it was a prerequisite of this study that the chemical target be tolerated at an acceptable volume of greater than 1% (10 g/L) by production strains. To evaluate acetoin and 23BD toxicity, we tested the growth of S. elongatus over 72 h in the presence of 23BD or acetoin (Fig. 2). Growth rates between 0 and 24 h decreased by 18% and 42% in the presence of 0.1 and 0.2 g/L acetoin, respectively (Fig. 2A). Growth in the presence of 0.2 and 1.0 g/L acetoin stopped after 24 h, but growth in the absence of acetoin reached a plateau at 48 h (Fig. 2A). These results indicate acute toxicity for this precursor. This finding is comparable to isobutyraldehyde and isobutanol, which prevent growth of S. elongatus at 1 g/L (14). Conversely, growth rates between 0 and 24 h in the presence of 10 and 30 g/L 23BD decreased by only 2.2% and 8.9%, respectively (Fig. 2B), surpassing our benchmark goal for product tolerance. Growth in the presence of 30 g/L 23BD stopped at 24 h, but that in the presence of 10 g/L 23BD reached a plateau at 48 h, similar to that seen in the absence of 23BD (Fig. 2B). These results indicate that 23BD is a suitable target for high titer and long-term cyanobacterial production, as long as high flux through acetoin can be maintained to prevent accumulation of the toxic intermediate.
Fig. 2.
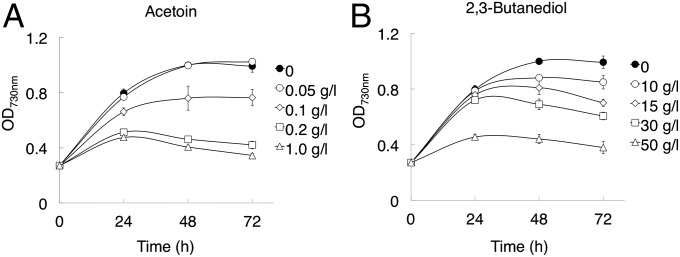
Effect of acetoin and 23BD on growth. (A) Time course for the growth of S. elongatus in the absence of acetoin (●) or in the presence of 0.05 g/L (○), 0.1 g/L (◇), 0.2 g/L (□), and 1.0 g/L (△) acetoin. (B) Time course for the growth of S. elongatus in the absence of 23BD (●) or in the presence of 10 g/L (○), 15 g/L (◇), 30 g/L (□), and 50 g/L (△) 23BD. Cells were incubated in BG-11 containing 50 mM NaHCO3 at 30 °C. Error bars are SD (n = 3).
Construction of the Acetoin Biosynthetic Pathway.
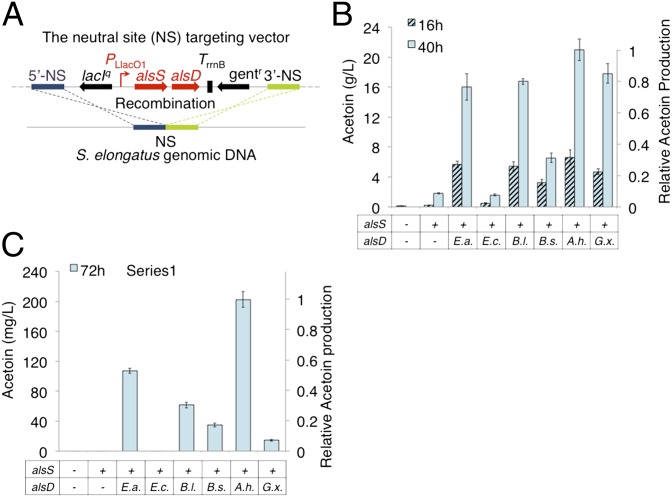
Acetoin can be produced by the decarboxylation of 2-acetolactate. In this pathway (Fig. 1) two pyruvate molecules are converted into 2-acetolactate by acetolactate synthase (ALS) encoded by alsS. 2-Acetolactate is then decarboxylated to yield acetoin by 2-acetolactate decarboxylase (ALDC) encoded by alsD. Pyruvate, the source of carbon for the pathway, is produced naturally through the fixation of three CO2 molecules in the Calvin-Benson-Bassham cycle (32). Conversion of pyruvate to 2-acetolactate occurs naturally during valine/leucine biosynthesis, albeit in low amounts (33). Previously alsS, which encodes ALS from Bacillus subtilis was overexpressed to increase carbon flux to 2-acetolactate for the production of isobutyraldehyde and was reported to have relatively high activity (14). To identify strong ALDC candidates, we used the bioinformatics tool, Braunschweig Enzyme Database (34) and a comprehensive literature review. We limited our search to O2-insensitive enzymes and looked for reports of strong acetoin production. We were further restricted by the need to match presequencing literature reports to chronologically consistent strain names, which now match currently available gene sequences. Based on these criteria six alsD were selected (Tables S1–S3).
To test acetoin production each alsD was overexpressed with alsS (B. subtilis) under the isopropyl-β-D-thiogalactoside (IPTG) -inducible promoter PLlacO1 (35) in E. coli (Fig. 3A). The cells were cultured in modified M9 medium (Methods), containing 50 g/L of glucose, at 30 °C for 16 and 40 h. A control strain expressing only alsS (B. subtilis) produced 0.2 g/L acetoin indicating that 2-acetolactate decomposes to acetoin in small amounts, which is consistent with previous observations (36, 37). When alsD was coexpressed more than 20 g/L of acetoin was produced, indicating that autodecarboxylation is not a major contributor to 2-acetolactate conversion (Fig. 3B). All ALDC except that from Enterobacter cloacae were active in E. coli, and displayed a pattern of activity that was consistent through 16 h and 40 h of production (Fig. 3B). The strain expressing alsD from Aeromonas hydrophila was the highest producer (21.0 g/L), followed by the strains expressing alsD from Gluconacetobacter xylinus (17.8 g/L), alsD from Bacillus licheniformis (16.7 g/L), and alsD from Enterobacter aerogenes (16.0 g/L)(Fig. 3B). The strain expressing codon optimized alsD (B. subtilis), which is the natural gene partner to the alsS (B. subtilis) used in the production operon, produced the least acetoin (6.6 g/L), which demonstrates that native pathways do not necessarily maintain their integrity when transferred to new hosts; screening of multiple candidates is necessary to find optimal genes for pathway optimization in each new host (Fig. 3B).
Fig. 3.
Acetoin production in modified strains. (A) Schematic representation of recombination to integrate alsS and alsD into the S. elongatus chromosome. (B) Acetoin production in modified E. coli. Cells were grown for 16 h (hashed) and 40 h (blue). (C) Acetoin production in modified S. elongatus. Cells were grown for 72 h. alsS indicates inclusion (+) of alsS (B. subtilis) or absence (−) of the gene. alsD indicates the source organism for alsD (Table S3). A. h., A. hydrophila; B. l., B. licheniformis; B. s., B. subtilis; E. a., E. aerogenes; E. c., E. cloacae; G. x., G. xylinus.
We obtained an estimate of activity for ALDC by coexpression with ALS in crude lysate from E. coli (Fig. S1). Our results show a range of activity similar to that of productivity in vivo. A significant lead in activity for ALDC (A. hydrophila) (369 nmol⋅min−1⋅mg−1) was seen relative to the moderate activity of ALDC (E. aerogenes, B. licheniformis, and G. xylinus, 152, 152, and 31 nmol⋅min−1⋅mg−1, respectively), and negligible activity of ALDC (E. cloacae and B. subtilis) (Fig. S1). Cell extracts of a strain expressing alsS in the absence of alsD, showed a background rate of 19 nmol⋅min−1⋅mg−1 for spontaneous 2-acetolactate conversion under assay conditions.
Acetoin Production in S. elongatus from CO2.
Following our screening strategy for pathway optimization, ALDC activity was compared in vivo in the photosynthetic cell environment of S. elongatus, based on production of acetoin during heterologous alsS and alsD expression. Each strain was cultured in 125-mL shake flasks with 25 mL BG-11 containing 50 mM NaHCO3 in constant light (55 μE⋅s−1⋅m−2) at 30 °C for 72 h (Fig. 3C). Strains expressing alsD from E. aerogenes, B. licheniformis, B. subtilis, A. hydrophila, and G. xylinus, produced 108 mg/L, 62 mg/L, 35 mg/L, 203 mg/L, and 14 mg/L, respectively (Fig. 3C). Control strains, and the strain expressing alsD (E. cloacae), did not produce a measureable amount of acetoin in this host. Based on these results, we had two alsD (from E. aerogenes and A. hydrophila) capable of moderate and high production of acetoin respectively in S. elongatus. To avoid excessive acetoin toxicity we chose alsD (E. aerogenes) as a starting point for secondary alcohol dehydrogenase (sADH) analysis.
Constructing the 23BD Biosynthetic Pathway.
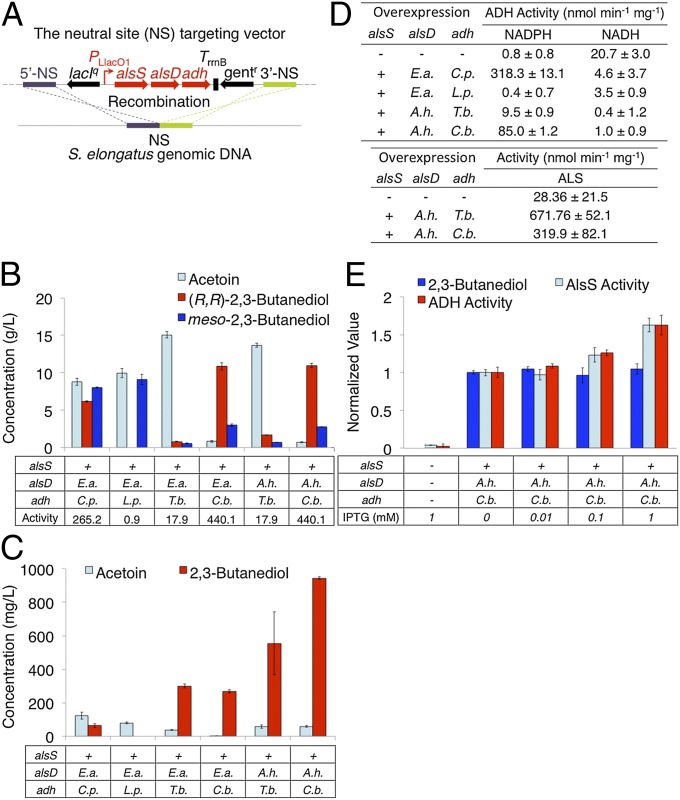
Acetoin can be reduced by an sADH to produce 23BD (Fig. 1). Identification of strong sADH candidates followed the same method used for ALDC; however, in addition to low oxygen sensitivity, two more criteria were added. First, we limited our search to NADPH-dependent sADH, as this cofactor is expected to have higher bioavailability during photosynthesis (22). Second, reduction of acetoin by sADH is a diastereoselective reaction, allowing us to choose enzymes to install either an R or S stereocenter. Two NADPH using sADH with R-installing reaction sites had been characterized previously in E. coli (38). The availability of sADH with S-installing reaction sites and NADPH as a cofactor, however, was limited. In the end we chose four sADH, two with R-installing reaction sites, and two with S-installing reaction sites (Table S3). Plasmids were constructed harboring alsS (B. subtilis), alsD (E. aerogenes), and each of the four adh under PLlacO1 (Fig. 4A) (Methods).
Fig. 4.
23BD production in modified strains. (A) Schematic representation of recombination to integrate alsS, alsD, and adh into the S. elongatus chromosome. (B) Production in modified E. coli. Cells were grown for 40 h. Bars represent acetoin (light blue), (R,R)-23BD (red), and meso-23BD (dark blue). alsS indicates inclusion (+) of alsS (B. subtilis). alsD and adh rows indicate the source organism for the gene (Table S3). Activity is that of sADH expressed in E. coli and measured in cell extract (nmol NADPH min−1⋅mg−1). (C) 23BD production in modified S. elongatus. Cells were grown for 72 h. (D) Specific activities of ALS and sADH in cell extracts from modified S. elongatus strains. (E) Effect of IPTG in modified S. elongatus. Cells were grown for 72 h after induction with the specified concentration of IPTG. Values are normalized to those from uninduced cultures. Bars represent 23BD production (dark blue), activity of sADH protein measured with NADPH as cofactor (red), and activity of ALS protein (light blue). Error indicates SD (n = 3). A. h., A. hydrophila; B. l., B. licheniformis; B. s., B. subtilis; C. p., C. parapsilosis; C. b., C. beijerinckii; E. a., E. aerogenes; E. c., E. cloacae; G. x., G. xylinus; L. p., L. pseudomesenteroides; T. b., T. brockii.
The resulting E. coli strains were cultured in modified M9 medium, containing 50 g/L glucose, at 30 °C for 40 h (Fig. 4B). The concentration of acetoin remained high for three of the four strains indicating that sADH activity is limiting in this cell environment. The fourth strain, expressing adh (Clostridium beijerinckii), maintained a relatively low acetoin concentration (less than 6% of total production) and produced 13.8 g/L total 23BD as a mixture of (R,R)-23BD and meso-23BD stereoisomers forming 74% and 21% of total production, respectively (Fig. 4B). The strains expressing adh (Thermoanaerobacter brockii) and adh (Candida parapsilosis) produced 2.4 g/L and 14.2 g/L 23BD, respectively, with both isomers formed in roughly equal amounts in each. High stereoselectivity was achieved in the strain expressing adh (Leuconostoc pseudomesenteroides), which produced 9.1 g/L meso-23BD exclusively. Enzyme activities measured from crude cell lysate were high for both sADH (C. parapsilosis) and sADH (C. beijerinckii), at 265 and 440 nmol⋅min−1⋅mg−1, respectively, when excess substrate was used (Fig. 4B). However, for the strain expressing adh (C. parapsilosis), accumulation of acetoin in the supernatant during production indicates that the enzyme turnover rate at the substrate concentrations present within the cell is comparable or slower than the rate of secretion. Relatively low activity of sADH (T. brockii) was consistent with accumulation of acetoin in the strain, indicating that sADH activity is a bottleneck for production in this E. coli strain (Fig. 4B). The major 23BD product of each of the adh expressing strains except the strain containing adh (C. parapsilosis) matched the stereochemistry predicted by previous characterization (38–40).
Production of 23BD in S. elongatus from CO2.
To screen the differences in 23BD productivity in S. elongatus, each of the plasmids used for 23BD production in E. coli was used for transformation of S. elongatus (Methods and Fig. S2). The engineered strains were cultured in 125-mL baffled shake flasks with 25 mL BG-11 containing 50 mM NaHCO3 in constant light (55 μE⋅s−1⋅m−2) at 30 °C.
23BD production was detected in three of four S. elongatus strains (Fig. 4C). Measurement of sADH performance in S. elongatus was made by comparison of acetoin and 23BD concentrations after 72 h of growth, using the less active ALDC (E. aerogenes) to lower toxicity in cases when acetoin conversion was low. The strain expressing adh (T. brockii) produced 301 mg/L (R,R)-23BD with trace amounts of meso-23BD but also allowed for accumulation of acetoin (Fig. 4C). The strain expressing adh (C. beijerinckii) produced 270 mg/L (R,R)-23BD (major) and undetectable levels of acetoin indicating high flux through the intermediate. The strain expressing adh (C. parapsilosis) produced 65 mg⋅L−1 23BD with meso-23BD as the predominant product and accumulated toxic levels of acetoin. The remaining S-installing sADH (L. pseudomensenteroides) was not active in S. elongatus, resulting only in accumulation of acetoin. Enzyme activities measured in crude cell lysate isolated during production showed a roughly ninefold higher activity for sADH (C. beijerinckii) than for sADH (T. brockii), 85.0 and 9.5 nmol⋅min−1⋅mg−1, respectively, which could explain the accumulation of acetoin in the less active strain (Fig. 4 C and D). Activity for sADH (C. parapsilosis) was roughly fivefold higher than sADH (C. beijerinckii); however, low production and acetoin accumulation was observed, similar to the result in E. coli. Enzyme activity could not be detected for the strain expressing adh (L. pseudomesenteroides), indicating that the sADH is responsible for lack of production (Fig. 4 C and D).
The two sADH with highest production and lowest acetoin accumulation were further tested with the more active ALDC encoded by alsD (A. hydrophila). Both strains increased production, yielding 568 mg⋅L−1 from adh (T. brockii) and 952 mg⋅L−1 from adh (C. beijerinckii), the latter of which is threefold higher than production with alsD (E. aerogenes) over 72 h (Fig. 4C). Both strains also showed increased acetoin concentrations, although neither reached toxic levels, accumulating 59 mg⋅L−1 and 61 mg⋅L−1, respectively.
To have an inducible expression system, lacIq, which encodes the E. coli lac repressor, was cloned upstream of PLlacO1 (Fig. 3A). The efficiency of LacI repression in the S. elongatus strain containing alsS (B. subtilis), alsD (A. hydrophila), and adh (C. beijerinckii) was investigated by testing 23BD production with or without various concentrations of IPTG (Fig. 4E). Interestingly, 23BD production without IPTG was similar to that with IPTG (Fig. 4E), suggesting that PLlacO1 was leaky in this construct. The promoter and coding region of lacIq were verified by Sanger sequencing. This phenomena has been reported with other IPTG-inducible promoters in S. elongatus PCC7942 (41–42) and Synechocystis sp. PCC6803 (43). However, ALS and sADH activities increased with elevated concentrations of IPTG (Fig. 4E). ALS and sADH activities both increased 1.2- and 1.6- fold with 0.1 and 1.0 mM IPTG, respectively, over that of cultures without IPTG induction (Fig. 4E). These results indicate that LacI repressed PLlacO1 in S. elongatus and IPTG induced expression from PLlacO1. From this finding we infer that leaky expression of genes under PLlacO1 without IPTG is significant, even though LacI repression is functioning.
To estimate the amount of installed enzyme translated in engineered S. elongatus, cell extracts of the engineered strain containing alsS (B. subtilis), alsD (A. hydrophila), and adh (C. beijerinckii) and nonengineered wild-type were analyzed with SDS/PAGE (Fig. S3). The bands of ALS were detected only in samples from the engineered strain. The amounts of ALS are estimated at about 0.21% without IPTG, and 0.28%, 0.37%, and 0.34% with 0.01, 0.1, and 1 mM IPTG, respectively, of total protein. The bands of ALDC and sADH were not visible on the gel, corresponding to less than 0.02% of total protein.
Long-Term Production of 23BD in S. elongatus.
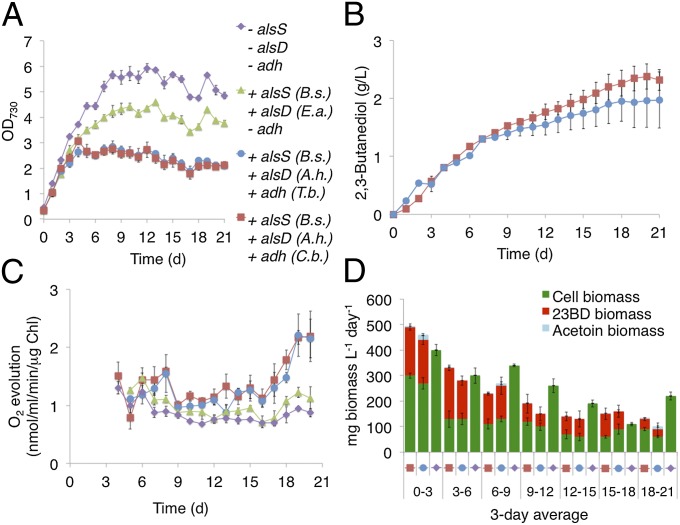
We verified stability of the highest producing strains by maintaining continuous production in 25-mL cultures at 30 °C in the presence of constant light (Fig. 5) (Methods). The strain containing adh (C. beijerinckii) reached a total yield of 2.38 g/L (R,R)-23BD and a maximum production rate of 9,847 μg⋅L−1⋅h−1 (3 d average) (Fig. 5B). Production was sustained for 21 d. The strain containing adh (T. brockii) showed similar results reaching a total yield of 1.97 g⋅L−1, achieving a maximum production rate of 7,757 μg⋅L−1⋅h−1 (3 d average), and sustaining production for 21 d (Fig. 5B). After 21 d, production in both cultures dropped off sharply and was not restored when cells were resuspended in fresh medium, indicating that changes in the culture population, such as spontaneous mutations that restore flux to metabolism, fundamentally impair production over time. Strains containing the 23BD biosynthetic pathway showed reduced growth compared with control strains, as expected, mirroring the rate of carbon redirection from central metabolism (Fig. 5 A, B, and D). A second control strain containing only alsS (B. subtilis) and alsD (E. aerogenes) produced acetoin up to toxic levels after the stationary phase was reached (Fig. S4) and showed impaired growth beyond what is attributable to carbon redirection.
Fig. 5.
Long-term 23BD production. (A–C) Summary of results for 23BD production in continuous cultures. Red: S. elongatus containing alsS (B. subtilis), alsD (A. hydrophila), and adh (C. beijerinckii). Blue: S. elongatus containing alsS (B. subtilis), alsD (A. hydrophila) and adh (T. brockii). Green: S. elongatus containing alsS (B. subtilis) and alsD (E. aerogenes). Purple: S. elongatus without alsS, alsD or adh. (A) Time courses for growth. (B) Total 23BD production. (C) Photosynthetic efficiency. (D) Total biomass production per day. Error bars indicate SD (n = 3).
Evaluating the Photosynthetic Efficiency of Production Strains.
Evolution of O2 from illuminated cells during continuous production was measured to verify whether the 23BD overproduction pathway could affect the photosynthetic system (Fig. 5C). Both strains expressing the 23BD biosynthetic pathway displayed a slightly higher rate of O2 evolution per microgram of chlorophyll compared with control strains (Fig. 5C). This rate increased during late stages of production. Both control strains, each with no production pathway, or only the acetoin production pathway expressed, displayed similar rates of O2 evolution (Fig. 5C). This trend follows the amount of fixed carbon diverted away from central metabolism, indicating that the burden placed on the cell by overproduction could stimulate a positive effect on the cells photosynthetic efficiency (Fig. 5 C and D).
Discussion
In this study, we are unique in describing production of 23BD and acetoin directly from CO2 and light, through engineering of the cyanobacterium S. elongatus. We systematically approached pathway design to match the production pathway to a photosynthetic host. Engineered strains achieved a production rate of 9,847 μg⋅L−1⋅h−1 and final titer of 2.38 g⋅L−1, with sustained production for 21 d. These values, achieved during continuous production from CO2 and light, compare favorably with other studies. The rate is 1.6-fold higher than that for isobutyraldehyde (6,230 μg⋅L−1⋅h−1), and significantly higher than other products overproduced from exogenous pathways (Fig. S5). The percentage of biomass produced as 23BD ranges from 30% to 60% (Fig. 5D), which compares favorably to the maximum of 80% achieved during endogenous sucrose overproduction (41).
To construct the 23BD pathway, we theorized that low toxicity would improve culture sustainability and production in S. elongatus. The negative effect of toxicity on pathway flux is reinforced by low production of acetoin, which is toxic above 0.1 g/L in S. elongatus (Fig. 2A), from the 23BD pathway without coexpression of adh (Fig. S4C). Addition of a strong sADH to the pathway to convert acetoin to 23BD increases total production 10-fold (Fig. 5B and Fig. S4C), even though reduction by sADH is not an irreversible step, but production of acetoin is. We also proposed that matching genes to their host could improve pathway function. All genes were screened for production in E. coli concurrently with cyanobacteria using identical operons. The patterns of production exhibited by the genes were different between hosts (Fig. 3 and 4). The productivities of strains expressing alsD (B. licheniformis) and alsD (G. xylinus) in S. elongatus were much lower (30% and 7% of top production, respectively) (Fig. 3C) than strains overexpressing the same genes in E. coli (80% and 85% of top production, respectively) (Fig. 3B). Conversely pathways expressing adh (T. brockii), which displayed severely attenuated production in E. coli achieved significant production in S. elongatus. Additionally the enzyme encoded by adh (L. pseudomesenteroides) was entirely inactive in S. elongatus despite production of 9.1 g/L meso-23BD in E. coli.
Production of 23BD was increased in strains expressing alsD (A. hydrophila), indicating that the second step of the pathway, catalyzed by ALDC (E. aerogenes) was limiting in the strain containing alsS, alsD (E. aerogenes), and adh (T. brockii or C. beijerinckii) (Fig. 4C). Gradient analysis of the IPTG expression system using a strain expressing alsS (B. subtilis), alsD (A. hydrophila), and adh (C. beijerinckii) (Fig. 4E), showed that even though enzyme concentrations in total protein increased, as measured by activity, the amount of 23BD produced did not increase. This finding implies that the amount of enzyme translated through leaking of PLlacO1 was sufficient to achieve the maximum production of 23BD (Fig. 4E). From these results, we assume that the supply of pyruvate from the Calvin-Benson-Bassham cycle (44) is limiting in this strain. Thus, further enzyme or pathway optimization would likely not be effective until the substrate pool of pyruvate from carbon fixation can be increased.
Using 23BD production as a model system allowed for the inclusion of stereoselectivity as part of our pathway design (Fig. 1). Chirality can be costly to install in chemical synthesis; however, biological control offers a much simpler route to chiral products. In all known cases in nature, acetoin is generated from 2-acetolactate containing an R-stereocenter (45), resulting in (S,S)-23BD not being observed. However, autodecarboxylation of 2-acetolactate, or enolate racemization of acetoin, could possibly form (S)-acetoin in the cell and result in (S,S)-23BD production in the presence of S-installing sADH. We designed two pathways, one for each possible stereocenter formed during reduction (Fig. 4 and Table S3). In S. elongatus both R-installing strains tested in long-term production consistently produced (R,R)-23BD as the major product (Fig. 5), although a trace amount of meso-23BD was observed in production by the strain expressing adh (T. brockii). The strain containing adh (C. parapsilosis) produced mixed isomers in E. coli and in S. elongatus (Fig. 4). These results indicate that sADH (C. parapsilosis) does not show stereoselectivity toward (R)-acetoin, although this enzyme catalyzes an S-installing reaction toward 2-hydroxyacetophenone (39). We assume that (R)-acetoin is too small to bind to the catalytic site stereospecifically. In this study no production of (S,S)-23BD was detected, indicating that degradation products do not contribute significantly to the pathway. With further engineering to remove endogenous S. elongatus sADH activity, observable in enzyme assays with NADH as a cofactor (Fig. 4D), it is likely that the purity of these chiral products can be increased.
During in vivo production acetoin is continuously secreted as evidenced by equilibration into culture supernatant (Fig. 4 and Fig. S4B). We can infer from this that enzymes with low velocity at intracellular acetoin concentrations or high Km may not be effective. Reported Km for sADH from C. beijerinckii and T. brockii are 8.3 mM and 0.23 mM, respectively (38); reported kcat are 8.2 and 0.91, respectively. The higher Km of sADH (C. beijerinckii) may explain why production from this strain surpassed that of the strain expressing adh (T. brockii) only after acetoin was increased by inclusion of alsD (A. hydrophila). This finding may also explain similar production from both strains in S. elongatus, but differing production in E. coli where acetoin is more abundant within the cell because of higher supernatant concentration.
Enzyme activities measured in crude S. elongatus cell lysate were roughly fourfold higher for sADH (C. parapsilosis) than for sADH (C. beijerinckii) at 318.3 and 85.0 nmol⋅min-1⋅mg-1, respectively (Fig. 4D), although the strain containing adh (C. parapsilosis) produced fourfold less 23BD than that of the strain containing adh (C. beijerinckii) (Fig. 4C). Accumulation of acetoin in the strain containing adh (C. parapsilosis) results in up to 120 mg/L during production. Assuming uniform diffusion this corresponds to 1.4 mM concentrations in the cell. The Km of sADH (C. parapsilosis) may be large, given that the native substrate of the enzyme has a phenyl ring adjacent to the reaction site and binding for acetoin without the phenyl ring may be inefficient (39). In this case the enzyme would have difficulty competing with diffusion of acetoin, yet would display high activity under assay conditions where acetoin concentration is high (25 mM).
During long-term production, evolution of O2 per microgram of chlorophyll increased in production strains relative to a control containing the recombination cassette but no alsS, alsD, or adh (Fig. 5C). This finding indicates that the stress imposed on metabolism by production elicits an increase in photosynthetic efficiency. Chlorophyll and O2 production have been seen to increase in roughly equal amounts during similar overproduction of sucrose in engineered S. elongatus (41).
For further improvements of the productivities, it is important to gain understanding of the engineered strains at the systems level using systematic and quantitative analysis methods. It is also important to increase the number of available parts, such as promoters and ribosome binding sites. These parts for pathway construction in S. elongatus are limited compared with model organisms such as E. coli. In this study we used a promoter, PLlacO1, because this promoter has been shown to be functional in S. elongatus (14). However, we showed that in the construct used in this study PLlacO1 was leaky and had a narrow expression range in S. elongatus (Fig. 4E). Additionally, it has been known that the composition of the cyanobacterial holopolymerase is quite different from those in most bacteria, thus E. coli promoters might perform differently when introduced into S. elongatus (11). To achieve efficient production, more controllable systems for expression of synthetic pathways would be desirable.
Defining the engineering principles for photosynthetic organisms is an important landmark in the search for sustainable technologies. Biological production of 23BD by heterotrophic microbes has attracted attention for many years because of the existence of natural fermentative producers, and the chemical’s potential as a versatile carbon feedstock for plastics, solvents, and fuel (24, 25). The biosynthetic production rate and titer presented here mark the second large increase in cyanobacterial yields in the last 3 y, because of increased understanding in pathway design.
Methods
See SI Methods for a more complete discussion.
Culture Conditions.
For acetoin and 23BD production in S. elongatus, cells in exponential phase were diluted to an OD730 of 0.1 in 25 mL BG-11 medium, including 50 mM NaHCO3, 10 mg/L thiamine, and 10 mg/L gentamicin in 125-mL baffled shake flasks. Cultures were grown to an OD730 of 0.3–0.5 before induction with 1 mM IPTG. Every 24 h 10% of the culture volume was removed, the pH was adjusted to 7.5 ± 0.4 with 10 N HCl, and volume was replaced with BG-11 containing 0.5 M NaHCO3, achieving a final concentration of 50 mM NaHCO3 in the culture.
Quantification of 23BD.
Supernatant samples from cultures were analyzed with gas chromatography (Shimadzu) equipped with a flame ionization detector and an HP-chiral 20b column (30 m, 0.32-mm internal diameter, 0.25-mm film thickness; Agilent Technologies). The stereoisomers were identified by matching retention time to standards for (R,R)-23BD, meso-23BD, and (S,S)-23BD.
Supplementary Material
Acknowledgments
We thank Jordan McEwen and Mike Connor for characterization of neutral sites used in this study. This work was supported by the Asahi Kasei Corporation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213024110/-/DCSupplemental.
References
- 1.McFarlane J, Robinson S. Survey of Alternative Feedstocks for Commodity Chemical Manufacturing. TN: Oak Ridge National Laboratory; 2007. [Google Scholar]
- 2. U.S. Energy Information Administration (2009) Annual Energy Review 2008. Available at http://www.eia.gov/totalenergy/data/annual/previous.cfm. Accessed December 21, 2012.
- 3.Raupach MR, et al. Global and regional drivers of accelerating CO2 emissions. Proc Natl Acad Sci USA. 2007;104(24):10288–10293. doi: 10.1073/pnas.0700609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herzog H, Golomb D. In: Encyclopedia of Energy. Cutler JC, editor. New York: Elsevier; 2004. pp. 277–287. [Google Scholar]
- 5.Ruffing AM. Engineered cyanobacteria: Teaching an old bug new tricks. Bioeng Bugs. 2011;2(3):136–149. doi: 10.4161/bbug.2.3.15285. [DOI] [PubMed] [Google Scholar]
- 6.Machado IM, Atsumi S. Cyanobacterial biofuel production. J Biotechnol. 2012;162(1):50–56. doi: 10.1016/j.jbiotec.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Ducat DC, Way JC, Silver PA. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011;29(2):95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Scharlemann JP, Laurance WF. Environmental science. How green are biofuels? Science. 2008;319(5859):43–44. doi: 10.1126/science.1153103. [DOI] [PubMed] [Google Scholar]
- 9.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 10.Golden SS, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 11.Heidorn T, et al. Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol. 2011;497:539–579. doi: 10.1016/B978-0-12-385075-1.00024-X. [DOI] [PubMed] [Google Scholar]
- 12.Huang HH, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38(8):2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keasling JD. Synthetic biology for synthetic chemistry. ACS Chem Biol. 2008;3(1):64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 14.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27(12):1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 15.Lan EI, Liao JC. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc Natl Acad Sci USA. 2012;109(16):6018–6023. doi: 10.1073/pnas.1200074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahama K, Matsuoka M, Nagahama K, Ogawa T. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J Biosci Bioeng. 2003;95(3):302–305. doi: 10.1016/s1389-1723(03)80034-8. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng. 2010;12(1):70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Zhang H, Zhang Y, Li Y, Ma Y. Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab Eng. 2012;14(4):394–400. doi: 10.1016/j.ymben.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Sheng J, Curtiss R., 3rd Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci USA. 2011;108(17):6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan X, et al. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng. 2011;13(2):169–176. doi: 10.1016/j.ymben.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Shen CR, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol. 2011;77(9):2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng. 2011;13(4):353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Bond-Watts BB, Bellerose RJ, Chang MC. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7(4):222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 24.Ji XJ, Huang H, Ouyang PK. Microbial 2,3-butanediol production: A state-of-the-art review. Biotechnol Adv. 2011;29(3):351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Celińska E, Grajek W. Biotechnological production of 2,3-butanediol—Current state and prospects. Biotechnol Adv. 2009;27(6):715–725. doi: 10.1016/j.biotechadv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Tran AV, Chambers RP. The dehydration of fermentative 2,3-butanediol into methyl ethyl ketone. Biotechnol Bioeng. 1987;29(3):343–351. doi: 10.1002/bit.260290308. [DOI] [PubMed] [Google Scholar]
- 27.van Haveren J, Scott EL, Sanders J. Bulk chemicals from biomass. Biofuels Bioprod Bioref. 2008;2(1):41–57. [Google Scholar]
- 28.Syu MJ. Biological production of 2,3-butanediol. Appl Microbiol Biotechnol. 2001;55(1):10–18. doi: 10.1007/s002530000486. [DOI] [PubMed] [Google Scholar]
- 29.Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329(5993):796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- 30.Greenwell HC, Laurens LM, Shields RJ, Lovitt RW, Flynn KJ. Placing microalgae on the biofuels priority list: A review of the technological challenges. J R Soc Interface. 2010;7(46):703–726. doi: 10.1098/rsif.2009.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eiteman MA, Gainer JL. In situ extraction versus the use of an external column in fermentation. Appl Microbiol Biotechnol. 1989;30(6):614–618. [Google Scholar]
- 32.Blankenship RE. Carbon Metabolism. Molecular Mechanisms of Photosynthesis. Oxford: Blackwell Science Ltd; 2002. pp. 172–203. [Google Scholar]
- 33.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 34.Scheer M, et al. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39(Database issue):D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25(6):1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aristidou AA, San KY, Bennett GN. Modification of central metabolic pathway in Escherichia coli to reduce acetate accumulation by heterologous expression of the Bacillus subtilis acetolactate synthase gene. Biotechnol Bioeng. 1994;44(8):944–951. doi: 10.1002/bit.260440810. [DOI] [PubMed] [Google Scholar]
- 37.Park HS, Xing R, Whitman WB. Nonenzymatic acetolactate oxidation to diacetyl by flavin, nicotinamide and quinone coenzymes. Biochim Biophys Acta. 1995;1245(3):366–370. doi: 10.1016/0304-4165(95)00103-4. [DOI] [PubMed] [Google Scholar]
- 38.Yan Y, Lee CC, Liao JC. Enantioselective synthesis of pure (R,R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases. Org Biomol Chem. 2009;7(19):3914–3917. doi: 10.1039/b913501d. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, et al. Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity. Protein Sci. 2008;17(8):1412–1423. doi: 10.1110/ps.035089.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rattray FP, Walfridsson M, Nilsson D. Purification and characterization of a diacetyl reductase from Leuconostoc pseudomesenteroides. Int Dairy J. 2000;10(11):781–789. [Google Scholar]
- 41.Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol. 2012;78(8):2660–2668. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278(21):19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 43.Ng WO, Zentella R, Wang Y, Taylor JS, Pakrasi HB. PhrA, the major photoreactivating factor in the cyanobacterium Synechocystis sp. strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch Microbiol. 2000;173(5-6):412–417. doi: 10.1007/s002030000164. [DOI] [PubMed] [Google Scholar]
- 44.Bassham JA, Benson AA, Calvin M. The path of carbon in photosynthesis. J Biol Chem. 1950;185(2):781–787. [PubMed] [Google Scholar]
- 45.Najmudin S, et al. Purification, crystallization and preliminary X-ray crystallographic studies on acetolactate decarboxylase. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 6):1073–1075. doi: 10.1107/s0907444903006978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.