Cancer cell lines (CCLs) are widely used to provide investigators with unlimited cell stocks, enabling experimental repetition and cumulative data assemblage. In numerous instances, cancer genes were first identified in CCL. Hence, the recent report in PNAS (1) that a classic multiple myeloma (MM) cell line, NCI-H929, bears cytogenetic rearrangement leading to expression of BCR–ABL1 fusion mRNA and protein—an oncogenomic change mainly restricted to chronic myeloid leukemia (CML)—and a subset of acute lymphoblastic, leukemia, is provocative. An unusual aspect of this study was the large discrepancy in BCR–ABL1 fusion gene expression levels in diverse NCI-H929 cell lines originally sourced from the American Type Culture Collection (ATCC) and the German Collection of Microorganisms and Cell Cultures (DSMZ), which the authors obtained from the Dana-Farber Cancer Institute. Based on their distinct phenotypes, the authors specifically excluded cross-contamination by BCR–ABL1–expressing K-562 CML cells, which they used as positive controls; this was despite observing identical BCR–ABL1 molecular alterations (e14a2) in both cell lines.
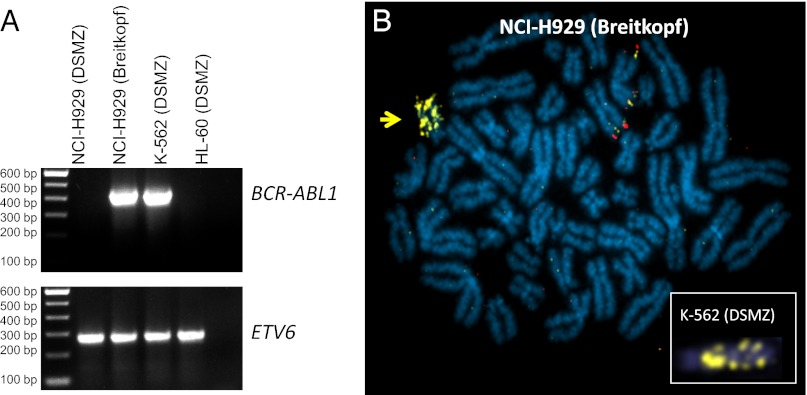
Because one of the BCR–ABL1–positive NCI-H929 samples purportedly originated from the DSMZ, we requested a sample for comparison, which the authors promptly supplied and we have now analyzed. RT-PCR confirmed high BCR–ABL1 expression therein, whereas reference DSMZ stocks lacked detectable transcript (Fig. 1A). FISH analysis of their sample revealed multiple copies of tandemly repeated BCR–ABL1 fusion (Fig. 1B), a configuration uniquely reported in K-562 cells (3). Short tandem repeat (STR) profiling of nine loci (Amelogenin, D5S818, D7S820, D13S317, D16S539, THO1, TPOX, vWA, CSF1) confirmed authenticity of DSMZ and original ATCC NCI-H929 stocks, whereas the authors’ sample matched K-562. Hence, we propose that cross-contamination by K-562 underlies BCR–ABL1 expression in the NCI-H929 MM cells.
Fig. 1.
Analysis of BCR–ABL1 in NCI-H929 cells. (A) RT-PCR confirms BCR-ABL1 transcript (e14a2) in K-562 and NCI-H929 [Breitkopf et al. (1)] but neither in NCI-H929 (DSMZ) nor HL-60 (negative control) with ETV6 as cDNA quality control. (B) Depiction of FISH in NCI-H929 [supplied by Breitkopf et al. (1)] using BAC clones flanking ABL1: upstream, RP11-202H3 (red); and downstream, RP11-83J21 (yellow). Note the tandemly repeated ABL1 rearrangement (arrow) diagnostic for K-562 (Inset). RT-PCR and FISH were performed as described previously (2).
Contradicting the authors’ claim that no recurrent chromosome translocations were associated with MM, at least five have been reported, effecting rearrangement of IGH with CCND1, CCND3, MAF, MAFB, or MMSET, the last of which was present in NCI-H929 cells. If, as proposed here, the investigation of BCR–ABL1 had been conducted in CML rather than MM cells, it would not have been the first occasion when cell line cross-contamination had misled investigators (4). These considerations apart, the authors’ demonstration of how directed proteomics may be applied to CCL in pursuit of new therapeutic targets highlights an original and interesting approach to drug discovery.
Cell line cross-contamination remains a chronic problem. Standardized PCR-based methods are now available for authenticating cell lines by reference to an interactive cell line database (www.dsmz.de/services/services-human-and-animal-cell-lines/online-str-analysis.html), although continually updated lists of false examples are published (5). It is now up to journals and grant agencies to encourage full exploitation of these resources.
Footnotes
The authors declare no conflict of interest.
References
- 1.Breitkopf SB, Yuan M, Pihan GA, Asara JM. Detection of a rare BCR-ABL tyrosine kinase fusion protein in H929 multiple myeloma cells using immunoprecipitation (IP)-tandem mass spectrometry (MS/MS) Proc Natl Acad Sci USA. 2012;109(40):16190–16195. doi: 10.1073/pnas.1212759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagel S, et al. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007;67(4):1461–1471. doi: 10.1158/0008-5472.CAN-06-2615. [DOI] [PubMed] [Google Scholar]
- 3.Drexler HG, MacLeod RA, Uphoff CC. Leukemia cell lines: In vitro models for the study of Philadelphia chromosome-positive leukemia. Leuk Res. 1999;23(3):207–215. doi: 10.1016/s0145-2126(98)00171-4. [DOI] [PubMed] [Google Scholar]
- 4.American Type Culture Collection Standards Development Organization Workgroup ASN-0002 Cell line misidentification: The beginning of the end. Nat Rev Cancer. 2010;10(6):441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 5.Capes-Davis A, et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127(1):1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]