Abstract
SMG1 is a member of the phosphoinositide kinase-like kinase family of proteins that includes ATM, ATR, and DNA-PK, proteins with known roles in DNA damage and cellular stress responses. SMG1 has a well-characterized role in nonsense-mediated decay as well as suggested roles in the DNA damage response, resistance to oxidative stress, regulation of hypoxic responses, and apoptosis. To understand the roles of SMG1 further, we generated a Genetrap Smg1 mouse model. Smg1 homozygous KO mice were early embryonic lethal, but Smg1 heterozygous mice showed a predisposition to a range of cancers, particularly lung and hematopoietic malignancies, as well as development of chronic inflammation. These mice did not display deficiencies in known roles of SMG1, including nonsense-mediated decay. However, they showed elevated basal tissue and serum cytokine levels, indicating low-level inflammation before the development of tumors. Smg1 heterozygous mice also showed evidence of oxidative damage in tissues. These data suggest that the inflammation observed in Smg1 haploinsufficiency contributes to susceptibility to cancer and that Smg1-deficient animals represent a model of inflammation-enhanced cancer development.
Keywords: PIKK, immune
SMG1 is an ∼400-kDa member of the phosphoinositide kinase-like kinase (PIKK) family of proteins (1). Other family members include ATM, ATR, and DNA-PK, which are known regulators of DNA damage and cellular stress responses. SMG1 has a well-characterized role in nonsense-mediated decay (NMD), a process that ensures the rapid degradation of mRNA containing premature termination codons (PTCs). It is important to eliminate these mRNAs to prevent production of truncated proteins that might interfere with the function of normal proteins in the cell. The importance of SMG1 activity for efficient NMD function is evident from markedly increased levels of PTC containing transcripts in the presence of kinase-dead SMG1 (1). Brumbaugh et al. (2) demonstrated that SMG1 is also functional in the genotoxic stress pathway. SMG1 was activated by UV and ionizing radiation (IR) to phosphorylate a p53 substrate as well as Upf1 in vitro. In cells treated with siRNA to Smg1, Chk2 and p53 were constitutively phosphorylated. Overall, the data suggested that SMG1 deficiency caused DNA damage and constitutive activation of ATM/ATR. SMG1 also plays a role in telomere stability by negatively regulating noncoding RNAs, known as telomeric repeat-containing RNA (TERRA), which associate with telomeres. Smg1 depletion increased the number of TERRA-positive chromosomes and resulted in telomere destabilization (3, 4). Additional less well-characterized roles for Smg1 have also been described. In Caenorhabditis elegans, using a candidate RNAi feeding screen for clones that lengthen lifespan, Masse et al. (5) reported that smg-1 inactivation increased lifespan. The effect of smg-1 inactivation on lifespan appeared to be unrelated to its NMD function but required the p53 tumor suppressor ortholog cep-1. Inactivation of smg-1 conferred resistance to oxidative stress, suggesting that the role of smg-1 in lifespan regulation was, at least in part, dependent on a function in oxidative stress resistance (5). Smg-1 in planarian worms can also act to regulate response to injury and growth via cross-talk with the PIKK family member mammalian target of rapamycin (6). Depletion of Smg1 in tumor cells markedly increased the extent of cell death induced by TNF-α or granzyme B (7, 8). SMG1 kinase activity was also induced in response to hypoxia, where it negatively regulated hypoxia-inducible factor 1α (HIF-1α), in part, by blocking MAPK activation (9). In addition, we recently showed that SMG1 had an essential role in the formation of stress granules (10). In short, SMG1 has known roles in NMD and genome maintenance and has been implicated in regulation of oxidative stress responses, apoptosis, hypoxia responses, and stress granule formation. However, for most of these pathways, the specific mechanism by which SMG1 regulation occurs is unknown, and very few SMG1 kinase substrates have been identified. More recently, McIlwain et al. (11) have demonstrated a critical role for Smg1 in mouse development, which was dependent on its role in NMD. They showed that Smg1-deficient mice died between embryonic day (E) 8 and E13 and that murine embryonic fibroblasts derived from KO embryos did not have an intact NMD pathway. To understand more fully the role of SMG1 in the stress response in vivo, we generated Smg1 KO (gt/gt) mice. The Smg1gt/gt mutant was embryonic lethal (E8.5), and Smg1 heterozygous (+/gt) mice showed evidence of haploinsufficiency. Smg1+/gt mice had a decreased lifespan in comparison to WT littermates, dying from a range of causes, including tumor development and inflammatory disorders.
Results
Generation of the Smg1+/gt Mice.
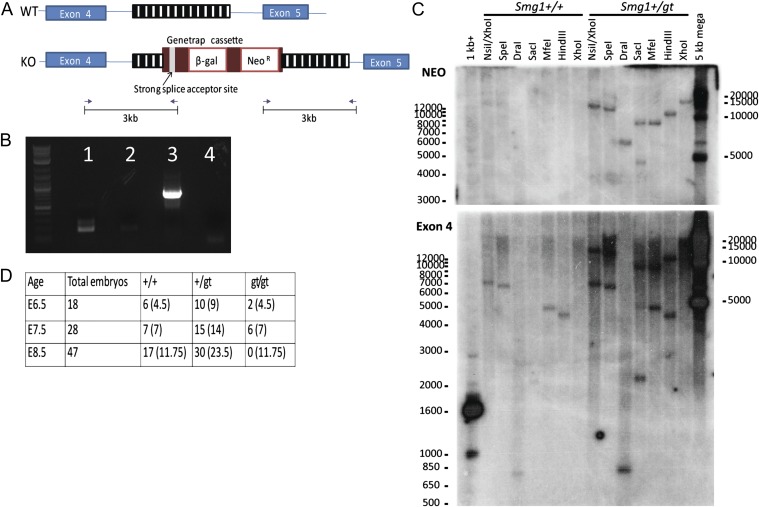
The loss-of-function mutation in Smg1 was produced by Genetrap mutagenesis in ES cells supplied by mutant mouse regional resource centers (MMRRC) (SIGTR ES cell line AG0297). These cells were characterized by the Genetrap Consortium as containing a single cassette inserted in intron 4 of Smg1. The cassette contains β-galactosidase and neomycin resistance genes as well as a strong splice acceptor site that disrupts normal mRNA expression (Fig. 1A and Fig. S1A). The cassette also encodes multiple stop codons in each reading frame, and transcripts from this system are likely to be unstable. The ES cells were injected into SV129 blastocysts to generate chimeric mice, crossed with C57BL/6 mice to generate Smg1 heterozygous mice. 5′ RACE using nested PCR primers in the cassette and commercial nested 5′ RACE primers produced only one product, supporting the presence of a single insertion site (Fig. 1B, lane 3). This product was purified from the agarose gel and directly sequenced (Fig. S1B), showing the expected coding sequence of Smg1 splicing into the Genetrap cassette. Interestingly, our Smg1 RACE product has a 5′ UTR shorter than predicted in the databases, explaining the shorter than expected size of the RACE product (∼700 vs. ∼900 bp). We further mapped the insertion site of the cassette by PCR assay and sequencing, and we showed that the cassette inserted ∼3 kb downstream of exon 4 within intron 4. We also mapped the junction with intron 4 downstream of the cassette and showed the insertion site to be 1.7 kb upstream of exon 5 in a repeat-rich region (Fig. S1A). Insertion of a single cassette in this site was confirmed by Southern blotting. Genomic DNA from Smg1+/+ or Smg1+/− mice was digested with a range of restriction enzymes, separated on an agarose gel, and then Southern-blotted using a probe either to exon 4/intron 4 of the Smg1 gene (exon 4) or to the cassette (Neo). For the Smg1+/gt samples incubated with the cassette (Neo) probe, in all but one digest, a single band was observed, indicating that there is a single integration site for the cassette (Fig. 1C, Upper). When incubated with the exon 4 probe, two bands (one representing the WT allele and the other representing the Genetrap insertion) were detected as expected (Fig. 1C, Lower). Bands detected were approximately the predicted size, given that parts of the cassette can be lost on integration (with the exception of the NsiI/XhoI products, where the size for both the exon 4 probe and the Neo probe suggests the loss of an XhoI site). Two bands were observed in SacI digests; however, given the single bands detected in all other samples, this is likely due to incomplete digestion of the DNA rather than a second insertion site. Combined, these data show that in the Smg1+/gt mice, there is a single Genetrap cassette inserted into intron 4 of the Smg1 gene.
Fig. 1.
Generation of Smg1 mice. (A) Schematic shows targeting of the Smg1 gene by insertion of a Genetrap cassette. Arrows indicate the primers used for PCR and sequencing to map the insertion site. (B) cDNA was isolated from Smg1+/gt and Smg1+/+ mice. 5′ RACE was performed on these samples, and products were visualized on an agarose gel. Lane 1: positive control for PCR, lane 2: negative control, lane 3: 5′ RACE sample, and lane 4: negative control for 5′ RACE. (C) Genomic DNA from Smg1+/gt and Smg1+/+ mice was used for Southern blotting. Multiple restriction digests, as indicated, were run for each sample and then separated on an agarose gel. DNA was transferred to a nylon membrane and incubated with a probe targeting either the neomycin gene in the Genetrap cassette (Upper; NEO) or intron4-exon 4 of the Smg1 gene (Lower; Exon 4). (D) Smg1gt/gt mice die around E8.5. Embryos were harvested and genotyped by real-time PCR assay at the indicated days after timed mating. Three litters were analyzed at each time point. Numbers in brackets indicate the expected number of embryos for each genotype.
Smg1+/gt × Smg1+/gt breedings produced pups in the ratio of 1:2 WT (+/+)/Smg1+/gt. No Smg1gt/gt mice were born, demonstrating that Smg1 is essential for development and viability. To determine the point of embryonic lethality for our model, the genotype of embryos ranging from E6.5 to E8.5 was determined by real-time PCR (Fig. 1D and Fig S1 C and D). At E6.5 and E7.5, Smg1gt/gt embryos showed similar morphology and structures as Smg1+/gt and Smg1+/+ littermates. However, no Smg1gt/gt embryos were detected at E8.5. Litters collected after this time point did not contain Smg1gt/gt embryos; however, blood-filled sacs were frequently observed, indicating that embryos were being absorbed. Therefore, we concluded that homozygous deletion of Smg1 is embryonic lethal between E7.5 and E8.5. This finding is consistent with the previously described model, although there were a small number of Smg1gt/gt embryos surviving until E12.5 in that study (11).
Smg1+/gt Mice Displayed Abnormal Growth.
Initial physical inspection revealed that a number of Smg1+/gt mice were substantially larger than their Smg1+/+ littermates (example of a growth curve is shown in Fig. S2A; P < 0.001) with some growing to 11 cm in length and weighing up to 70 g (vs. an average of 40 g for WT mice). Additionally, a number of mice had deformed teeth. Teeth in the Smg1+/gt mice were often substantially overgrown (Fig. S2B and Movies S1 and S2). Craniofacial measurements revealed that the GoPg/Nba angle (the angle formed by the plane of the mandible to the plane of the cranial base) was significantly less in the Smg1+/gt mice compared with controls, indicating that the mandible was prognathic (displaced forward) and that the tooth-related deformities may be skeletally based (Table S1). We investigated whether large mice represented the extreme end of a spectrum by analyzing the bone structure and insulin-like growth factor-1 (IGF-1) levels of growing mice. IGF-1 is the growth factor responsible for the most variability in the size of mammals (12). We measured IGF-1 serum levels in 10 Smg1+/gt and Smg1+/+ mice at 4–6 wk and 12–16 wk after birth. We found no significant difference in IGF-1 levels between Smg1+/gt and Smg1+/+ mice, nor did any individual Smg1+/gt mice examined have significantly elevated IGF-1 levels (Fig. S2C). A subset of these mice also underwent dual-energy X-ray absorptiometry analysis to quantify their bone mineral density, lean mass, and fat mass, which all showed no difference between the Smg1+/gt and Smg1+/+ mice (Fig. S2 D and E). H&E staining of femurs and tibias from these mice showed normal bone development in Smg1+/gt mice at 5.5 wk and 15.5 wk of age (Fig. S2F). These data suggest that Smg1 heterozygosity does not affect long bone development or IGF-1 regulation.
Smg1+/gt Mice Have a Shortened Lifespan Associated with Higher Tumor Incidence.
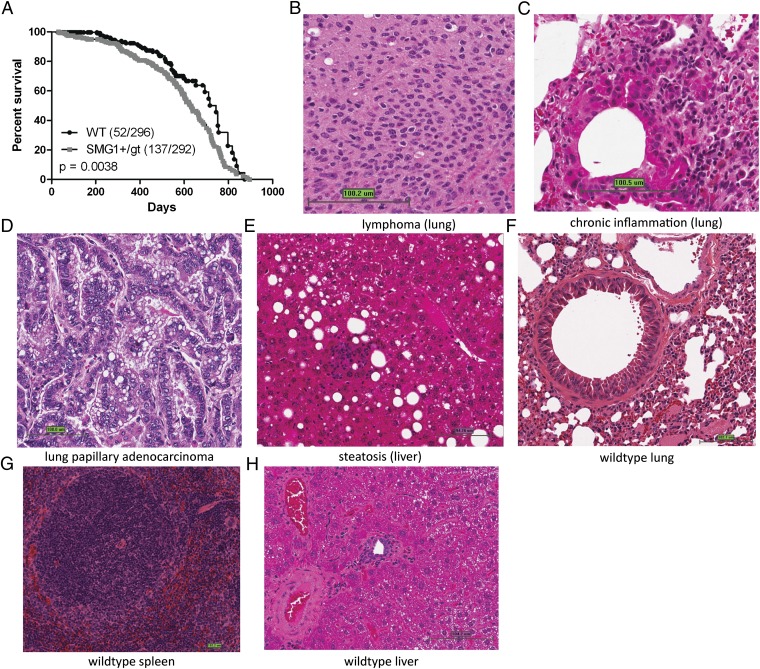
As the mice aged, it also became apparent that Smg1+/gt mice had a significantly shorter lifespan than Smg1+/+ littermates (P = 0.0038) (Fig. 2A). The major observable defects at the autopsy of the Smg1+/gt mice were kidney abnormalities (24%) and enlarged organs, particularly the spleen (36%) and liver (26%) (Table S2). To characterize the diseases present in the Smg1+/gt mice further, we performed histological and pathological examination on a range of tissues collected at autopsy. The results from this analysis are summarized in Table 1, where asterisks denote pathological findings increased in Smg1+/gt mice (detailed information is provided in Table S3). Key histological phenotypes observed included steatosis, chronic inflammation, and cancer (examples of pathological images are shown in Fig. 2 B–E, compared with WT tissue in Fig. 2 F–H). The observed chronic inflammation appeared mainly in the kidneys and lung, where inflammation was likely to decrease tissue functionality and, consequently, may affect the mouse lifespan. There were two particularly interesting facets to the cancer development in Smg1+/gt mice. One was the high percentage of hematopoietic malignancies (39% vs. 8% in Smg1+/+ mice), and the second was the development of a single histological form of lung tumor, papillary adenocarcinoma (14% vs. 2% in Smg1+/+ mice), a cancer type that is found in humans in nonsmokers with lung cancer (13). To determine whether the hematopoietic malignancies were of the same subtype, sections were stained for B220, CD3, Bcl-2, and myeloperoxidase. This staining showed that the lymphomas were predominantly but not exclusively B-cell non-Hodgkin lymphomas of follicular center origin, due to both B220 and Bcl-2 positivity (Fig. S3). This outcome is consistent with the morphology of the tumors.
Fig. 2.
Smg1 heterozygous mice have decreased viability compared with WT littermates. (A) Kaplan–Meier survival plot shows the significantly decreased viability of Smg1+/gt mice compared with Smg1+/+ littermates. Statistical significance was determined using Prism 5 software (GraphPad Software). H&E staining of paraffin-embedded sections shows key pathological findings observed in Smg1+/gt mice [lymphoma (B), chronic inflammation (C), papillary adenocarcinoma (D), and steatosis (E)] and WT tissues for comparison [lung (F), spleen (G), and liver (H)].
Table 1.
Results from analysis of tissues from Smg1+/gt and Smg1+/+ mice
| No. of mice |
% of total |
|||
| Pathology | +/+ | +/gt | +/+ | +/gt |
| Extramedullary hematopoiesis | 13 | 14 | 27 | 24 |
| Chronic inflammation* | 16 | 29 | 34 | 49 |
| Adenocarcinoma* | 1 | 8 | 2 | 14 |
| Lymphoma* | 4 | 23 | 8 | 39 |
| Other solid tumors | 1 | 3 | 2 | 5 |
| Hyperplasia | 33 | 34 | 38 | 32 |
| Steatosis/hepatitis* | 5 | 16 | 11 | 27 |
| Total number analyzed | 47 | 59 | ||
*Denotes pathological findings increased in Smg1+/gt mice.
Smg1+/gt Mice Showed Normal NMD.
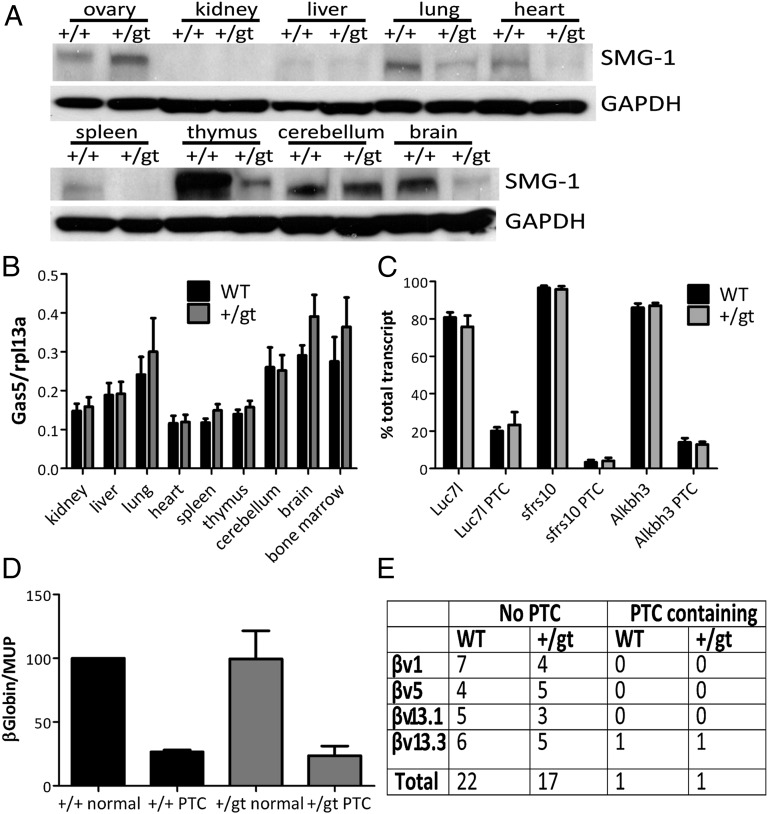
To determine whether SMG1 protein levels were altered in Smg1+/gt mice, we performed Western blotting on protein extracts from a range of tissues from Smg1+/gt and Smg1+/+ mice. Fig. 3A shows that SMG1 protein levels were markedly reduced in the spleen, lung, heart, thymus, and brain samples from a Smg1+/gt mouse compared with a WT littermate. The decreased protein level in these tissues was also observed in another three pairs of mice. These data indicated that Smg1+/gt mice may have sufficiently low protein levels to be adversely affecting SMG1 function. Because the Smg1+/gt mice frequently showed abnormalities in the kidney and liver, we performed immunoprecipitation analysis on extracts from these tissues to confirm SMG1 expression. Fig. S4A shows that SMG1 protein could also be detected in the liver and kidney; in the liver, Smg1+/gt mice had lower levels of SMG1 than WT littermates, but levels were similar in kidney samples from Smg1+/gt and Smg1+/+ animals. Because the major observed phenotypes in the kidney were chronic inflammation and lymphoma (Table S3), it is likely that decreased SMG1 levels in hematopoietic cells (as demonstrated by the strong decrease in the thymus and spleen) are likely to account for the phenotype observed here. We also examined the level of Genetrap fused mRNA using semiquantitative PCR (Fig. S4B) and full-length Smg1 mRNA using real-time PCR (Fig. S4C) in the kidney compared with the spleen. Genetrap fused mRNA levels were similar between spleen and kidney samples in the two tested samples, and although full-length Smg1 mRNA levels were slightly lower in Smg1+/gt animals, the levels in the spleen and kidney were again similar, suggesting that SMG1 protein levels may be partially determined posttranscriptionally. We next aimed to determine whether the decrease in protein level affected SMG1 function in NMD. To gauge NMD levels initially, we examined the level of Gas5 transcript [a known NMD target (14)] in a range of tissues by quantitative real-time PCR. If NMD was defective in Smg1+/gt mice, there should be an increase in the amount of Gas5 transcript. Gas5 levels in the tissues examined did not show a significant difference between Smg1+/+ and Smg1+/gt animals (Fig. 3B). The levels of two other known NMD-targeted transcripts [Atf4 and Map13k4 (15)] were also measured in tissues with low SMG1 expression (Fig. S4D). The expression levels of these transcripts were not different between Smg1+/+ and Smg1+/gt mice. McIlwain et al. (11) previously showed a requirement for Smg1 in murine NMD by comparing levels of alternatively spliced transcripts, where only one splice variant contains a PTC, and is therefore regulated by NMD. To compare their findings with our model, we measured levels of three of the same alternatively spliced transcripts. We established crisis-derived murine embryonic fibroblast (MEF) lines from Smg1+/gt and Smg1+/+ mice (Fig. S4E). Due to the low level of endogenous SMG1 protein in MEFs, we performed immunoprecipitation analyses to confirm reduced SMG1 protein levels in the cell lines derived from Smg1+/gt mice. The expected reduction in protein level was also observed in MEFs (Fig. S4F). If Smg1+/gt mice were defective in NMD, an increase in the PTC containing splice variant would be observed. However, we saw no difference in the levels of splice variants between cells from Smg1+/gt and Smg1+/+ mice (Fig. 3C). Given that Smg1+/gt mice showed hematopoietic abnormalities and cancer development on aging, we also measured the levels of these transcripts in splenocytes and bone marrow from two pairs of young (∼12 wk) and older (∼9 mo) mice. Again, we saw no significant difference in the transcript levels between Smg1+/gt and Smg1+/+ animals (Fig. S4G). To investigate NMD levels in MEFs from Smg1+/gt and Smg1+/+ animals further, we used a plasmid-based assay commonly utilized in the literature to measure NMD (16, 17). In this assay, cells are transfected with a plasmid encoding normal β-globin or β-globin containing a PTC. The level of expression of each of these transcripts is normalized to a control gene, murine urinary tract protein, to correct for differences in transfection efficiency, and the level of normal transcript is then compared with the mutated transcript. There was no significant difference in the level of PTC containing transcript in Smg1+/+ and Smg1+/gt MEFs (Fig. 3D), again suggesting that Smg1+/gt cells do not have a deficiency in NMD. In some cell types, NMD appears to work more effectively than in other cells, particularly B and T cells, where NMD is thought to be 10- to 100-fold more effective (18). This is thought to be due to an increased requirement for transcript screening of B-cell and T-cell receptor chains after antigen-driven selection. Therefore, it was possible that in most cells, the lower levels of SMG1 protein levels observed were sufficient to sustain normal NMD but that levels may not be sufficient in cells requiring more efficient NMD. To measure NMD in T cells, we isolated thymocytes from Smg1+/+ and Smg1+/gt mice and amplified regions of the T-cell receptor β-chain as described previously (18). These PCR products were then cloned, sequenced, and analyzed for the presence of PTC. Again, we found no difference in PTC levels between Smg1+/gt and Smg1+/+ mice (Fig. 3E). Combined, these assays show that Smg1+/gt mice do not have a substantial defect in NMD.
Fig. 3.
Smg1+/gt mice do not show a deficiency in NMD. (A) SMG1 Western blotting shows reduced levels of SMG1 protein expression in the lung, heart, spleen, thymus, and brain compared with Smg1+/+ levels. Tissues were harvested, total protein was extracted, and SMG1 expression was measured by Western blotting. GAPDH was used to show loading. (B) Real-time PCR quantification of endogenous NMD target transcript Gas5 in the indicated tissues. Gas5 expression is shown relative to control gene rpl13a. Data are pooled from five independent experiments and expressed as mean ± SEM. (C) Quantification of NMD-regulated alternative splicing in MEFs. Transcripts for luc7l, sfrs10, and alkbh3 were amplified by PCR, and samples were separated by electrophoresis. Products from the PTC-containing transcripts are a different size from the major non-PTC transcript. The amount of product for each splice variant was determined by ethidium bromide incorporation. Data shown are pooled from three independent pairs of MEFs and expressed as mean ± SEM. (D) Smg1+/gt and Smg1+/+ MEFs were transfected with plasmid containing normal β-globin (normal) or β-globin with an introduced PTC and a control for transfection efficiency, murine urinary tract protein (MUP). Twenty-four hours after transfection, samples were analyzed for β-globin mRNA expression by PCR. The amount of PCR product was determined by ethidium bromide incorporation. Data are normalized to Smg1+/+ normal MEFs and are pooled from analysis of three pairs of MEFs and expressed as mean ± SEM. (E) Indicated T-cell receptor β-chains (1, 5, 13.3, and 13.1) were amplified from thymocytes from Smg1+/gt and Smg1+/+ mice. PCR products were cloned, sequenced, and analyzed for inclusion of a PTC in the transcript. No difference in the frequency of PTC inclusion was observed between transcripts from Smg1+/gt and Smg1+/+ mice.
Smg1+/gt Mice Showed Normal Stress Responses Compared with WT Littermates.
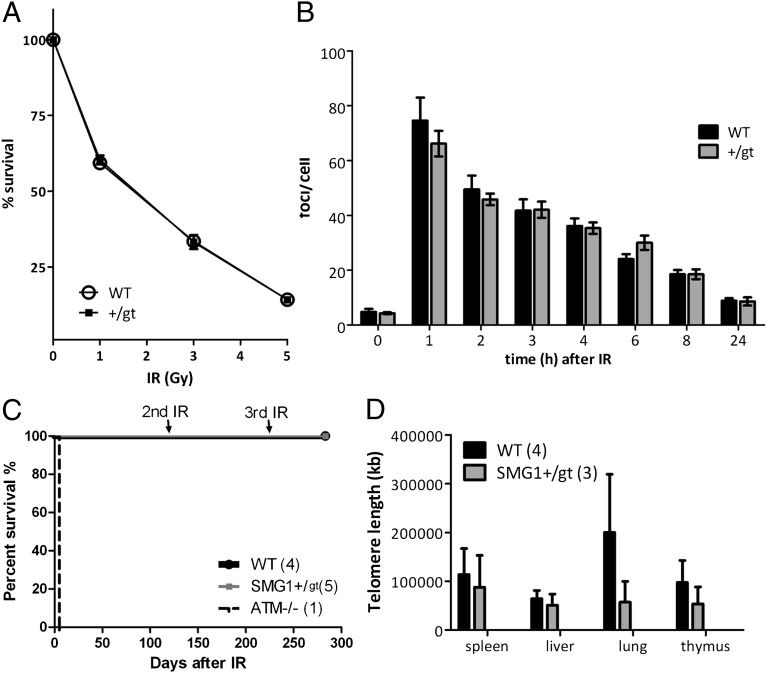
In a previously published study, SMG1-deficient U2OS cells displayed spontaneous DNA damage, enhanced radiosensitivity, and primarily G2/M checkpoint arrest after 10-Gy irradiation (2), and we examined these parameters in the Smg1+/gt mice and/or primary MEFs. When MEFs from WT and Smg1+/gt mice were exposed to IR and cell survival was determined by colony formation assay, no difference was observed between WT and Smg1+/gt MEFs (Fig. 4A). Following 10-Gy irradiation of Smg1+/gt and Smg1+/+ MEFs, we measured the appearance and disappearance of γH2AX (Millipore) foci (19) to monitor DNA damage response and repair. There was no difference in the appearance of γH2AX foci or in the kinetics of their resolution, indicating that there was no defect in the DNA double-strand break response in Smg1+/gt MEFs (Fig. 4B). In addition, Smg1+/gt and Smg1+/+ mice were treated with two 3-Gy doses of IR, 4 mo apart, followed by 5 Gy of IR another 3 mo later, and survival was monitored. At 3 mo after the final dose of IR, 100% of mice (Smg1+/gt and Smg1+/+) were still alive (Fig. 4C). These data clearly suggest that Smg1+/gt mice are not abnormally radiosensitive. Furthermore, Smg1+/gt MEFs were treated with 5-Gy IR or 100 μM H2O2 (which causes both DNA damage and oxidative stress) for 24 h and were then analyzed by flow cytometry for cell cycle progression and apoptosis. No differences were observed between Smg1+/gt and Smg1+/+ MEFs (Fig. S5 A and B), suggesting that one allele of Smg1 is sufficient for normal cell cycle inhibition and apoptosis after induced DNA damage. We also examined whether the amount of oxidative stress in splenocytes in response to H2O2 differed between Smg1+/gt and Smg1+/+ mice. Splenocytes were treated with 250 μM H2O2 for 4 h or 24 h, and the levels of cytosolic reactive oxygen species (ROS) [2′,7′-dichlorodihydrofluorescein diacetate (DCFDA); Molecular Probes] or mitochondrial ROS (MitoSox; Molecular Probes) were measured by flow cytometry (Fig. S5C). No significant difference in inducible or basal ROS levels was observed. Because SMG1-deficient U2OS cells have been shown to have an increased basal level of phosphorylated p53 (2), we examined p53 phosphorylation in the Smg1+/gt and Smg1+/+ MEFs. Strong p53 phosphorylation at Ser18 was observed at 1 h and 2 h post-IR in both Smg1+/gt and Smg1+/+ samples, but no significant basal phosphorylation was detected in either genotype (Fig. S5D). The p21 protein levels were also examined and showed no difference between Smg1+/gt and Smg1+/+ samples over the 4-h period (Fig. S5D). These results suggest that normal p53 signaling occurs in the Smg1+/gt mice, possibly due to compensation of other factors that are involved in regulating p53 Ser18 phosphorylation. To determine whether DNA damage was accumulating in vivo, γH2AX staining was performed on tissues from Smg1+/gt and Smg1+/+ mice (Fig. S5E). Although γH2AX-positive cells were detected in some Smg1+/gt mice, not all samples showed γH2AX signal (variation of signal is shown in the three Smg1+/gt panels in Fig. S5E). These data suggest that accumulation of DNA damage in these mice is not ubiquitous and that there is not likely to be a major defect in DNA damage responses in Smg1+/gt mice.
Fig. 4.
Smg1+/gt mice do not show defects in DNA damage responses. (A) Immortalized MEF survival after the indicated dose of IR was determined by colony formation assay. Data points show the average of two independent experiments, and the error bars, which may fall within the size of the symbol, indicate the standard error. (B) γH2AX foci formation and resolution were measured by immunofluorescence at the indicated time points after 10-Gy irradiation in Smg1+/gt and Smg1+/+ MEFs. The number of foci per cell was counted for a minimum of 150 cells at each time point. Data shown are pooled from three independent experiments and expressed as mean ± SEM. (C) Smg1+/gt and Smg1+/+ littermates were exposed to two 3-Gy doses of IR 4 mo apart, followed by 5 Gy of IR another 3 mo later, and survival was monitored. An ATM−/− mouse was included to ensure irradiation was effective. Smg1+/gt mice did not show sensitivity to IR. (D) Genomic DNA was isolated from the indicated tissues from Smg1+/gt and Smg1+/+ mice at 9 mo of age. Telomere length was measured by real-time PCR. Data shown are pooled from the indicated number of replicates for each genotype. Data are expressed as mean ± SEM. No significant difference was observed between genotypes.
SMG1 has been proposed to regulate TERRA, noncoding RNA that associates with telomeres. Depletion of Smg1 showed increased TERRA foci and TERRA-positive cells, as well as increased telomere-free chromosomal ends (3). Here, telomere length from the liver, spleen, lung, and thymus of Smg1+/gt and Smg1+/+ mice 9 mo of age were measured by quantitative PCR assay. No significant difference in telomere length was observed between Smg1+/gt and Smg1+/+ mice (Fig. 4D), indicating that one allele of Smg1 is sufficient to maintain telomere stability at this age. Furthermore, we examined HIF-1α stability in response to hypoxia in MEFs from Smg1+/gt and Smg1+/+ mice. HIF-1α was stabilized equally in MEFs from the two mouse strains (Fig. S5F). Finally, MEFs from Smg1+/+ and Smg1+/gt mice showed equal induction of stress granules in response to sodium arsenite treatment (Fig. S5G). The combination of these data indicates that Smg1 haploinsufficiency does not affect SMG1 function in DNA damage responses, cell cycle, and apoptosis regulation in response to DNA damage or oxidative stress, telomere maintenance, or stress granule formation.
Composition of the Hematopoietic Compartment of Smg1+/gt Mice Is Normal.
Given the presence of inflammation and increased levels of hematopoietic malignancies in Smg1+/gt mice, we next determined whether there was a difference in the cell composition of key lymphoid organs. We first measured the number of T and B cells and granulocytes in the thymus and lymph nodes of 6- to 8-wk-old mice to investigate the composition of the developing immune system. Fig. S6 shows that there was no significant difference in these populations. We then went on to examine splenic and bone marrow cell populations from mice aged from 6 wk to 9 mo. There was no significant difference observed between cell populations from Smg1+/gt and Smg1+/+ mice (Fig. S6). Neither was there a significant difference between the cell populations when the ages were analyzed separately. In summary, there is no major defect in the composition of the hematopoietic compartment of Smg1+/gt mice.
Smg1+/gt Mice Show Signs of Constant Low-Level Inflammation.
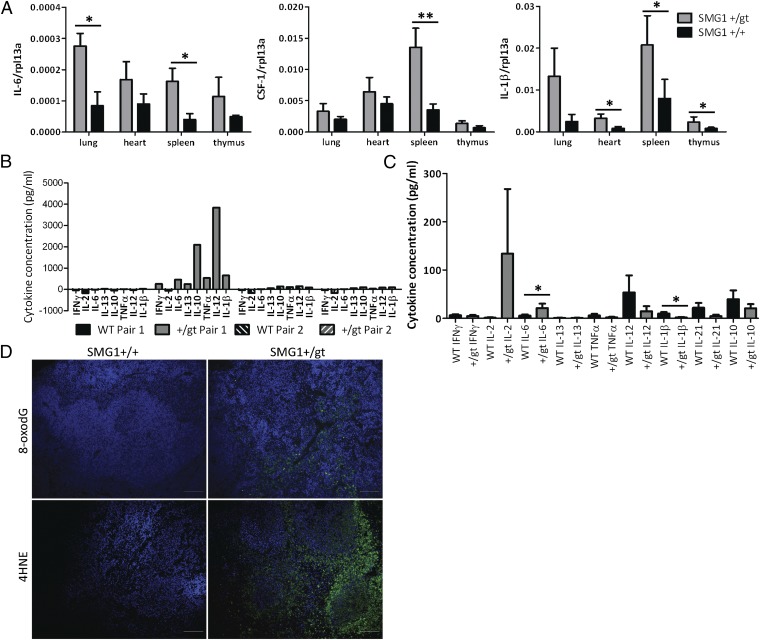
Because we had observed higher than normal levels of inflammation in Smg1+/gt mice, we examined tissue cytokine levels in Smg1+/gt and Smg1+/+ animals. We measured levels of IL-6, IL-1β, and Csf-1 mRNA in tissues in which SMG1 protein levels were significantly decreased in heterozygous mice (Fig. 5A). Although the basal levels for each of these cytokines was quite low and variable between the animals, there was a clear trend toward increased cytokine levels in Smg1+/gt mice. Statistically significant increases in IL-6 production were observed in the lung and spleen; in Csf-1 in the spleen; and in IL-1β in the heart, spleen, and thymus. These data suggest that low-level inflammation may be occurring in Smg1+/gt mice. We also examined the level of serum cytokines in Smg1+/gt and Smg1+/+ mice to see if this reflected the expression pattern in tissues. Some mice showed significantly elevated levels of cytokine production (Fig. 5B). When the mice with very high cytokine production were removed from the analysis, there was a trend for a number of cytokines to be increased in Smg1+/gt mice (Fig. 5C). However, only IL-6 was statistically significantly increased in serum. Interestingly, IL-1β was statistically significantly lower in Smg1+/gt mice compared with Smg1+/+ littermates. This suggests that although IL-1β mRNA was increased in tissues, the IL-1β was not released as an active cytokine into the serum or the increase may be tissue- or microenvironment-specific. Because tissues showed signs of increased cytokine production, it was possible that levels of ROS or reactive nitrogen species (RNS) were also increased. We examined hyperplasic spleens from Smg1+/gt mice to determine whether there was evidence of damage caused by ROS/RNS. Tissues from these animals and Smg1+/+ mice were stained with antibodies against 8-oxo-2′-deoxyguanosine (8oxodG; Trevigen), a product caused by oxidative damage to DNA, and 4-hydroxynonenal (Chemicon), a product of lipid peroxidation. All tested Smg1+/gt spleen samples showed evidence of ROS/RNS-mediated cellular damage, but Smg1+/+ spleens did not (Fig. 5D). These data provide evidence that ROS and RNS are elevated in the Smg1+/gt mouse spleen to a level that can cause cellular damage but not extensive DNA double-strand breaks (Fig. S5E). Interestingly, we did not find elevated basal levels of ROS production when we examined total splenocytes (Fig. S5C), nor did we see evidence of oxidative damage in prediseased lung, kidney, or liver samples. This suggests that a subpopulation of splenocytes may be responsible for the elevated ROS/RNS production. Collectively, these findings indicate that low-level inflammation is occurring in Smg1+/gt mice before development of either chronic inflammation or cancer. Therefore, Smg1 deficiency may lead to low-level inflammation that can either progress to chronic inflammation or enhance cancer development.
Fig. 5.
Smg1+/gt mice have elevated basal levels of cancer-related cytokines. (A) RNA was isolated from the indicated tissues, cDNA synthesis was performed, and cytokine mRNA levels were measured by real-time PCR. Expression levels are expressed relative to the control gene rpl13a. Data presented are pooled from a minimum of five pairs of Smg1+/gt and Smg1+/+ littermates analyzed in independent experiments. Data are expressed as mean ± SEM. A Student t test was performed to determine statistical significance: *P < 0.05; **P < 0.01. (B) Serum cytokine levels were measured by cytokine bead array in Smg1+/gt and Smg1+/+ mice. (C) Serum cytokine levels for mice not showing extreme cytokine production (as in B) were pooled and analyzed. Data shown are from a minimum of seven Smg1+/gt or Smg1+/+ mice. Data are expressed as mean ± SEM. A Student t test was performed to determine statistical significance: *P < 0.05. (D) Oxidative damage to tissues was measured by immunofluorescent staining for 8oxodG or 4-hydroxynonenal (4HNE; green), and tissues were counterstained with DAPI (blue) to show nuclei. Tissues from Smg1+/gt spleens of mice with hyperplasia were compared with those of Smg1+/+ mice of a similar age. (Scale bar = 100 μM.)
Discussion
SMG1 has been implicated in a wide range of functions, including regulation of NMD, genome maintenance, apoptosis, and hypoxia responses. In our model, Smg1gt/gt mice died at E8.5, similar to the findings for Smg1-deficient mouse mortality in the study by McIlwain et al. (11) and mortality observed in Upf1 and Upf2 KO mice, suggesting that the process of NMD may be essential for early development (20, 21). However, in contrast to McIlwain et al. (11), we observed a striking phenotype in Smg1+/gt mice. The difference in these studies may be explained by the fact that our mice are maintained on a 129/C57 mixed background, whereas the mice used by McIlwain et al. (11) were backcrossed to C57/Bl6 mice, and other genes may be contributing to the penetrance of the Smg1+/gt phenotype. Alternatively, the position of the Genetrap cassette may alter the phenotype. In our mice, the cassette is inserted into intron 4 of the Smg1 gene, whereas the model used by McIlwain et al. (11) has the cassette inserted into intron 12. It is conceivable that a product produced may have some function, because there is an additional 1,000 bp transcribed in the mouse used in the study by McIlwain et al. (11) compared with our model. In support of this possibility, there are annotated transcripts in the Ensembl database that terminate before intron 12 and would be unaffected in the model of McIlwain et al. (11) but disrupted in ours (ENSMUST00000180009 and ENSMUST00000179847) (www.ensembl.org). Alternatively, it is possible that our Genetrap cassette was affecting surrounding genes. We examined the genes within 1 Mb of the Smg1 gene. Dysregulation of these genes would not be predicted to result in a phenotype similar to that of the Smg1+/gt mice. Therefore, the phenotype observed here is most likely due to the loss of expression of SMG1.
Our Smg1+/gt mice had a shortened lifespan compared with their WT littermates. Smg1+/gt mice frequently died from cancer and exhibited increased inflammation related to organ dysfunction, particularly in the kidneys and lungs. Smg1+/gt mice showed no major defects in any of the well-characterized roles of SMG1. The most well-established role for SMG1 is in the process of NMD. Here, we examined four assays measuring NMD in a range of tissues or isolated cells; surprisingly, none of these assays showed evidence of any defect in NMD in Smg1+/gt mice. Similarly, we found no defect in DNA damage responses in Smg1+/gt mice despite SMG1 having a characterized role in these pathways (1, 4, 5, 7–10). These data suggest that expression of Smg1 from a single allele is sufficient for full functionality of these pathways in the situations studied. However, we acknowledge that in a specific subset of cells or under particular circumstances, Smg1 haploinsufficiency may result in defects in NMD or DNA damage response pathways. Here, we could find no evidence of widespread deficiency in the characterized roles of SMG1. Furthermore, we analyzed IGF-1 levels and bone growth in Smg1+/gt and Smg1+/+ mice and detected no defect in these pathways in Smg1+/gt mice. Given the high levels of chronic inflammation observed in the tissues of Smg1+/gt mice (Table S3), we examined the level of cytokine production in the tissues and blood of Smg1+/gt mice before onset of disease. At the mRNA level, we detected elevated IL-6, Csf-1, and IL-1β in the tissues and an elevated level of IL-6 protein in the serum of healthy Smg1+/gt mice. This kind of low-level inflammation may develop into the chronic inflammation observed in the Smg1+/gt mice but has also been implicated in increasing cancer susceptibility or progression (22–24). Notably, we did not see elevated serum levels of IL-1β but, in fact, saw less IL-1β in Smg1+/gt mice. This suggests that the precursor form of IL-1β is not being processed to allow secretion into the serum or that the increased IL-1β production is restricted to tissues or is microenvironment-dependent.
Two pathways, termed intrinsic and extrinsic, have been proposed to explain the link between inflammation and cancer development (24). In the intrinsic pathway, mutations within tumor cells result in secretion of chemokines that recruit macrophages to the tumor. These macrophages, termed tumor-associated macrophages, are manipulated by the tumor cells to express proinflammatory proteins causing the development of an inflammatory microenvironment, termed cancer-related inflammation (CRI), in which the tumor cells thrive. Key cytokines involved in CRI are IL-6, TNF-α, and IL-1β (24). These cytokines can act as growth factors for the tumors and can increase proliferation and expression of antiapoptotic proteins in the tumor cells. Only a slight elevation of these factors is required to increase tumor progression (22). On the other hand, in the extrinsic pathway, inflammation occurs before development of neoplastic cells and increases cancer susceptibility. Inflammatory sites may have high local concentrations of ROS/RNS. ROS/RNS may then cause damage to cells, resulting in malignant transformation (22, 24, 25). Additionally, cytokines can act to support the growth of these newly transformed cells, and the established protumorigenic microenvironment acts in a similar manner as described for the intrinsic pathway. In the Smg1+/gt mice, we see evidence of increased basal cytokine production, cellular damage, and increased death due to cancer development. However, we see varying levels of endogenous DNA damage. This variation could indicate differences in ROS/RNS production between animals or that DNA damage repair pathways in the Smg1+/gt mice are largely able to repair any damage caused. Therefore, it is possible that mutations contributing to the increased cancer incidence may be accumulating during the life of the animals. Particularly interesting is that the increased cytokine levels and cellular oxidative damage occur before the development of tumors in these mice (Fig. 5). These findings suggest that Smg1+/gt mice show low-level inflammation, which, with age, can either progress to chronic inflammatory disorders or lead to the development of inflammation-enhanced cancers. Although SMG1 does not have a currently described role in regulation of inflammatory responses, it has been suggested to regulate MAPK activation (9). MAPKs are important in regulation of multiple immune regulatory pathways, and studies to determine the role of SMG1 in these pathways are ongoing. Alternatively, SMG1 may be important in determining how tumor cells respond to inflammatory mediators, potentially by regulation of apoptosis. Although we saw no difference in the apoptotic response of normal cells to IR, it is possible that apoptosis/survival in tumor cells is regulated differently and that SMG1 is important for that process.
The two key types of tumor detected in Smg1+/gt mice were lymphomas and lung adenocarcinomas. There is evidence to link Smg1 mutations to these cancers in human studies as well. Lung adenocarcinomas are the predominant type of cancer in nonsmokers. Ding et al. (26) identified Smg1 as a significantly mutated gene in lung adenocarcinoma. In addition to the lung, the catalogue of somatic mutations in cancer (COSMIC) database shows that mutations in Smg1 have been associated with breast, kidney, and stomach cancer (www.sanger.ac.uk/genetics/CGP/cosmic) (27). Additionally, in the human protein atlas studies (www.proteinatlas.org), SMG1 expression was not detected in any malignant lymphoma samples (28). These findings suggest that SMG1 may normally act to suppress tumor formation and its deficiency leads to increased risk for a number of different cancers, particularly lung and hematopoietic cell cancers. However, the story may not be that simple; a kinome-wide screen identified SMG1 as a kinase essential for multiple myeloma survival. Smg1 knockdown by siRNA decreased survival of myeloma but not control cell lines (29), and SMG1 mRNA has been shown to be up-regulated in acute myeloid leukemia (30). Furthermore, in a number of treatment studies, SMG1 expression or enzymatic activity has been required for an effective response to anticancer treatments, including antioxidant therapy and radiotherapy (31–34). Combined, these studies suggest that SMG1 may play different roles in cancer progression depending on the stage of disease, the type of cancer, and the therapy used. Also, a decrease in functional SMG1 may cause a different phenotype to complete loss of expression or inhibition of SMG1 enzymatic activity. Further studies will be required to understand the intricacies of SMG1 function and its place in cancer biology.
In summary, we describe here a unique mouse model of Smg1 deficiency. These mice are prone to development of tumors and chronic inflammation. These are significant findings because they indicate that SMG1 participates in the regulation of inflammation and cancer development, findings that are supported by data from human genetic databases showing mutations in human cancers that reflect the type of tumors observed in the mouse model. Consequently, we suggest that SMG1 is a cancer susceptibility gene that may play a role in the pathogenesis of both lung and hematopoietic cancers.
Materials and Methods
Basic methodology is described here; more details are provided in SI Materials and Methods.
Generation of Smg1+/gt Mice, Animal Husbandry.
The loss-of-function mutation in Smg1 was produced by Genetrap mutagenesis in ES cells supplied by MMRRC (SIGTR ES cell line AG0297). The ES cells were injected into SV129 blastocysts to generate chimeric mice crossed with C57BL/6 mice to generate Smg1 heterozygous (Smg1+/gt) mice. All animal experiments were approved by the Animal Ethics Committee at the Queensland Institute of Medical Research (QIMR).
Isolation and Culture of Murine Embryonic Fibroblasts.
E14.5 embryos were isolated with the brain and liver removed, diced, and trypsinized, and MEFs were plated on 0.1% (wt/vol) gelatin-coated T25 flasks. MEFs were cultured at 37 °C with 5% (vol/vol) CO2 in DMEM (Gibco) supplemented with 12% (vol/vol) heat-inactivated FCS, 100 units/mL penicillin/streptomycin (Gibco), 1% (vol/vol) glutamate (Gibco), 1× nonessential amino acid (Gibco), and β-mercaptoethanol (Sigma). Experiments on primary MEFs were performed at passages 4–5 before they underwent senescence. Clonogenic survival assay was performed on immortalized MEFs as described (35).
Histology.
Tissues were isolated and immediately placed in 10% formalin. Samples were processed at the QIMR histology facility. The sections were analyzed by H&E staining or immunohistochemistry. Images were scanned using an Aperio scanner. Histology was examined by one of the authors (C.S.L.).
Western Blotting.
MEFs were grown to 70% confluency in T75 flasks. Cells were exposed to γ-irradiation for MEFs at a dose of 5 Gy. Cells were harvested at 0, 1, 2, and 4 h postirradiation in Western lysis buffer [2% (wt/vol) SDS, 50 mM Tris (pH 6.8), 1× phosphatase inhibitors] and sonicated. Western blotting using home-poured 4.2% (wt/vol) SDS/PAGE or Invitrogen gradient gels was performed as described previously (36).The primary antibodies used were rabbit antiphospho p53 (Ser15; Cell Signaling) at a 1:1,000 dilution, mouse anti-p21 (Pharmingen) at a 1:1,000 dilution, and mouse anti-GAPDH (Millipore) at a 1:1,000 dilution. The anti-SMG1 antibodies have been described previously (10). Appropriate secondary HRP-conjugated antibodies were used at a 1:5,000 ratio.
Cellular Damage Assays.
γH2AX analysis and immunofluorescence were performed as described previously (19). The 8oxodG staining (Trevigen) for oxidative stress on tissue sections was performed according to the manufacturer’s instructions. Splenocytes were isolated from Smg1+/+ and Smg1+/gt mice that were ∼9 mo of age and cultured in 250 μM H2O2 or 10 μM Rotenone (Sigma) for 4 or 24 h. ROS measurements of treated splenocytes were determined by preloading of 10 μM DCFDA (Molecular Probes) or 5 μM MitoSox (Molecular Probes) for 30 min at 37 °C and then analyzed on FACSCalibur (BD Biosciences) and CellQuest (BD Biosciences) instruments. Trypan blue survival assay was performed as described previously (37).
Flow Cytometry
Flow cytometry was performed using a FACSCalibur or FACSCanto instrument (BD Biosciences). Annexin V (BD Pharmingen) apoptotic analysis was performed according to the manufacturer’s instructions. Propidium iodide (Sigma) cell cycle analysis was performed as described (38). Measurement of serum cytokines levels was performed using cytokine bead array as described previously (39). Staining for cell surface markers was also performed as described previously (40, 41).
Quantitative PCR Assay.
The telomere lengths from tissue samples were quantitated by real-time PCR assay as described previously (42). All other real-time PCR assays (Table S4) were performed as described previously (43) using an ABI7900 instrument (Applied Biosystems).
NMD Assays.
NMD assays were performed as described previously, including alternative splicing (11), β-globin plasmid-based assay (16, 17), and T-cell receptor analysis (44). β-globin plasmids were kindly supplied by Lynne Maquat (University of Rochester, Rochester, NY).
Supplementary Material
Acknowledgments
We thank the QIMR animal house staff, histology staff, and FACS staff for their assistance. We also thank Penny Groves for assistance with cytokine bead arrays, Amity Roberts and Shazrul Fazry for advice regarding telomere length quantification assays, Patricia Lusby for assistance with histological analysis, Yi Chieh Lim for assistance with flow cytometry, and Drs. Slavica Vuckovic and Ingrid Winkler for assistance with hypoxia response assays. This study was supported by Australian Research Council Grant DP1092486, a National Health and Medical Research Council (NHMRC) Peter Doherty Research Fellowship, and QIMR salary funding (to T.L.R.). M.F.L. is supported by a NHMRC Research Fellowship. This study was also supported by NHMRC Project Grants 455941 and 631484 (to A.R.P. and L.J.R.) and NHMRC CDA Award 519744 (to A.R.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.G.K. is a guest editor invited by the Editorial Board.
See Author Summary on page 1151 (volume 110, number 4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215696110/-/DCSupplemental.
References
- 1.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15(17):2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumbaugh KM, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14(5):585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 4.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res. 2008;122(3-4):194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 5.Masse I, et al. A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PLoS ONE. 2008;3(10):e3354. doi: 10.1371/journal.pone.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Estévez C, et al. SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 2012;8(3):e1002619. doi: 10.1371/journal.pgen.1002619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meslin F, et al. hSMG-1 is a granzyme B-associated stress-responsive protein kinase. J Mol Med (Berl) 2011;89(4):411–421. doi: 10.1007/s00109-010-0708-0. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira V, et al. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 2008;283(19):13174–13184. doi: 10.1074/jbc.M708008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen RQ, et al. Kinome siRNA screen identifies SMG-1 as a negative regulator of hypoxia-inducible factor-1alpha in hypoxia. J Biol Chem. 2009;284(25):16752–16758. doi: 10.1074/jbc.M109.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JA, et al. A novel role for hSMG-1 in stress granule formation. Mol Cell Biol. 2011;31(22):4417–4429. doi: 10.1128/MCB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McIlwain DR, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci USA. 2010;107(27):12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stratikopoulos E, Szabolcs M, Dragatsis I, Klinakis A, Efstratiadis A. The hormonal action of IGF1 in postnatal mouse growth. Proc Natl Acad Sci USA. 2008;105(49):19378–19383. doi: 10.1073/pnas.0809223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samet JM, et al. Lung cancer in never smokers: Clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15(18):5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita A, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23(9):1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 16.Belgrader P, Cheng J, Zhou X, Stephenson LS, Maquat LE. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14(12):8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106(5):607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury D, Novina CD. RNAi and RNA-based regulation of immune system function. Adv Immunol. 2005;88:267–292. doi: 10.1016/S0065-2776(05)88008-3. [DOI] [PubMed] [Google Scholar]
- 19.Harris JL, et al. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet. 2009;18(21):4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 20.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22(10):1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medghalchi SM, et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10(2):99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 22.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 25.Hofseth LJ, Hussain SP, Wogan GN, Harris CC. Nitric oxide in cancer and chemoprevention. Free Radic Biol Med. 2003;34(8):955–968. doi: 10.1016/s0891-5849(02)01363-1. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 29.Tiedemann RE, et al. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115(8):1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neben K, et al. Distinct gene expression patterns associated with FLT3- and NRAS-activating mutations in acute myeloid leukemia with normal karyotype. Oncogene. 2005;24(9):1580–1588. doi: 10.1038/sj.onc.1208344. [DOI] [PubMed] [Google Scholar]
- 31.Erker L, et al. Cancer chemoprevention by the antioxidant tempol acts partially via the p53 tumor suppressor. Hum Mol Genet. 2005;14(12):1699–1708. doi: 10.1093/hmg/ddi181. [DOI] [PubMed] [Google Scholar]
- 32.Cheung HH, et al. SMG1 and NIK regulate apoptosis induced by Smac mimetic compounds. Cell Death Dis. 2011;2:e146. doi: 10.1038/cddis.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia QS, et al. Human SMG-1 is involved in gemcitabine-induced primary microRNA-155/BIC up-regulation in human pancreatic cancer PANC-1 cells. Pancreas. 2011;40(1):55–60. doi: 10.1097/MPA.0b013e3181e89f74. [DOI] [PubMed] [Google Scholar]
- 34.Gubanova E, et al. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18(5):1257–1267. doi: 10.1158/1078-0432.CCR-11-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang N, et al. Isolation of full-length ATM cDNA and correction of the ataxia-telangiectasia cellular phenotype. Proc Natl Acad Sci USA. 1997;94(15):8021–8026. doi: 10.1073/pnas.94.15.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts TL, Dunn JA, Sweet MJ, Hume DA, Stacey KJ. The immunostimulatory activity of phosphorothioate CpG oligonucleotides is affected by distal sequence changes. Mol Immunol. 2011;48(8):1027–1034. doi: 10.1016/j.molimm.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Gatei M, et al. ATM protein-dependent phosphorylation of Rad50 protein regulates DNA repair and cell cycle control. J Biol Chem. 2011;286(36):31542–31556. doi: 10.1074/jbc.M111.258152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol. 2002;22(4):1049–1059. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apte SH, Groves PL, Roddick JS, P da Hora V, Doolan DL. High-throughput multi-parameter flow-cytometric analysis from micro-quantities of plasmodium-infected blood. Int J Parasitol. 2011;41(12):1285–1294. doi: 10.1016/j.ijpara.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Apte SH, Baz A, Groves P, Kelso A, Kienzle N. Interferon-gamma and interleukin-4 reciprocally regulate CD8 expression in CD8+ T cells. Proc Natl Acad Sci USA. 2008;105(45):17475–17480. doi: 10.1073/pnas.0809549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill GR, et al. SOCS3 regulates graft-versus-host disease. Blood. 2010;116(2):287–296. doi: 10.1182/blood-2009-12-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. Biotechniques. 2008;44(6):807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- 43.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 44.Obata F, et al. A single universal primer for the T-cell receptor (TCR) variable genes enables enzymatic amplification and direct sequencing of TCR beta cDNA of various T-cell clones. Hum Immunol. 1993;36(3):163–167. doi: 10.1016/0198-8859(93)90120-p. [DOI] [PubMed] [Google Scholar]